Characterisation of Orthohantavirus Serotypes in Human Infections in Kazakhstan
Abstract
1. Introduction
2. Materials and Methods
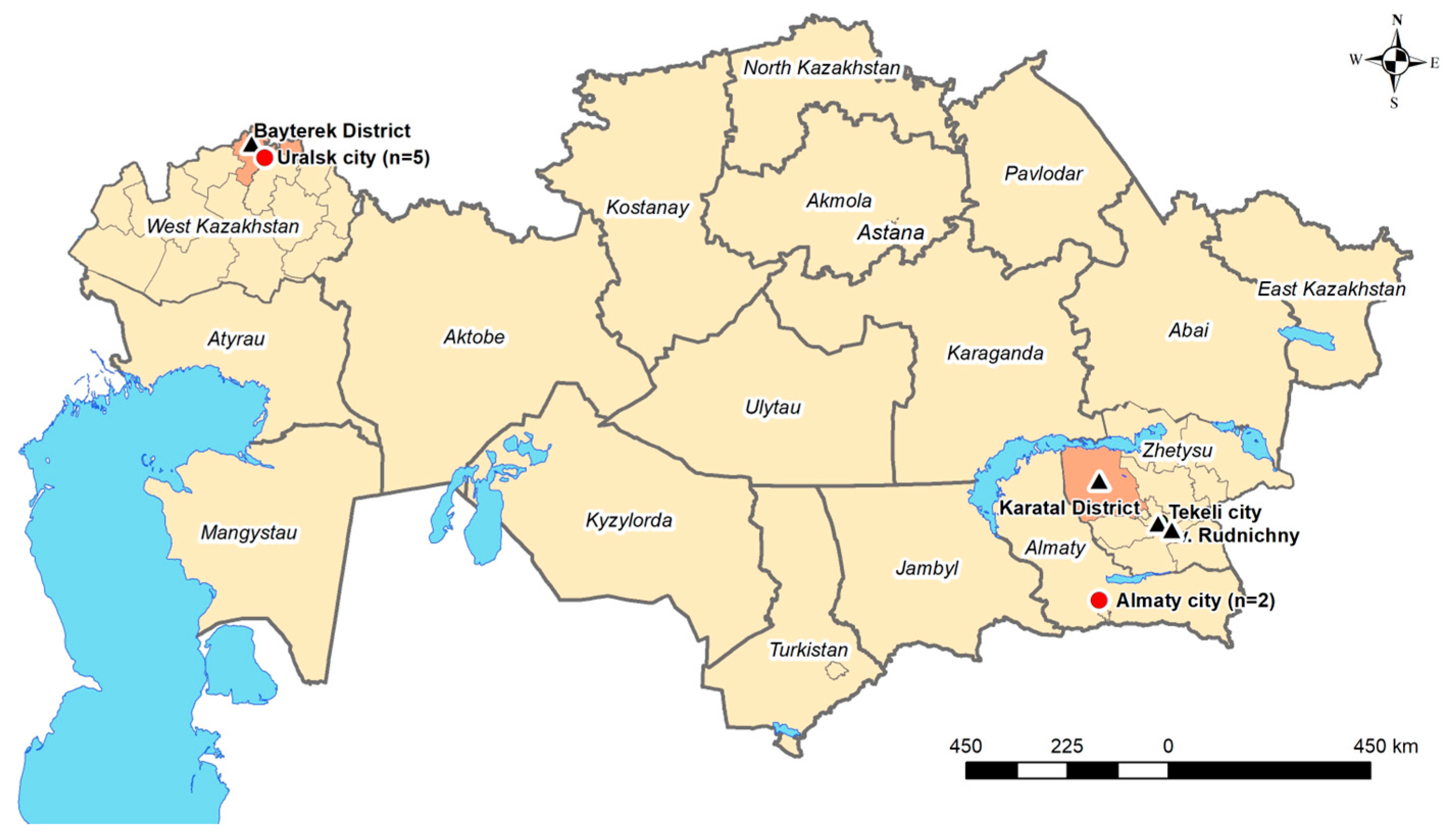
2.1. Study Setting and Sample Collection
2.2. ELISA Screening
| 1st and 2nd serum IgM− and IgG− | negative | No infection |
| 1st and/or 2nd serum IgM+ 1st and 2nd serum IgG− | IgM-positive | Acute infection |
| 1st and 2nd serum IgM− 1st and/or 2nd serum IgG+ | IgG-positive | Previous infection or cross reactive |
| 1st IgM+ and 2nd IgG++ | Both IgM- and IgG-positive | Acute infection |
2.3. Immunoblot Testing
2.4. RT-PCR
| Primer Name | Sequence (5′–3′) | Direction |
| Pan-Hanta 1a-fw | TgATgCATATTgTgTgCAgAC | Forward |
| Pan-Hanta 1b-fw | TgATgCATACTgTgTgCAAAC | Forward |
| Pan-Hanta 1c-fw | CAgTATgATgCATACTgTgTCCAA | Forward |
| Pan-Hanta 1d-fw | TgATgCCTATTgTgTTCAgAC | Forward |
| Pan-Hanta 1a-rev | CTTgCTCTgTTTTgAATCTCA | Reverse |
| Pan-Hanta 1b-rev | CTTgCTCggTgTTgAATCgCA | Reverse |
| Pan-Hanta 1c-rev | CCTgTTCTgTATTAAATCTCA | Reverse |
| Pan-Hanta 1d-rev | CTTgTTCAgTCTTgAATCTCA | Reverse |
2.5. Ethics Approval
2.6. Data Analysis
3. Results
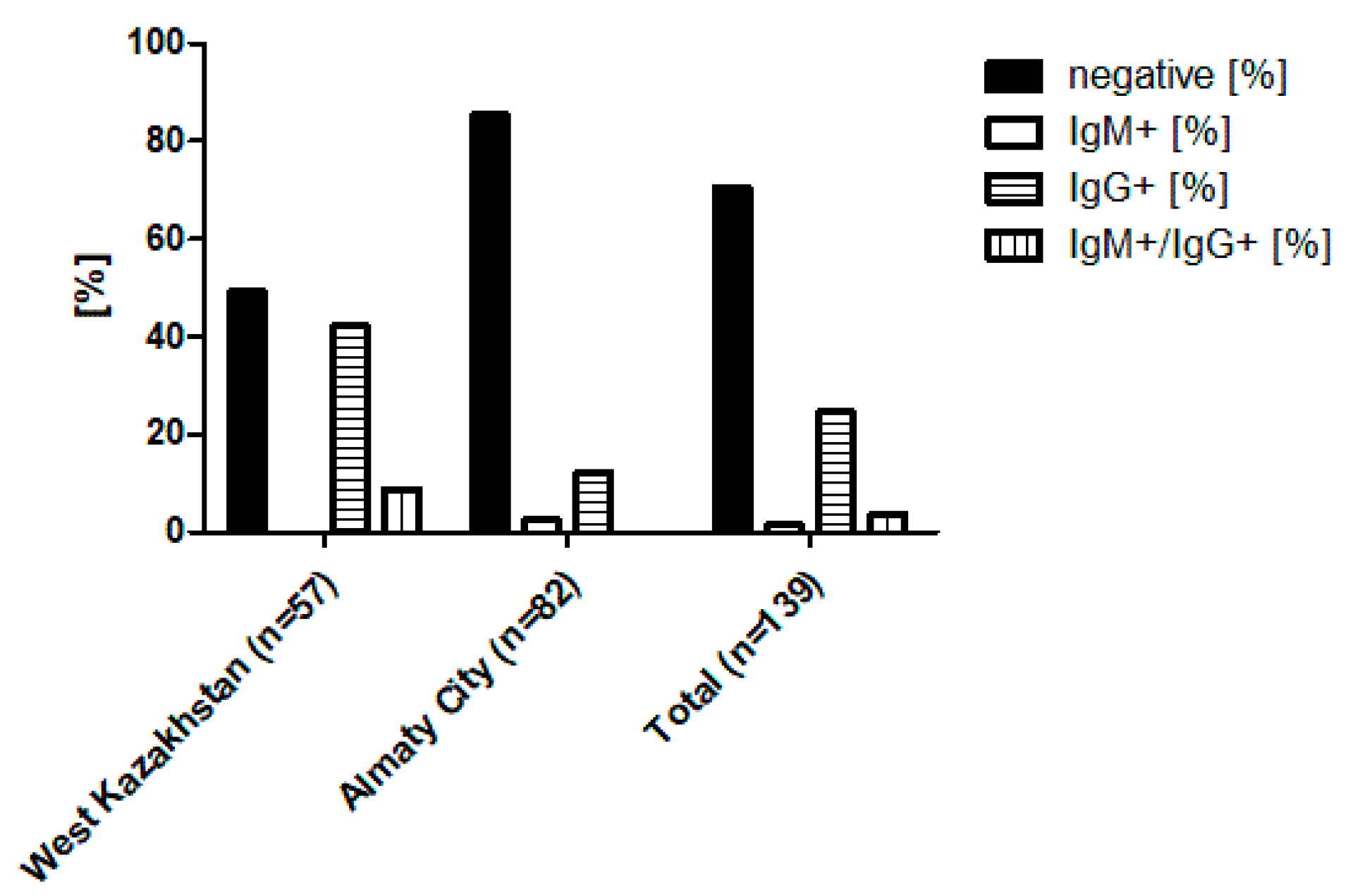
3.1. Sample Overview and ELISA-Based Screening Results
3.2. Serotype Differentiation by Immunoblot
3.3. Molecular Confirmation of Acute Cases, Demographic, and Clinical Features
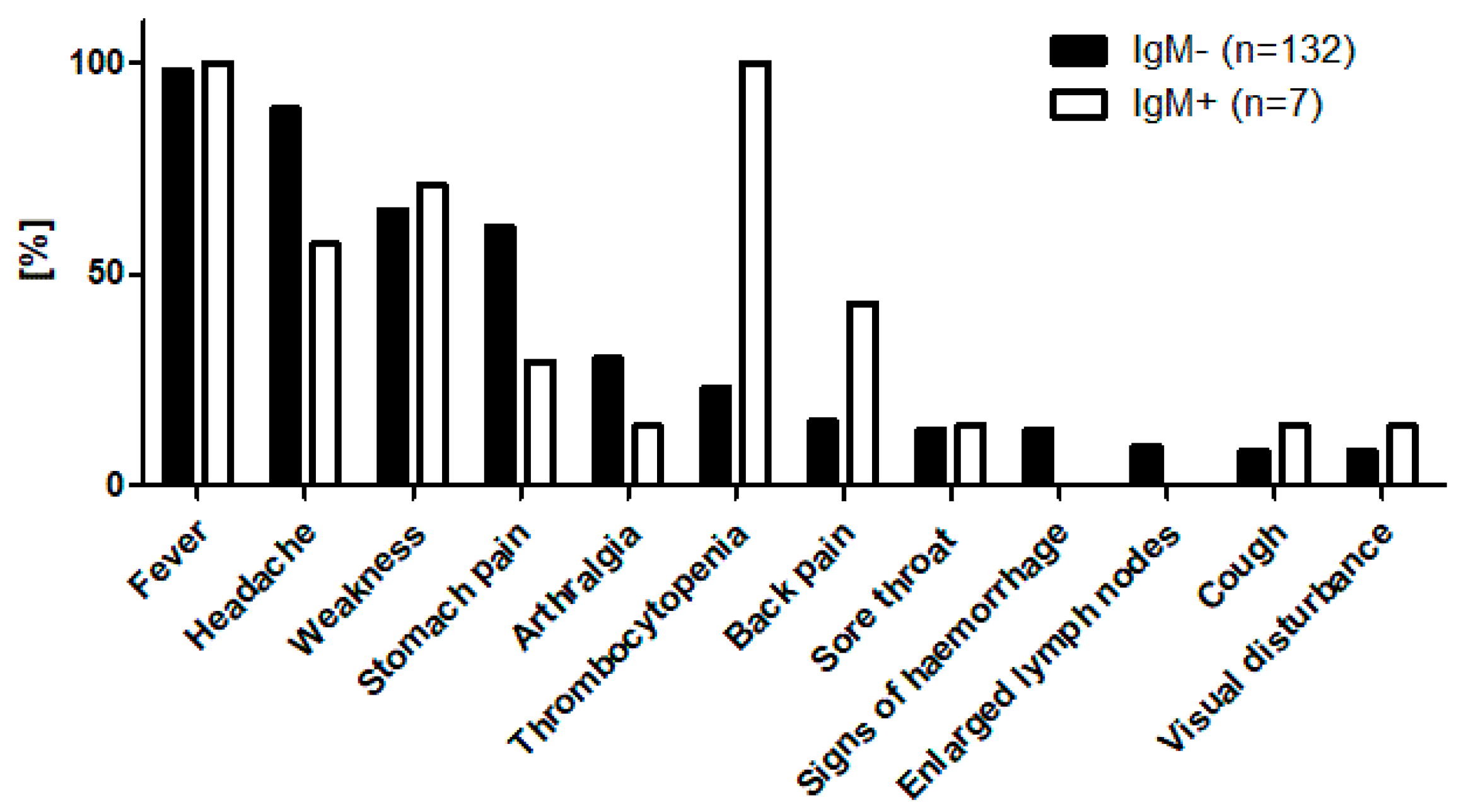
3.4. Clinical Profile of IgG-Positive Individuals
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HFRS | haemorrhagic fever with renal syndrome |
| HCPS | hantavirus cardiopulmonary syndrome |
| IFA | immunofluorescence assays |
| ELISA | enzyme-linked immunosorbent assay |
| RT-PCR | Reverse transcriptase polymerase chain reaction |
| FRNT | focus reduction neutralisation test |
| HIN | Hospital Identification Number |
| RNA | Ribonucleic acid |
References
- Vaheri, A.; Henttonen, H.; Mustonen, J. Hantavirus research in finland: Highlights and perspectives. Viruses 2021, 13, 1452. [Google Scholar] [CrossRef] [PubMed]
- Holmes, E.C.; Zhang, Y.Z. The evolution and emergence of hantaviruses. Curr. Opin. Virol. 2015, 10, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Kruger, D.H.; Figueiredo, L.T.M.; Song, J.W.; Klempa, B. Hantaviruses-Globally emerging pathogens. J. Clin. Virol. 2015, 64, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Avšič-Županc, T.; Saksida, A.; Korva, M. Hantavirus infections. Clin. Microbiol. Infect. 2019, 21, e6–e16. [Google Scholar] [CrossRef]
- He, J.; Wang, Y.; Mu, D.; Xu, Z.; Qian, Q.; Chen, G.; Wen, L.; Yin, W.; Li, S.; Zhang, W.; et al. The impacts of climatic factors and vegetation on hemorrhagic fever with renal syndrome transmission in China: A study of 109 counties. Int. J. Environ. Res. Public Health 2019, 16, 3434. [Google Scholar] [CrossRef]
- Vaheri, A.; Henttonen, H.; Voutilainen, L.; Mustonen, J.; Sironen, T.; Vapalahti, O. Hantavirus infections in Europe and their impact on public health. Rev. Med. Virol. 2013, 23, 35–49. [Google Scholar] [CrossRef]
- Zou, L.X.; Chen, M.J.; Sun, L. Haemorrhagic fever with renal syndrome: Literature review and distribution analysis in China. Int. J. Infect. Dis. 2016, 43, 95–100. [Google Scholar] [CrossRef]
- Jiang, H.; Du, H.; Wang, L.M.; Wang, P.Z.; Bai, X.F. Hemorrhagic Fever with Renal Syndrome: Pathogenesis and Clinical Picture. Front. Cell. Infect. Microbiol. 2016, 6, 1–11. [Google Scholar] [CrossRef]
- Grazhdanov, A.; Zakharov, A.; Biryukov, A. First cases of Hemorrhagic fever with renal syndrome in Kazakhstan. J. Quar. Zoonotic Infect. Kaz. 2001, 3, 94–98. [Google Scholar]
- Grazhdanov, A.K.; Ayazbaev, T.Z.; Toporkov, A.V.; Bidashko, F.G.; Zakharov, A.V.; Belonozhkina, L.B.; Pak, M.V.; Andryushchenko, A.V. Concerning the Allocation of Emerging Natural Foci of the Currently Important Infectious Diseases in the West of Kazakhstan. Probl. Part. Danger. Infect. 2014, 2, 20–24. [Google Scholar] [CrossRef]
- Tukhanova, N.; Shin, A.; Turebekov, N.; Nurmakhanov, T.; Abdiyeva, K.; Shevtsov, A.; Yerubaev, T.; Tokmurziyeva, G.; Berdibekov, A.; Sutyagin, V.; et al. Molecular Characterisation and Phylogeny of Tula Virus in Kazakhstan. Viruses 2022, 14, 1258. [Google Scholar] [CrossRef] [PubMed]
- Plyusnina, A.; Laakkonen, J.; Niemimaa, J.; Henttonen, H.; Plyusnin, A. New Genetic Lineage of Tula Hantavirus in Microtus arvalis obscurus in Eastern Kazakhstan. Open Virol. J. 2008, 2, 32–36. [Google Scholar] [CrossRef] [PubMed]
- Schultze, D.; Fierz, W.; Matter, H.C.; Bankoul, S.; Niedrig, M.; Schmiedl, A. Cross-sectional survey on hantavirus seroprevalence in Canton St. Gallen, Switzerland. Swiss Med. Wkly. 2007, 137, 21–26. [Google Scholar] [PubMed]
- Brockmann, J.; Kleines, M.; Ghaffari Laleh, N.; Kather, J.N.; Wied, S.; Floege, J.; Braun, G.S. A simple clinical score to reduce unnecessary testing for Puumala hantavirus. PLoS ONE 2024, 19, e0304500. [Google Scholar] [CrossRef]
- Mossbrugger, I.; Felder, E.; Gramsamer, B.; Wölfel, R. EvaGreen based real-time RT-PCR assay for broad-range detection of hantaviruses in the field. J. Clin. Virol. 2013, 58, 334–335. [Google Scholar] [CrossRef]
- Tkachenko, E.; Kurashova, S.; Balkina, A.; Ivanov, A.; Egorova, M.; Leonovich, O.; Popova, Y.; Teodorovich, R.; Belyakova, A.; Tkachenko, P.; et al. Cases of Hemorrhagic Fever with Renal Syndrome in Russia during 2000–2022. Viruses 2023, 15, 1537. [Google Scholar] [CrossRef]
- Zelena, H.; Mrazek, J.; Kuhn, T. Tula Hantavirus Infection in Czech Republic. Emerg. Infect. Dis. 2013, 19, 1873–1876. [Google Scholar] [CrossRef]
- Schultze, D.; Lundkvist, Å.; Blauenstein, U.; Heyman, P. Tula virus infection associated with fever and exanthema after a wild rodent bite. Eur. J. Clin. Microbiol. Infect. Dis. 2002, 21, 304–306. [Google Scholar] [CrossRef]
- Hofmann, J.; Kramer, S.; Herrlinger, K.R.; Jeske, K.; Kuhns, M.; Weiss, S.; Ulrich, R.G.; Krüger, D.H. Tula virus as causative agent of hantavirus disease in immunocompetent person, Germany. Emerg. Infect. Dis. 2021, 27, 1232–1234. [Google Scholar] [CrossRef]
- Engler, O.; Klingstrom, J.; Aliyev, E.; Niederhauser, C.; Fontana, S.; Strasser, M.; Portmann, J.; Signer, J.; Bankoul, S.; Frey, F.; et al. Seroprevalence of hantavirus infections in Switzerland in 2009: Difficulties in determining prevalence in a country with low endemicity. Eurosurveillance 2013, 18, 20660. [Google Scholar] [CrossRef]
- Bergstedt Oscarsson, K.; Brorstad, A.; Baudin, M.; Lindberg, A.; Forssén, A.; Evander, M.; Eriksson, M.; Ahlm, C. Human Puumala hantavirus infection in northern Sweden; increased seroprevalence and association to risk and health factors. BMC Infect. Dis. 2016, 16, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Brorstad, A.; Oscarsson, K.B.; Ahlm, C. Early diagnosis of Hantavirus infection by family doctors can reduce inappropriate antibiotic use and hospitalization. Scand. J. Prim. Health Care 2010, 28, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Gozalan, A.; Kalaycioglu, H.; Uyar, Y.; Sevindi, D.F.; Turkyilmaz, B.; Cakir, V.; Cindemir, C.; Unal, B.; Yağçi-Çağlayik, D.; Korukluoglu, G.; et al. Human puumala and dobrava hantavirus infections in the black sea region of Turkey: A cross-sectional study. Vector-Borne Zoonotic Dis. 2013, 13, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Mattar, S.; Garzon, D.; Tadeu, L.; Faccini-Martínez, A.A.; Mills, J.N. Serological diagnosis of hantavirus pulmonary syndrome in a febrile patient in Colombia. Int. J. Infect. Dis. 2014, 25, 201–203. [Google Scholar] [CrossRef]
- Pettersson, L.; Klingström, J.; Hardestam, J.; Lundkvist, Å.; Ahlm, C.; Evander, M. Hantavirus RNA in saliva from patients with hemorrhagic fever with renal syndrome. Emerg. Infect. Dis. 2008, 14, 406–411. [Google Scholar] [CrossRef]
- Seo, J.W.; Kim, C.M.; Yun, N.R.; Lee, Y.M.; Panchali, M.J.; Kim, D.M. Utility of Nested Reverse-Transcriptase Polymerase Chain Reaction of Clinical Specimens for Early Diagnosis of Hemorrhagic Fever with Renal Syndrome. Am. J. Trop. Med. Hyg. 2021, 105, 1285–1289. [Google Scholar] [CrossRef]
| Sample ID | Age | Sex | ELISA | Immunoblot IgM Strip Intensity Rating | RT-qPCR (Positive Samples) | Initial Hospital Diagnosis | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| IgM 1st Serum | IgG 2nd Serum | PuN | HaN | DobN | SeoN | SinN | |||||
| URA 007 | 38 |  | pos | pos | ++ | +/− | +/− | − | +/− | urine | HFRS |
| URA 016 | 28 |  | pos | pos | ++ | − | − | − | +/− | urine | HFRS |
| BUR 001 | 62 |  | pos | pos | ++ | − | − | − | +/− | serum | HFRS |
| BUR 003 | 34 |  | pos | pos | ++ | − | − | − | +/− | serum | Fever of unknown origin |
| BUR 024 | 33 |  | pos | pos | ++ | − | − | − | +/− | serum | HFRS |
| ALM 028 | 32 |  | pos | neg | − | − | − | − | − | negative | Enterovirus infection |
| ALB 003 | 29 |  | pos | neg | ++ | − | − | − | +/− | urine | Acute tubulointerstitial nephritis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tukhanova, N.; Shin, A.; Bakuli, A.; Yeraliyeva, L.; Maikanov, N.; Froeschl, G.; Zhumadilova, Z.; Tokmurziyeva, G.; Wagner, E.; Essbauer, S.; et al. Characterisation of Orthohantavirus Serotypes in Human Infections in Kazakhstan. Viruses 2025, 17, 925. https://doi.org/10.3390/v17070925
Tukhanova N, Shin A, Bakuli A, Yeraliyeva L, Maikanov N, Froeschl G, Zhumadilova Z, Tokmurziyeva G, Wagner E, Essbauer S, et al. Characterisation of Orthohantavirus Serotypes in Human Infections in Kazakhstan. Viruses. 2025; 17(7):925. https://doi.org/10.3390/v17070925
Chicago/Turabian StyleTukhanova, Nur, Anna Shin, Abhishek Bakuli, Lyazzat Yeraliyeva, Nurbek Maikanov, Guenter Froeschl, Zauresh Zhumadilova, Gulnara Tokmurziyeva, Edith Wagner, Sandra Essbauer, and et al. 2025. "Characterisation of Orthohantavirus Serotypes in Human Infections in Kazakhstan" Viruses 17, no. 7: 925. https://doi.org/10.3390/v17070925
APA StyleTukhanova, N., Shin, A., Bakuli, A., Yeraliyeva, L., Maikanov, N., Froeschl, G., Zhumadilova, Z., Tokmurziyeva, G., Wagner, E., Essbauer, S., & Peintner, L. (2025). Characterisation of Orthohantavirus Serotypes in Human Infections in Kazakhstan. Viruses, 17(7), 925. https://doi.org/10.3390/v17070925







