Population Genetic Structure of Citrus Tatter Leaf Virus in Zhejiang Province, China
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Genetic Diversity Estimates of CTLV Populations
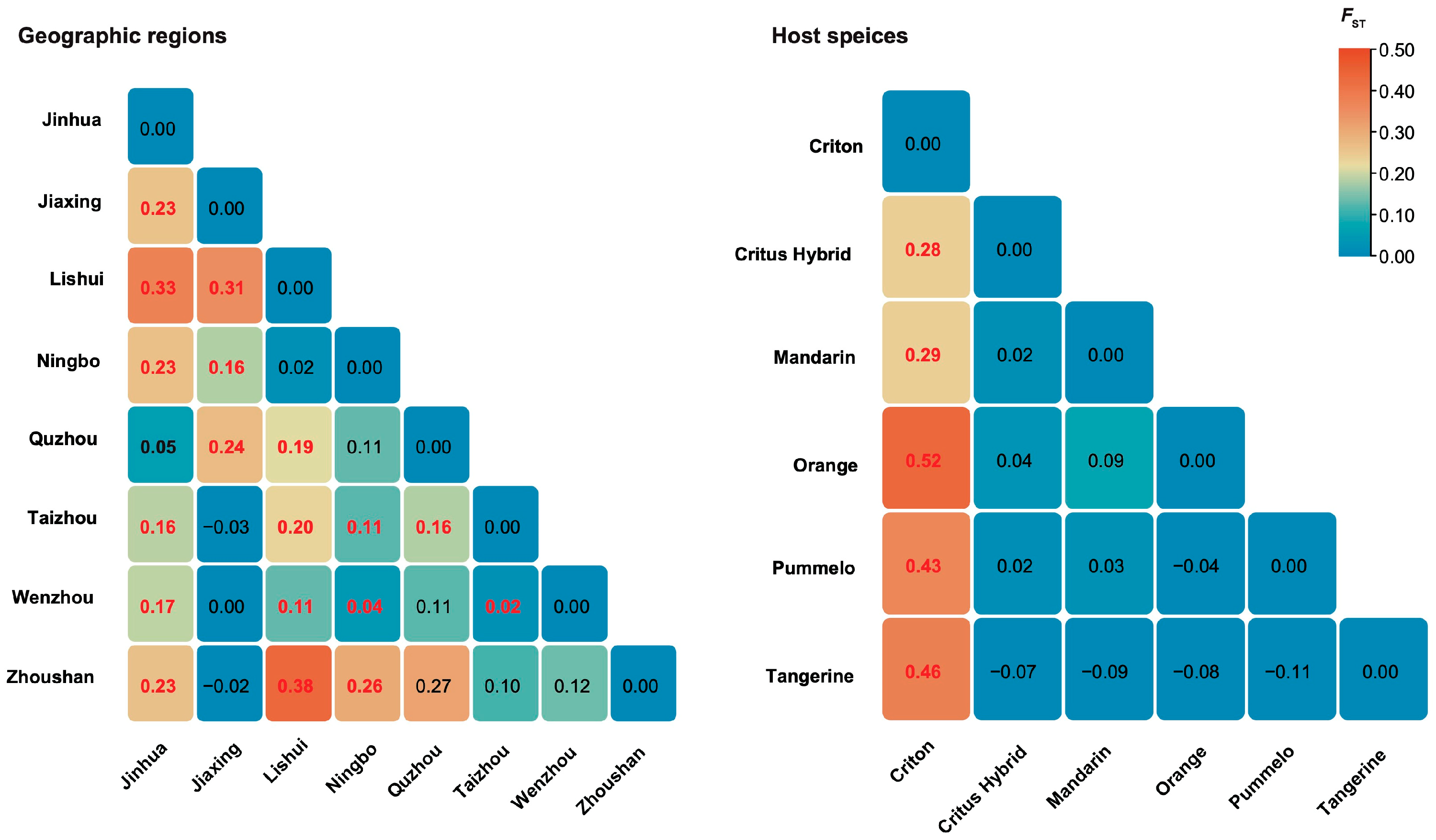
3.2. Genetic Differentiation Between CTLV Populations
3.3. Adaptative Evolution of CTLV
3.4. Demographic History of CTLV Population
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vashisth, T.; Kadyampakeni, D. Chapter 49—Diagnosis and management of nutrient constraints in citrus. In Fruit Crops; Srivastava, A.K., Hu, C., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 723–737. [Google Scholar]
- Huang, Z.; Li, Z.; Yao, L.; Yuan, Y.; Hong, Z.; Huang, S.; Wang, Y.; Ye, J.; Zhang, L.; Ding, J. Geographical distribution and potential distribution prediction of thirteen species of Citrus L. in China. Environ. Sci. Pollut. Res. 2024, 31, 6558–6571. [Google Scholar] [CrossRef]
- Wallace, J.M.; Drake, R.J. Tatter leaf, a previously undesoribed virus effect on citrus. Plant Dis. Report. 1962, 46, 211–212. [Google Scholar]
- Tan, S.H.; Osman, F.; Bodaghi, S.; Dang, T.; Greer, G.; Huang, A.; Hammado, S.; Abu-Hajar, S.; Campos, R.; Vidalakis, G. Full genome characterization of 12 citrus tatter leaf virus isolates for the development of a detection assay. PLoS ONE 2019, 14, e0223958. [Google Scholar] [CrossRef] [PubMed]
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2018, 46, D708–D717. [Google Scholar] [CrossRef] [PubMed]
- Tatineni, S.; Afunian, M.R.; Hilf, M.E.; Gowda, S.; Dawson, W.O.; Garnsey, S.M. Molecular characterization of citrus tatter leaf virus historically associated with Meyer lemon trees: Complete genome sequence and development of biologically active In Vitro transcripts. Phytopathology 2009, 99, 423–431. [Google Scholar] [CrossRef]
- Komatsu, K.; Hirata, H.; Fukagawa, T.; Yamaji, Y.; Okano, Y.; Ishikawa, K.; Adachi, T.; Maejima, K.; Hashimoto, M.; Namba, S. Infection of capilloviruses requires subgenomic RNAs whose transcription is controlled by promoter-like sequences conserved among flexiviruses. Virus Res. 2012, 167, 8–15. [Google Scholar] [CrossRef]
- Po, D.W.; Zhou, H.F.; Yao, Y. Seventy years of fruit industry development in Zhejiang Province. South China Fruits. 2021, 50, 177–183. [Google Scholar]
- Zhang, T.M.; Liang, X.Y.; Roistacher, C.N. Occurrence and detection of citrus tatter leaf virus (CTLV) in Huangyan, Zhejiang Province, China. Plant Dis. 1988, 72, 543–545. [Google Scholar] [CrossRef]
- Xie, P.; Jin, X.; He, X. Identification of citrus tatter leaf in the south of Zhejiang Province. Acta Phytopathol. Sin. 1992, 22, 5–6. [Google Scholar]
- Gao, F.; Zou, W.; Xie, L.; Zhan, J. Adaptive evolution and demographic history contribute to the divergent population genetic structure of potato virus Y between China and Japan. Evol. Appl. 2017, 10, 379–390. [Google Scholar] [CrossRef]
- Shokri, S.; Shujaei, K.; Gibbs, A.J.; Hajizadeh, M. Evolution and biogeography of apple stem grooving virus. Virol. J. 2023, 20, 105. [Google Scholar] [CrossRef]
- Lu, L.M.; Liu, S.M.; An, B.J.; Du, D.C.; Hu, X.R.; Pu, Z.X.; Lv, J. Occurrence and distribution of citrus tatter leaf disease in Zhejiang and genetic diversity analysis of its pathogen. J. Fruit Sci. 2025, 42, 1269–1280. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Xiang, C.-Y.; Gao, F.; Jakovlić, I.; Lei, H.-P.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.-T.; Zhang, D. Using PhyloSuite for molecular phylogeny and tree-based analyses. iMeta 2023, 2, e87. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Gao, F.; Jin, J.; Zou, W.; Liao, F.; Shen, J. Geographically driven adaptation of chilli veinal mottle virus revealed by genetic diversity analysis of the coat protein gene. Arch. Virol. 2016, 161, 1329–1333. [Google Scholar] [CrossRef]
- Pfeifer, B.; Wittelsbürger, U.; Ramos-Onsins, S.E.; Lercher, M.J. PopGenome: An efficient Swiss army knife for population genomic analyses in R. Mol. Biol. Evol. 2014, 31, 1929–1936. [Google Scholar] [CrossRef]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef]
- Parker, J.; Rambaut, A.; Pybus, O.G. Correlating viral phenotypes with phylogeny: Accounting for phylogenetic uncertainty. Infect. Genet. Evol. 2008, 8, 239–246. [Google Scholar] [CrossRef]
- Gao, F.; Chen, C.; Arab, D.A.; Du, Z.; He, Y.; Ho, S.Y.W. EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evol. 2019, 9, 3891–3898. [Google Scholar] [CrossRef]
- Yang, Z.; Nielsen, R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 2002, 19, 908–917. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wong, W.S.; Nielsen, R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005, 22, 1107–1118. [Google Scholar] [CrossRef] [PubMed]
- Murrell, B.; Wertheim, J.O.; Moola, S.; Weighill, T.; Scheffler, K.; Kosakovsky Pond, S.L. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012, 8, e1002764. [Google Scholar] [CrossRef] [PubMed]
- Delport, W.; Poon, A.F.; Frost, S.D.; Kosakovsky Pond, S.L. Datamonkey 2010: A suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 2010, 26, 2455–2457. [Google Scholar] [CrossRef]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef]
- Zhan, J. Population Genetics of Plant Pathogens. In eLS; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2016. [Google Scholar]
- Ohshima, K.; Tomitaka, Y.; Wood, J.T.; Minematsu, Y.; Kajiyama, H.; Tomimura, K.; Gibbs, A.J. Patterns of recombination in turnip mosaic virus genomic sequences indicate hotspots of recombination. J. Gen. Virol. 2007, 88 Pt 1, 298–315. [Google Scholar] [CrossRef]
- Cuevas, J.; Delaunay, A.; Rupar, M.; Jacquot, E.; Elena, S.F. Molecular evolution and phylogeography of potato virus Y based on the CP gene. J. Gen. Virol. 2012, 93 Pt 11, 2496–2501. [Google Scholar] [CrossRef][Green Version]
- Zhang, W.; Sun, X.; Wei, X.; Gao, Y.; Song, J.; Bai, Y. Geography-driven evolution of potato virus A revealed by genetic diversity analysis of the complete genome. Front. Microbiol. 2021, 12, 738646. [Google Scholar] [CrossRef]
- Kim, J.; Lal, A.; Kil, E.J.; Kwak, H.R.; Yoon, H.S.; Choi, H.S.; Kim, M.; Ali, M.; Lee, S. Adaptation and codon-usage preference of apple and pear-Infecting apple stem grooving viruses. Microorganisms 2021, 9, 1111. [Google Scholar] [CrossRef]
- Rohožková, J.; Navrátil, M. P1 peptidase—A mysterious protein of family Potyviridae. J. Biosci. 2011, 36, 189–200. [Google Scholar] [CrossRef] [PubMed]



| Population | n | s | h | Hd | π | Identity (%) | |
|---|---|---|---|---|---|---|---|
| Region | |||||||
| Jinhua | 12 | 125 | 12.000 | 1.000 ± 0.034 | 0.058 ± 0.011 | 89.36–99.86 | |
| Jiaxing | 4 | 94 | 4.000 | 1.000 ± 0.177 | 0.072 ± 0.018 | 89.36–99.86 | |
| Lishui | 18 | 121 | 9.000 | 0.804 ± 0.091 | 0.049 ± 0.011 | 89.36–100 | |
| Ningbo | 40 | 151 | 23.000 | 0.928 ± 0.031 | 0.065 ± 0.005 | 89.08–100 | |
| Quzhou | 4 | 63 | 4.000 | 1.000 ± 0.177 | 0.054 ± 0.014 | 92.02–99.72 | |
| Taizhou | 75 | 198 | 48.000 | 0.981 ± 0.006 | 0.077 ± 0.002 | 87.82–100 | |
| Wenzhou | 25 | 158 | 23.000 | 0.993 ± 0.013 | 0.073 ± 0.003 | 89.36–100 | |
| Zhoushan | 3 | 57 | 3.000 | 1.000 ± 0.272 | 0.054 ± 0.024 | 92.02–99.72 | |
| Host | |||||||
| Criton | 8 | 53 | 8.000 | 1.000 ± 0.063 | 0.024 ± 0.009 | 93.42–99.86 | |
| Citrus hybrid | 138 | 214 | 86.000 | 0.976 ± 0.008 | 0.075 ± 0.001 | 87.68–100 | |
| Orange | 2 | 65 | 2.000 | 1.000 ± 0.500 | 0.091 ± 0.046 | 90.09 | |
| Pummelo | 4 | 108 | 4.000 | 1.000 ± 0.177 | 0.083 ± 0.016 | 89.78–93.70 | |
| Tangerine | 3 | 90 | 3.000 | 1.000 ± 0.272 | 0.086 ± 0.024 | 90.76–92.44 | |
| Total | 181 | 231 | 118.000 | 0.984 ± 0.005 | 0.076 ± 0.001 | ||
| Statistic | Observed Mean (95% HPD) | Null Mean (95% HPD) | p-Value |
|---|---|---|---|
| Region | |||
| AI | 7.77 (6.68–8.89) | 15.49 (14.52–16.51) | <0.001 *** |
| PS | 64.26 (61.00–67.00) | 95.30 (92.02–98.40) | <0.001 *** |
| MC (Jinhua) | 6.97 (7.00–7.00) | 1.25 (1.00–2.00) | 0.010 ** |
| MC (Jiaxing) | 1.00 (1.00–1.00) | 1.02 (1.00–1.04) | 1.000 ns |
| MC (Lishui) | 2.54 (2.00–4.00) | 1.43 (1.00–2.04) | 0.110 ns |
| MC (Ningbo) | 3.31 (3.00–5.00) | 2.28 (1.89–3.01) | 0.100 ns |
| MC (Quzhou) | 1.01 (1.00–1.00) | 1.02 (1.00–1.07) | 1.000 ns |
| MC (Taizhou) | 7.13 (7.00–8.00) | 3.60 (2.88–5.09) | 0.010 ** |
| MC (Wenzhou) | 2.22 (2.00–3.00) | 1.83 (1.17–2.35) | 0.440 ns |
| MC (Zhoushan) | 1.99 (2.00–2.00) | 1.00 (1.00–1.01) | 0.010 ** |
| Host | |||
| AI | 3.97 (3.21–4.75) | 8.06 (7.24–8.97) | <0.001 *** |
| PS | 32.13 (30.00–34.00) | 41.30 (39.58–42.64) | <0.001 *** |
| MC (Criton) | 6.94 (7.00–7.00) | 1.08 (1.00–1.31) | 0.010 ** |
| MC (Citrus hybrid) | 25.13 (11.00–31.00) | 8.55 (6.57–10.78) | 0.020 * |
| MC (Mandarin) | 3.01 (3.00–3.00) | 1.82 (1.14–2.61) | 0.020 * |
| MC (Orange) | n/a | n/a | n/a |
| MC (Pummelo) | 1.00 (1.00–1.00) | 1.04 (1.00–1.12) | 1.000 ns |
| MC (Tangerine) | 1.00 (1.00–1.00) | 1.01 (1.00–1.00) | 1.000 ns |
| Model | np | ln L | 2 Δl = 2 × (ln L1 − ln L2) | LRT p-Value | Positively Selected Sites (BEB: Pr (ω > 1)> 0.5) [ ωML] | |
|---|---|---|---|---|---|---|
| (a) | ||||||
| M1a | 362 | −5349.129 | Not allowed | |||
| M2a | 364 | −5349.129 | (M1a vs. M2a) 0.000 | 1.000 ns | [] | |
| M7 | 362 | −5361.708 | Not allowed | |||
| M8 | 364 | −5344.074 | (M7 vs. M8) 35.267 | <0.001 *** | 27 [1.450], 38 [1.399], 47 [1.364], 103 [1.200], 137 [1.342] | |
| Site | α | β− | β+ | LRT | Substitution | |
| From | To | |||||
| (b) | ||||||
| 27 | 1.94 | 0.00 | 45.71 | 0.050 | GCA(Ala) | GAA (Glu), GGA (Gly), ACA(Thr) |
| 38 | 6.48 | 0.00 | 86.08 | 0.010 | GGA (Gly) | AAA(Lys), AGA(Arg), AGC(Ser), AGT(Ser), TCA(Ser) |
| 47 | 0.00 | 0.00 | 7.80 | 0.010 | GGC (Gly) | AGC (Ser), GAC (Asp) |
| 106 | 0.00 | 0.00 | 5.32 | 0.030 | GCC(Ala) | GTC(Val), TCC(Ser) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, L.; Liu, S.; Pu, Z.; An, B.; Du, D.; Hu, X.; Lv, J.; Huang, Z. Population Genetic Structure of Citrus Tatter Leaf Virus in Zhejiang Province, China. Viruses 2025, 17, 909. https://doi.org/10.3390/v17070909
Lu L, Liu S, Pu Z, An B, Du D, Hu X, Lv J, Huang Z. Population Genetic Structure of Citrus Tatter Leaf Virus in Zhejiang Province, China. Viruses. 2025; 17(7):909. https://doi.org/10.3390/v17070909
Chicago/Turabian StyleLu, Lianming, Shunmin Liu, Zhanxu Pu, Baoju An, Danchao Du, Xiurong Hu, Jia Lv, and Zhendong Huang. 2025. "Population Genetic Structure of Citrus Tatter Leaf Virus in Zhejiang Province, China" Viruses 17, no. 7: 909. https://doi.org/10.3390/v17070909
APA StyleLu, L., Liu, S., Pu, Z., An, B., Du, D., Hu, X., Lv, J., & Huang, Z. (2025). Population Genetic Structure of Citrus Tatter Leaf Virus in Zhejiang Province, China. Viruses, 17(7), 909. https://doi.org/10.3390/v17070909





