The Differential Expression of the JAK/STAT Pathway in Breast Cancer Cells Transfected with Human Papillomavirus Oncogenes
Abstract
1. Introduction
2. Materials and Methods
2.1. Vector Construction, Cell Culture, and Transfection
2.2. RNA Extraction, cDNA Synthesis, and JAK/STAT Expression Analysis
2.3. Statistical Analysis
3. Results
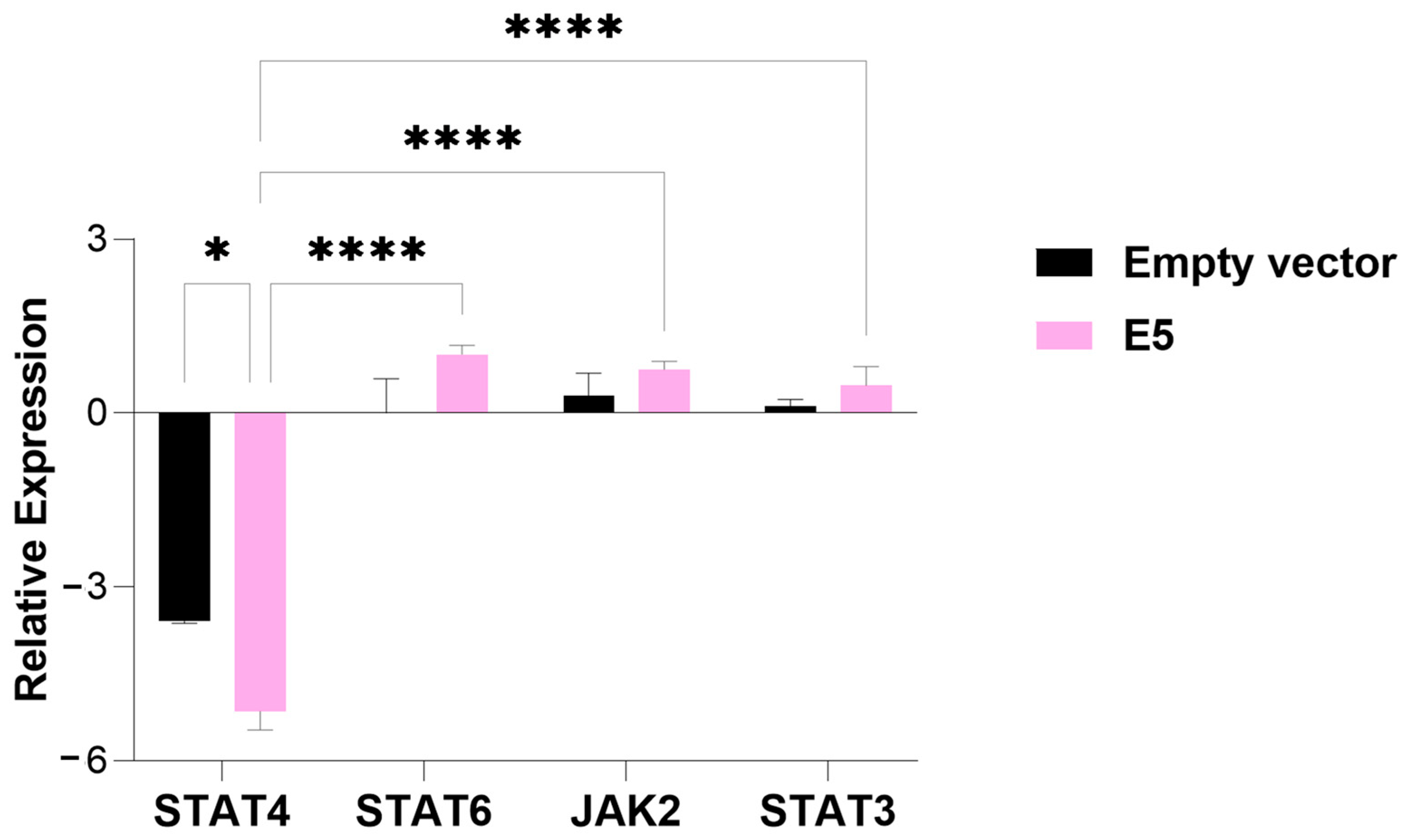
3.1. Modulation of Gene Expression of JAK/STAT in Presence of HPV E5 Oncogene
3.2. Modulation of Gene Expression of JAK/STAT in Presence of HPV E6 Oncogene
3.3. Modulation of Gene Expression of JAK/STAT in Presence of HPV E7 Oncogene
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO—World Health Organization Breast Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/breast-cancer (accessed on 4 April 2025).
- Derakhshan, F.; Reis-Filho, J.S. Pathogenesis of Triple-Negative Breast Cancer. Annu. Rev. Pathol. Mech. Dis. 2022, 17, 181–204. [Google Scholar] [CrossRef]
- Johnson, K.S.; Conant, E.F.; Soo, M.S. Molecular Subtypes of Breast Cancer: A Review for Breast Radiologists. J. Breast Imaging 2021, 3, 12–24. [Google Scholar] [CrossRef]
- Zhang, X. Molecular Classification of Breast Cancer: Relevance and Challenges. Arch. Pathol. Lab. Med. 2023, 147, 46–51. [Google Scholar] [CrossRef]
- Huber-Keener, K.J. Cancer Genetics and Breast Cancer. Best Pract. Res. Clin. Obstet. Gynaecol. 2022, 82, 3–11. [Google Scholar] [CrossRef]
- Kudela, E.; Kudelova, E.; Kozubík, E.; Rokos, T.; Pribulova, T.; Holubekova, V.; Biringer, K. HPV-Associated Breast Cancer: Myth or Fact? Pathogens 2022, 11, 1510. [Google Scholar] [CrossRef]
- Lawson, J.S.; Heng, B. Viruses and Breast Cancer. Cancers 2010, 2, 752–772. [Google Scholar] [CrossRef]
- Purrahman, D.; Avarvand, A.Y.; Paradowska-Gorycka, A.; Saki, N.; Karimpourian, H.; Jodat, H.; Mahmoudian-Sani, M.-R. Association of Human Papillomavirus with Breast Cancer: A New Perspective on an Old Debate. Future Oncol. 2022, 18, 2483–2494. [Google Scholar] [CrossRef]
- Salman, N.A.; Davies, G.; Majidy, F.; Shakir, F.; Akinrinade, H.; Perumal, D.; Ashrafi, G.H. Association of High Risk Human Papillomavirus and Breast Cancer: A UK Based Study. Sci. Rep. 2017, 7, 43591. [Google Scholar] [CrossRef]
- Scarth, J.A.; Patterson, M.R.; Morgan, E.L.; Macdonald, A. The Human Papillomavirus Oncoproteins: A Review of the Host Pathways Targeted on the Road to Transformation. J. Gen. Virol. 2021, 102, 001540. [Google Scholar] [CrossRef]
- Scott-Wittenborn, N.; Fakhry, C. Epidemiology of HPV Related Malignancies. Semin. Radiat. Oncol. 2021, 31, 286–296. [Google Scholar] [CrossRef]
- De Carolis, S.; Storci, G.; Ceccarelli, C.; Savini, C.; Gallucci, L.; Sansone, P.; Santini, D.; Seracchioli, R.; Taffurelli, M.; Fabbri, F.; et al. HPV DNA Associates with Breast Cancer Malignancy and It Is Transferred to Breast Cancer Stromal Cells by Extracellular Vesicles. Front. Oncol. 2019, 9, 860. [Google Scholar] [CrossRef] [PubMed]
- Kroupis, C.; Markou, A.; Vourlidis, N.; Dionyssiou-Asteriou, A.; Lianidou, E.S. Presence of High-Risk Human Papillomavirus Sequences in Breast Cancer Tissues and Association with Histopathological Characteristics. Clin. Biochem. 2006, 39, 727–731. [Google Scholar] [CrossRef]
- Bejarano, L.; Jordāo, M.J.C.; Joyce, J.A. Therapeutic Targeting of the Tumor Microenvironment. Cancer Discov. 2021, 11, 933–959. [Google Scholar] [CrossRef] [PubMed]
- Hinshaw, D.C.; Shevde, L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef]
- Lebre, M.C.; Van Der Aar, A.M.G.; Van Baarsen, L.; Van Capel, T.M.M.; Schuitemaker, J.H.N.; Kapsenberg, M.L.; De Jong, E.C. Human Keratinocytes Express Functional Toll-Like Receptor 3, 4, 5, and 9. J. Investig. Dermatol. 2007, 127, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Cai, X.; Shen, F.; Ma, F. HPV Post-Infection Microenvironment and Cervical Cancer. Cancer Lett. 2021, 497, 243–254. [Google Scholar] [CrossRef]
- Brooks, A.J.; Putoczki, T. JAK-STAT Signalling Pathway in Cancer. Cancers 2020, 12, 1971. [Google Scholar] [CrossRef]
- Hu, X.; Li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT Signaling Pathway: From Bench to Clinic. Signal Transduct. Target. Ther. 2021, 6, 402. [Google Scholar] [CrossRef]
- Erdogan, F.; Radu, T.B.; Orlova, A.; Qadree, A.K.; De Araujo, E.D.; Israelian, J.; Valent, P.; Mustjoki, S.M.; Herling, M.; Moriggl, R.; et al. JAK-STAT Core Cancer Pathway: An Integrative Cancer Interactome Analysis. J. Cell. Mol. Med. 2022, 26, 2049–2062. [Google Scholar] [CrossRef]
- Santos, D.L.; São Marcos, B.D.F.; De Sousa, G.F.; Cruz, L.C.D.O.; Barros, B.R.D.S.; Nogueira, M.C.D.B.L.; Oliveira, T.H.D.A.; Silva, A.J.D.; Santos, V.E.P.; De Melo, C.M.L.; et al. Immunological Response against Breast Lineage Cells Transfected with Human Papillomavirus (HPV). Viruses 2024, 16, 717. [Google Scholar] [CrossRef]
- Bustin, S.A.; Wittwer, C.T. MIQE: A Step Toward More Robust and Reproducible Quantitative PCR. Clin. Chem. 2017, 63, 1537–1538. [Google Scholar] [CrossRef] [PubMed]
- Leitão, M.D.C.G.; Coimbra, E.C.; Lima, R.D.C.P.D.; Guimarães, M.D.L.; Heráclio, S.D.A.; Silva Neto, J.D.C.; De Freitas, A.C. Quantifying mRNA and MicroRNA with qPCR in Cervical Carcinogenesis: A Validation of Reference Genes to Ensure Accurate Data. PLoS ONE 2014, 9, e111021. [Google Scholar] [CrossRef] [PubMed]
- Usui, T.; Nishikomori, R.; Kitani, A.; Strober, W. GATA-3 Suppresses Th1 Development by Downregulation of Stat4 and Not Through Effects on IL-12Rbeta2 Chain or T-Bet. Immunity 2003, 18, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Lee, C.-J.; Choi, J.-H.; Kim, J.-H.; Kim, J.-W.; Kim, J.-Y.; Nam, J.-S. The JAK2/STAT3/CCND2 Axis Promotes Colorectal Cancer Stem Cell Persistence and Radioresistance. J. Exp. Clin. Cancer Res. CR 2019, 38, 399. [Google Scholar] [CrossRef]
- Zhao, M.; Tan, Y.; Peng, Q.; Huang, C.; Guo, Y.; Liang, G.; Zhu, B.; Huang, Y.; Liu, A.; Wang, Z.; et al. IL-6/STAT3 Pathway Induced Deficiency of RFX1 Contributes to Th17-Dependent Autoimmune Diseases via Epigenetic Regulation. Nat. Commun. 2018, 9, 583. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- De Gregorio, V.; Urciuolo, F.; Netti, P.A.; Imparato, G. In Vitro Organotypic Systems to Model Tumor Microenvironment in Human Papillomavirus (HPV)-Related Cancers. Cancers 2020, 12, 1150. [Google Scholar] [CrossRef]
- Sher, G.; Salman, N.A.; Kulinski, M.; Fadel, R.A.; Gupta, V.K.; Anand, A.; Gehani, S.; Abayazeed, S.; Al-Yahri, O.; Shahid, F.; et al. Prevalence and Type Distribution of High-Risk Human Papillomavirus (HPV) in Breast Cancer: A Qatar Based Study. Cancers 2020, 12, 1528. [Google Scholar] [CrossRef] [PubMed]
- Tosi, A.; Parisatto, B.; Menegaldo, A.; Spinato, G.; Guido, M.; Del Mistro, A.; Bussani, R.; Zanconati, F.; Tofanelli, M.; Tirelli, G.; et al. The Immune Microenvironment of HPV-Positive and HPV-Negative Oropharyngeal Squamous Cell Carcinoma: A Multiparametric Quantitative and Spatial Analysis Unveils a Rationale to Target Treatment-Naïve Tumors with Immune Checkpoint Inhibitors. J. Exp. Clin. Cancer Res. 2022, 41, 279. [Google Scholar] [CrossRef]
- Kim, S.-H.; Juhnn, Y.-S.; Kang, S.; Park, S.-W.; Sung, M.-W.; Bang, Y.-J.; Song, Y.-S. Human Papillomavirus 16 E5 Up-Regulates the Expression of Vascular Endothelial Growth Factor through the Activation of Epidermal Growth Factor Receptor, MEK/ERK1,2 and PI3K/Akt. Cell. Mol. Life Sci. 2006, 63, 930–938. [Google Scholar] [CrossRef]
- Ren, S.; Gaykalova, D.A.; Guo, T.; Favorov, A.V.; Fertig, E.J.; Tamayo, P.; Callejas-Valera, J.L.; Allevato, M.; Gilardi, M.; Santos, J.; et al. HPV E2, E4, E5 Drive Alternative Carcinogenic Pathways in HPV Positive Cancers. Oncogene 2020, 39, 6327–6339. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.L.; Woodby, B.L.; Ulicny, J.; Raikhy, G.; Orr, A.W.; Songock, W.K.; Bodily, J.M. Human Papillomavirus 16 E5 Inhibits Interferon Signaling and Supports Episomal Viral Maintenance. J. Virol. 2020, 94, e01582-19. [Google Scholar] [CrossRef]
- Fu, C.; Jiang, L.; Hao, S.; Liu, Z.; Ding, S.; Zhang, W.; Yang, X.; Li, S. Activation of the IL-4/STAT6 Signaling Pathway Promotes Lung Cancer Progression by Increasing M2 Myeloid Cells. Front. Immunol. 2019, 10, 2638. [Google Scholar] [CrossRef]
- Li, B.H.; Yang, X.Z.; Li, P.D.; Yuan, Q.; Liu, X.H.; Yuan, J.; Zhang, W.J. IL-4/Stat6 Activities Correlate with Apoptosis and Metastasis in Colon Cancer Cells. Biochem. Biophys. Res. Commun. 2008, 369, 554–560. [Google Scholar] [CrossRef] [PubMed]
- Seif, F.; Khoshmirsafa, M.; Aazami, H.; Mohsenzadegan, M.; Sedighi, G.; Bahar, M. The Role of JAK-STAT Signaling Pathway and Its Regulators in the Fate of T Helper Cells. Cell Commun. Signal. 2017, 15, 23. [Google Scholar] [CrossRef]
- LaCasse, C.J.; Janikashvili, N.; Larmonier, C.B.; Alizadeh, D.; Hanke, N.; Kartchner, J.; Situ, E.; Centuori, S.; Har-Noy, M.; Bonnotte, B.; et al. Th-1 Lymphocytes Induce Dendritic Cell Tumor Killing Activity by an IFN-γ-Dependent Mechanism. J. Immunol. 2011, 187, 6310–6317. [Google Scholar] [CrossRef]
- Vikas, P.; Borcherding, N.; Zhang, W. The Clinical Promise of Immunotherapy in Triple-Negative Breast Cancer. Cancer Manag. Res. 2018, 10, 6823–6833. [Google Scholar] [CrossRef] [PubMed]
- Münger, K.; Howley, P.M. Human Papillomavirus Immortalization and Transformation Functions. Virus Res. 2002, 89, 213–228. [Google Scholar] [CrossRef]
- Bacon, C.M.; Petricoin, E.F.; Ortaldo, J.R.; Rees, R.C.; Larner, A.C.; Johnston, J.A.; O’Shea, J.J. Interleukin 12 Induces Tyrosine Phosphorylation and Activation of STAT4 in Human Lymphocytes. Proc. Natl. Acad. Sci. USA 1995, 92, 7307–7311. [Google Scholar] [CrossRef]
- Sugimoto, N.; Nakahira, M.; Ahn, H.-J.; Micallef, M.; Hamaoka, T.; Kurimoto, M.; Fujiwara, H. Differential Requirements for JAK2 and TYK2 in T Cell Proliferation and IFN-Gamma Production Induced by IL-12 Alone or Together with IL-18. Eur. J. Immunol. 2003, 33, 243–251. [Google Scholar] [CrossRef]
- Chen, X.; Liu, Y.; Luo, X.; Pan, T.; Zhang, T.; Hu, L.; Wu, B.; Liu, W.; Wei, F. HPV16 E6-Induced M2 Macrophage Polarization in the Cervical Microenvironment via Exosomal miR-204-5p. Sci. Rep. 2024, 14, 23725. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.P.; Thorpe, J.D.; Kortum, A.N.; Coy, C.M.; Cheng, W.-Y.; Ou Yang, T.-H.; Anastassiou, D.; Beatty, J.D.; Urban, N.D.; Blau, C.A. JAK2 Expression Is Associated with Tumor-Infiltrating Lymphocytes and Improved Breast Cancer Outcomes: Implications for Evaluating JAK2 Inhibitors. Cancer Immunol. Res. 2014, 2, 301–306. [Google Scholar] [CrossRef]
- Chen, M.; Pockaj, B.; Andreozzi, M.; Barrett, M.T.; Krishna, S.; Eaton, S.; Niu, R.; Anderson, K.S. JAK2 and PD-L1 Amplification Enhance the Dynamic Expression of PD-L1 in Triple-Negative Breast Cancer. Clin. Breast Cancer 2018, 18, e1205. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wan, F.; Wang, L.; Peng, C.; Huang, R.; Peng, F. STAT4 Facilitates PD-L1 Level via IL-12R/JAK2/STAT3 Axis and Predicts Immunotherapy Response in Breast Cancer. MedComm 2023, 4, e464. [Google Scholar] [CrossRef]
- Gooch, J.L.; Christy, B.; Yee, D. STAT6 Mediates Interleukin-4 Growth Inhibition in Human Breast Cancer Cells. Neoplasia 2002, 4, 324–331. [Google Scholar] [CrossRef]
- Wang, S.; Yu, L.; Shi, W.; Li, X.; Yu, L. Prognostic Roles of Signal Transducers and Activators of Transcription Family in Human Breast Cancer. Biosci. Rep. 2018, 38, BSR20171175. [Google Scholar] [CrossRef] [PubMed]
- Venuti, A.; Donzelli, S.; Nisticò, P.; Blandino, G.; Ciliberto, G. Does Interleukin-6 Bridge SARS-CoV-2 with Virus-Associated Cancers? J. Immunother. Precis. Oncol. 2021, 4, 79–85. [Google Scholar] [CrossRef]
- Núñez-Marrero, A. Assessing the Role of the Interleukin-12/STAT4 Axis in Breast Cancer by a Bioinformatics Approach. Int. J. Sci. Basic Appl. Res. 2019, 48, 38–52. [Google Scholar]
- Zhou, F.; Chen, J.; Zhao, K.-N. Human Papillomavirus 16-Encoded E7 Protein Inhibits IFN-γ-Mediated MHC Class I Antigen Presentation and CTL-Induced Lysis by Blocking IRF-1 Expression in Mouse Keratinocytes. J. Gen. Virol. 2013, 94, 2504–2514. [Google Scholar] [CrossRef]
- Morgan, E.L.; Macdonald, A. Autocrine STAT3 Activation in HPV Positive Cervical Cancer through a Virus-Driven Rac1—NFκB—IL-6 Signalling Axis. PLOS Pathog. 2019, 15, e1007835. [Google Scholar] [CrossRef]
- Hillmer, E.J.; Zhang, H.; Li, H.S.; Watowich, S.S. STAT3 Signaling in Immunity. Cytokine Growth Factor Rev. 2016, 31, 1–15. [Google Scholar] [CrossRef]
- Levy, D.E.; Lee, C. What Does Stat3 Do? J. Clin. Investig. 2002, 109, 1143–1148. [Google Scholar] [CrossRef] [PubMed]
- Ben-Baruch, A. Host Microenvironment in Breast Cancer Development: Inflammatory Cells, Cytokines and Chemokines in Breast Cancer Progression: Reciprocal Tumor-Microenvironment Interactions. Breast Cancer Res. 2003, 5, 31–36. [Google Scholar] [CrossRef]
- Oh, K.; Lee, O.-Y.; Park, Y.; Seo, M.W.; Lee, D.-S. IL-1β Induces IL-6 Production and Increases Invasiveness and Estrogen-Independent Growth in a TG2-Dependent Manner in Human Breast Cancer Cells. BMC Cancer 2016, 16, 724. [Google Scholar] [CrossRef] [PubMed]
- Kettner, N.M.; Vijayaraghavan, S.; Durak, M.G.; Bui, T.; Kohansal, M.; Ha, M.J.; Liu, B.; Rao, X.; Wang, J.; Yi, M.; et al. Combined Inhibition of STAT3 and DNA Repair in Palbociclib-Resistant ER-Positive Breast Cancer. Clin. Cancer Res. 2019, 25, 3996–4013. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Guan, Y.-Q.; Liu, J.-M. The Role of STAT-6 as a Key Transcription Regulator in HeLa Cell Death Induced by IFN-γ/TNF-α Co-Immobilized on Nanoparticles. Biomaterials 2014, 35, 5016–5027. [Google Scholar] [CrossRef]
- Zhang, W.J.; Li, B.H.; Yang, X.Z.; Li, P.D.; Yuan, Q.; Liu, X.H.; Xu, S.B.; Zhang, Y.; Yuan, J.; Gerhard, G.S.; et al. IL-4-Induced Stat6 Activities Affect Apoptosis and Gene Expression in Breast Cancer Cells. Cytokine 2008, 42, 39–47. [Google Scholar] [CrossRef]


| Gene | Sequence | T.a | Reference |
|---|---|---|---|
| EEF1A1 F | GTTGCGGTGGGTGTCATCA | 60 °C | Leitão et al. (2014) [23] |
| EEF1A1 R | GAGTGGGGTGGCAGGTATT | 60 °C | Leitão et al. (2014) [23] |
| GAPDH F | GAAGGTGGGGCTCATTTG | 60 °C | Leitão et al. (2014) [23] |
| GAPDH R | TTAAAAGCAGCCCTGGTG | 60 °C | Leitão et al. (2014) [23] |
| STAT4 F | CCTGGGTGGACCAATCTGAA | 60 °C | Usui et al. (2003) [24] |
| STAT4 R | CTCGCAGGATGTCAGCGAA | 60 °C | Usui et al. (2003) [24] |
| JAK2 F | TCTGGGGAGTATGTTGCAGAA | 60 °C | Park et al. (2019) [25] |
| JAK2 R | AGACATGGTTGGGTGGATACC | 60 °C | Park et al. (2019) [25] |
| STAT6 F | CAAAGCCCTAGTGCTGAAGAG | 60 °C | Park et al. (2019) [25] |
| STAT6 R | CTCCTGCTGTAGCTGGGAATA | 60 °C | Park et al. (2019) [25] |
| STAT 3 F | GGAGGAGGCATTCGGAAAG | 60 °C | Zhao et al. (2018) [26] |
| STAT3 R | TCGTTGGTGTCACACACAGAT | 60 °C | Zhao et al. (2018) [26] |
| Gene | Quantification Cycle (Qc) |
|---|---|
| E5 | 25 |
| E6 | 22 |
| E7 | 29 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leão, S.L.; Parente da Silva, G.R.; dos Santos, D.L.; São Marcos, B.d.F.; Bezerra Fontes, P.H.; de Oliveira Isídio, B.E.; Simões, I.S.; Genn Barros, E.F.; Lussón, D.B.; Crispim, J.G.; et al. The Differential Expression of the JAK/STAT Pathway in Breast Cancer Cells Transfected with Human Papillomavirus Oncogenes. Viruses 2025, 17, 880. https://doi.org/10.3390/v17070880
Leão SL, Parente da Silva GR, dos Santos DL, São Marcos BdF, Bezerra Fontes PH, de Oliveira Isídio BE, Simões IS, Genn Barros EF, Lussón DB, Crispim JG, et al. The Differential Expression of the JAK/STAT Pathway in Breast Cancer Cells Transfected with Human Papillomavirus Oncogenes. Viruses. 2025; 17(7):880. https://doi.org/10.3390/v17070880
Chicago/Turabian StyleLeão, Stephanie Loureiro, Gabriel Rômulo Parente da Silva, Daffany Luana dos Santos, Bianca de França São Marcos, Pedro Henrique Bezerra Fontes, Beatriz Eda de Oliveira Isídio, Isabelle Silva Simões, Elisa Fotin Genn Barros, David Beltrán Lussón, Joelson Germano Crispim, and et al. 2025. "The Differential Expression of the JAK/STAT Pathway in Breast Cancer Cells Transfected with Human Papillomavirus Oncogenes" Viruses 17, no. 7: 880. https://doi.org/10.3390/v17070880
APA StyleLeão, S. L., Parente da Silva, G. R., dos Santos, D. L., São Marcos, B. d. F., Bezerra Fontes, P. H., de Oliveira Isídio, B. E., Simões, I. S., Genn Barros, E. F., Lussón, D. B., Crispim, J. G., Leal, L. R. S., Silva, A. J. D., Pereira Santos, V. E., & de Freitas, A. C. (2025). The Differential Expression of the JAK/STAT Pathway in Breast Cancer Cells Transfected with Human Papillomavirus Oncogenes. Viruses, 17(7), 880. https://doi.org/10.3390/v17070880







