The Use of Intrinsic Disorder and Phosphorylation by Oncogenic Viral Proteins to Dysregulate the Host Cell Cycle Through Interaction with pRb
Abstract
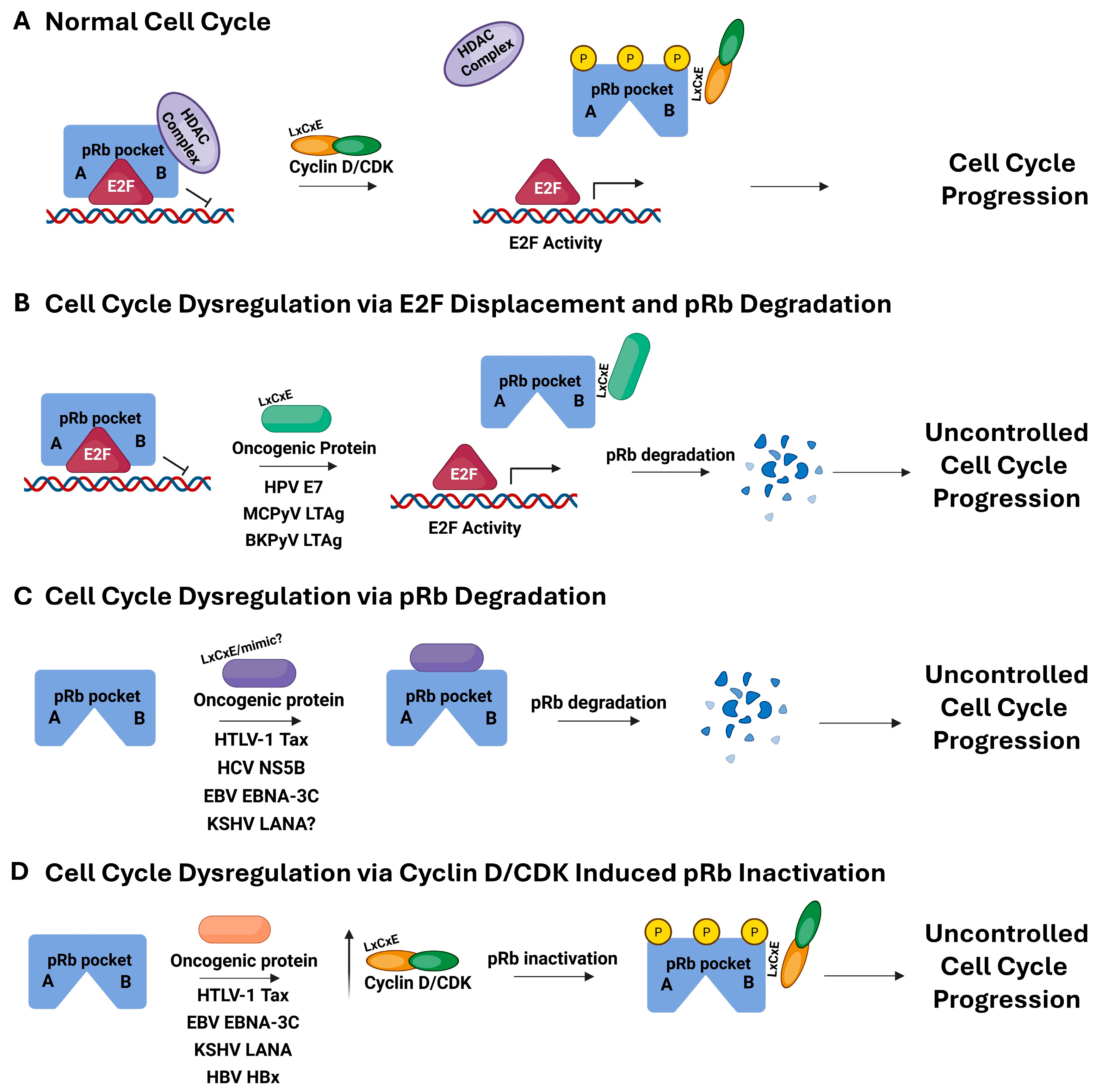
1. Introduction
2. LxCxE-Mediated pRb Interaction: Human Papillomavirus, Merkel Cell Polyomavirus, and BK Polyomavirus
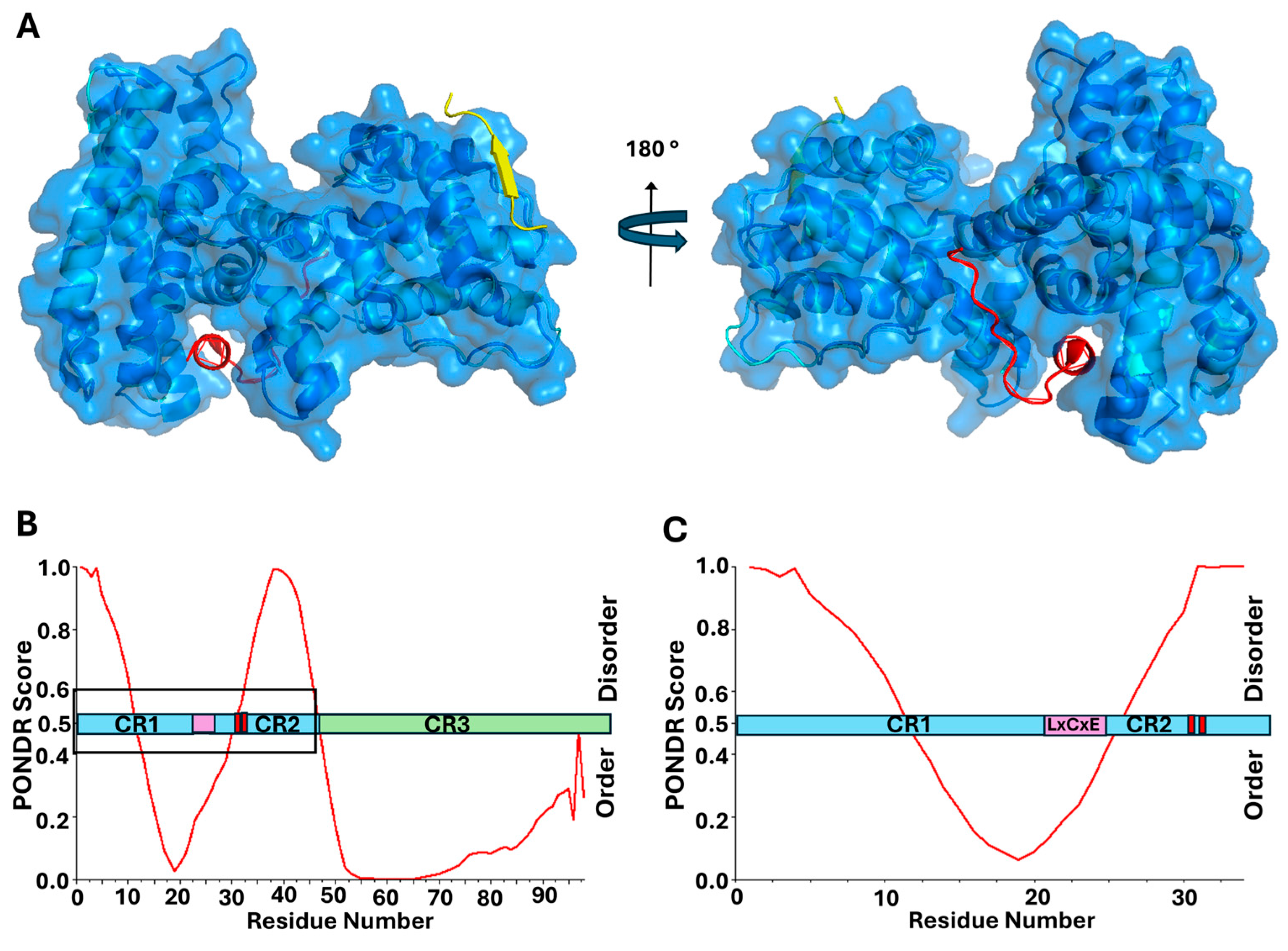
2.1. Human Papillomavirus (HPV) E7 Directly Binds and Deactivates pRb Through LxCxE Motif
2.2. Polyomaviruses
2.3. Merkel Cell Polyomavirus (MCPyV) LTAg Protein Directly Binds and Deactivates pRb Through LxCxE Motif
2.4. BK Polyomavirus (BKPyV) LTAg Protein Directly Binds and Deactivates pRb Through LxCxE Motif
3. LxCxE-Mimic Strategies: Human T-Cell Lymphotropic Virus Type 1, Hepatitis C Virus, and Epstein–Barr Virus
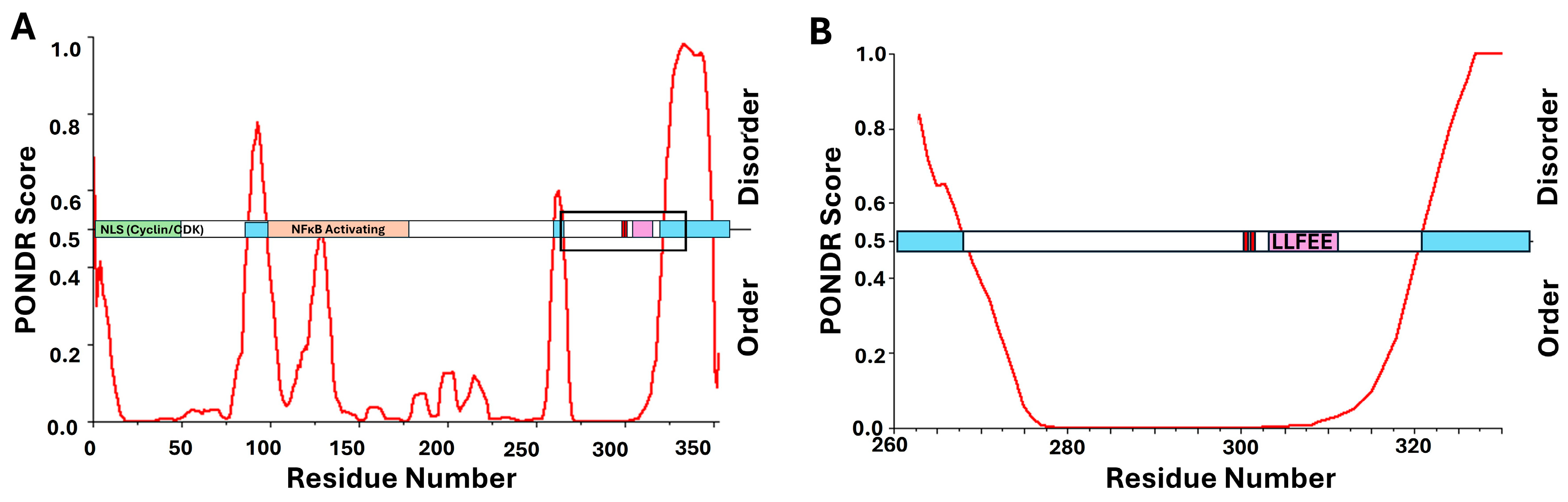
3.1. Human T-Cell Lymphotropic Virus Type 1 (HTLV-1) Tax Directly Inhibits pRb Function Through LxCxE-Mimic Motif
3.2. Hepatitis C Virus (HCV) NS5B Directly Binds and Inhibits pRb Function Through LxCxE-Mimic Motif
3.3. Epstein–Barr Virus (EBV) EBNA-3C Directly Binds pRb Through a Potential LxCxE-Mimic Domain
4. LxCxE-Independent or Indirect pRb Interaction: Kaposi Sarcoma-Associated Herpesvirus and Hepatitis B Virus
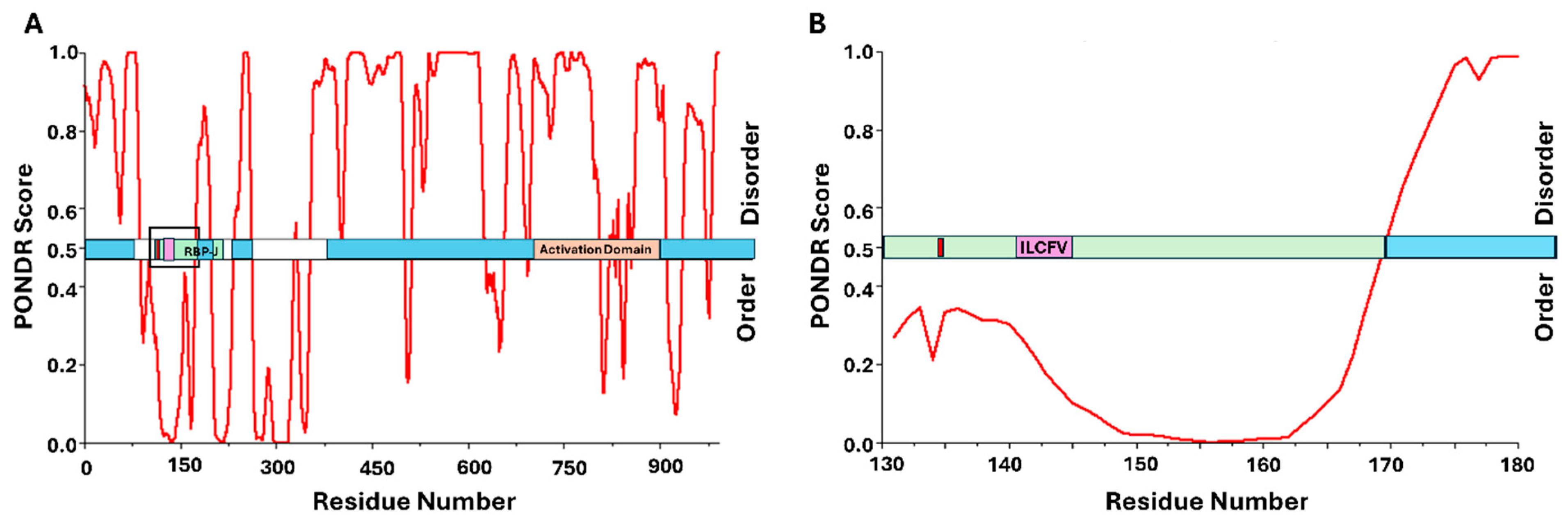
4.1. Kaposi Sarcoma-Associated Herpesvirus (KSHV) LANA Directly Binds pRb Through Its C-Terminal Region
4.2. Hepatitis B Virus (HBV) HBx Indirectly Inhibits pRb Activity with the Potential for Direct Interaction
5. Conclusions
5.1. Viruses and Cancer Biology
5.2. Viral Detection and Prevention
6. Materials and Methods
6.1. PONDR® VLXT
6.2. NETPHOS-3.1
6.3. Protein Sequence Alignment
6.4. ACCESSION Numbers
Funding
Acknowledgments
Conflicts of Interest
References
- Weiss, R.A.; Vogt, P.K. 100 years of Rous sarcoma virus. J. Exp. Med. 2011, 208, 2351–2355. [Google Scholar] [CrossRef] [PubMed]
- Moore, P.S.; Chang, Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat. Rev. Cancer 2010, 10, 878–889. [Google Scholar] [CrossRef]
- Church, C.D.; Nghiem, P. How does the Merkel polyomavirus lead to a lethal cancer? Many answers, many questions, and a new mouse model. J. Investig. Dermatol. 2015, 135, 1221–1224. [Google Scholar] [CrossRef]
- Gupta, G.; Kuppachi, S.; Kalil, R.S.; Buck, C.B.; Lynch, C.F.; Engels, E.A. Treatment for presumed BK polyomavirus nephropathy and risk of urinary tract cancers among kidney transplant recipients in the United States. Am. J. Transplant. 2018, 18, 245–252. [Google Scholar] [CrossRef]
- Starrett, G.J.; Buck, C.B. The case for BK polyomavirus as a cause of bladder cancer. Curr. Opin. Virol. 2019, 39, 8–15. [Google Scholar] [CrossRef]
- Martinez, M.P.; Al-Saleem, J.; Green, P.L. Comparative virology of HTLV-1 and HTLV-2. Retrovirology 2019, 16, 21. [Google Scholar] [CrossRef] [PubMed]
- Tessier, T.M.; Dodge, M.J.; MacNeil, K.M.; Evans, A.M.; Prusinkiewicz, M.A.; Mymryk, J.S. Almost famous: Human adenoviruses (and what they have taught us about cancer). Tumour Virus Res. 2021, 12, 200225. [Google Scholar] [CrossRef]
- Vangipuram, R.; Tyring, S.K. AIDS-Associated Malignancies. In HIV/AIDS-Associated Viral Oncogenesis; Cancer Treatment and Research; Springer: Berlin/Heidelberg, Germany, 2019; Volume 177, pp. 1–21. [Google Scholar] [CrossRef]
- Elkhalifa, A.M.E.; Nabi, S.U.; Shah, O.S.; Bashir, S.M.; Muzaffer, U.; Ali, S.I.; Wani, I.A.; Alzerwi, N.A.N.; Elderdery, A.Y.; Alanazi, A.; et al. Insight into Oncogenic Viral Pathways as Drivers of Viral Cancers: Implication for Effective Therapy. Curr. Oncol. 2023, 30, 1924–1944. [Google Scholar] [CrossRef] [PubMed]
- Zella, D.; Gallo, R.C. Viruses and Bacteria Associated with Cancer: An Overview. Viruses 2021, 13, 1039. [Google Scholar] [CrossRef]
- Gravitt, P.E.; Winer, R.L. Natural History of HPV Infection across the Lifespan: Role of Viral Latency. Viruses 2017, 9, 267. [Google Scholar] [CrossRef]
- Berman, T.A.; Schiller, J.T. Human papillomavirus in cervical cancer and oropharyngeal cancer: One cause, two diseases. Cancer 2017, 123, 2219–2229. [Google Scholar] [CrossRef] [PubMed]
- Chaiwongkot, A.; Vinokurova, S.; Pientong, C.; Ekalaksananan, T.; Kongyingyoes, B.; Kleebkaow, P.; Chumworathayi, B.; Patarapadungkit, N.; Reuschenbach, M.; von Knebel Doeberitz, M. Differential methylation of E2 binding sites in episomal and integrated HPV 16 genomes in preinvasive and invasive cervical lesions. Int. J. Cancer 2013, 132, 2087–2094. [Google Scholar] [CrossRef]
- Jeon, S.; Allen-Hoffmann, B.L.; Lambert, P.F. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 1995, 69, 2989–2997. [Google Scholar] [CrossRef]
- Hottiger, M.O.; Nabel, G.J. Viral replication and the coactivators p300 and CBP. Trends Microbiol. 2000, 8, 560–565. [Google Scholar] [CrossRef] [PubMed]
- Polansky, H.; Schwab, H. Dynamics of sequestering the limiting p300/CBP, viral cis-regulatory elements, and disease. J. Biosci. 2020, 45, 79. [Google Scholar] [CrossRef]
- Groeneveldt, C.; van Hall, T.; van der Burg, S.H.; Ten Dijke, P.; van Montfoort, N. Immunotherapeutic Potential of TGF-beta Inhibition and Oncolytic Viruses. Trends Immunol. 2020, 41, 406–420. [Google Scholar] [CrossRef]
- Lazo, P.A.; Santos, C.R. Interference with p53 functions in human viral infections, a target for novel antiviral strategies? Rev Med Virol. 2011, 21, 285–300. [Google Scholar] [CrossRef]
- Balasubramaniam, S.D.; Balakrishnan, V.; Oon, C.E.; Kaur, G. Key Molecular Events in Cervical Cancer Development. Medicina 2019, 55, 384. [Google Scholar] [CrossRef] [PubMed]
- Ludlow, J.W.; Skuse, G.R. Viral oncoprotein binding to pRB, p107, p130, and p300. Virus Res. 1995, 35, 113–121. [Google Scholar] [CrossRef]
- Schaal, D.L.; Amucheazi, A.A.; Jones, S.C.; Nkadi, E.H.; Scott, R.S. Epstein-Barr virus replication within differentiated epithelia requires pRb sequestration of activator E2F transcription factors. J. Virol. 2024, 98, e0099524. [Google Scholar] [CrossRef]
- Spurgeon, M.E.; Cheng, J.; Ward-Shaw, E.; Dick, F.A.; DeCaprio, J.A.; Lambert, P.F. Merkel cell polyomavirus large T antigen binding to pRb promotes skin hyperplasia and tumor development. PLoS Pathog. 2022, 18, e1010551. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Galvez, C.C.; Ordaz-Favila, J.C.; Villar-Calvo, V.M.; Cancino-Marentes, M.E.; Bosch-Canto, V. Retinoblastoma: Review and new insights. Front. Oncol. 2022, 12, 963780. [Google Scholar] [CrossRef]
- Seville, L.L.; Shah, N.; Westwell, A.D.; Chan, W.C. Modulation of pRB/E2F functions in the regulation of cell cycle and in cancer. Curr. Cancer Drug Targets 2005, 5, 159–170. [Google Scholar] [CrossRef]
- Stewart, C.L.; Soria, A.M.; Hamel, P.A. Integration of the pRB and p53 cell cycle control pathways. J. Neurooncol. 2001, 51, 183–204. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, L. Modeling cell cycle control and cancer with pRB tumor suppressor. In Cell Cycle Regulation; Results and Problems in Cell Differentiation; Springer: Berlin/Heidelberg, Germany, 2006; Volume 42, pp. 227–256. [Google Scholar] [CrossRef]
- Wang, C.; Fu, M.; Mani, S.; Wadler, S.; Senderowicz, A.M.; Pestell, R.G. Histone acetylation and the cell-cycle in cancer. Front. Biosci. 2001, 6, 610–629. [Google Scholar] [CrossRef] [PubMed]
- Dick, F.A.; Rubin, S.M. Molecular mechanisms underlying RB protein function. Nat. Rev. Mol. Cell Biol. 2013, 14, 297–306. [Google Scholar] [CrossRef]
- Hassler, M.; Singh, S.; Yue, W.W.; Luczynski, M.; Lakbir, R.; Sanchez-Sanchez, F.; Bader, T.; Pearl, L.H.; Mittnacht, S. Crystal structure of the retinoblastoma protein N domain provides insight into tumor suppression, ligand interaction, and holoprotein architecture. Mol. Cell 2007, 28, 371–385. [Google Scholar] [CrossRef]
- Lee, C.; Chang, J.H.; Lee, H.S.; Cho, Y. Structural basis for the recognition of the E2F transactivation domain by the retinoblastoma tumor suppressor. Genes Dev. 2002, 16, 3199–3212. [Google Scholar] [CrossRef]
- Xiao, B.; Spencer, J.; Clements, A.; Ali-Khan, N.; Mittnacht, S.; Broceno, C.; Burghammer, M.; Perrakis, A.; Marmorstein, R.; Gamblin, S.J. Crystal structure of the retinoblastoma tumor suppressor protein bound to E2F and the molecular basis of its regulation. Proc. Natl. Acad. Sci. USA 2003, 100, 2363–2368. [Google Scholar] [CrossRef]
- Connell-Crowley, L.; Harper, J.W.; Goodrich, D.W. Cyclin D1/Cdk4 regulates retinoblastoma protein-mediated cell cycle arrest by site-specific phosphorylation. Mol. Biol. Cell 1997, 8, 287–301. [Google Scholar] [CrossRef]
- Chan, H.M.; Smith, L.; La Thangue, N.B. Role of LXCXE motif-dependent interactions in the activity of the retinoblastoma protein. Oncogene 2001, 20, 6152–6163. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Cho, Y. Interactions of SV40 large T antigen and other viral proteins with retinoblastoma tumour suppressor. Rev. Med. Virol. 2002, 12, 81–92. [Google Scholar] [CrossRef]
- Singh, M.; Krajewski, M.; Mikolajka, A.; Holak, T.A. Molecular determinants for the complex formation between the retinoblastoma protein and LXCXE sequences. J. Biol. Chem. 2005, 280, 37868–37876. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, A.; Gavin, M.R.; Luo, R.X.; Dean, D.C. Role of the LXCXE binding site in Rb function. Mol. Cell. Biol. 2000, 20, 6799–6805. [Google Scholar] [CrossRef] [PubMed]
- Putta, S.; Alvarez, L.; Ludtke, S.; Sehr, P.; Muller, G.A.; Fernandez, S.M.; Tripathi, S.; Lewis, J.; Gibson, T.J.; Chemes, L.B.; et al. Structural basis for tunable affinity and specificity of LxCxE-dependent protein interactions with the retinoblastoma protein family. Structure 2022, 30, 1340–1353.e3. [Google Scholar] [CrossRef]
- Engeland, K. Cell cycle regulation: p53-p21-RB signaling. Cell Death Differ. 2022, 29, 946–960. [Google Scholar] [CrossRef]
- Fischer, M.; Schade, A.E.; Branigan, T.B.; Muller, G.A.; DeCaprio, J.A. Coordinating gene expression during the cell cycle. Trends Biochem. Sci. 2022, 47, 1009–1022. [Google Scholar] [CrossRef]
- Ovrebo, J.I.; Ma, Y.; Edgar, B.A. Cell growth and the cell cycle: New insights about persistent questions. Bioessays 2022, 44, e2200150. [Google Scholar] [CrossRef]
- Tamrakar, S.; Rubin, E.; Ludlow, J.W. Role of pRB dephosphorylation in cell cycle regulation. Front. Biosci. 2000, 5, 121–137. [Google Scholar] [CrossRef]
- Gonzalez, S.L.; Stremlau, M.; He, X.; Basile, J.R.; Munger, K. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J. Virol. 2001, 75, 7583–7591. [Google Scholar] [CrossRef]
- Kehn, K.; Fuente Cde, L.; Strouss, K.; Berro, R.; Jiang, H.; Brady, J.; Mahieux, R.; Pumfery, A.; Bottazzi, M.E.; Kashanchi, F. The HTLV-I Tax oncoprotein targets the retinoblastoma protein for proteasomal degradation. Oncogene 2005, 24, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Munakata, T.; Nakamura, M.; Liang, Y.; Li, K.; Lemon, S.M. Down-regulation of the retinoblastoma tumor suppressor by the hepatitis C virus NS5B RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA 2005, 102, 18159–18164. [Google Scholar] [CrossRef] [PubMed]
- Parker, G.A.; Crook, T.; Bain, M.; Sara, E.A.; Farrell, P.J.; Allday, M.J. Epstein-Barr virus nuclear antigen (EBNA)3C is an immortalizing oncoprotein with similar properties to adenovirus E1A and papillomavirus E7. Oncogene 1996, 13, 2541–2549. [Google Scholar]
- Haller, K.; Wu, Y.; Derow, E.; Schmitt, I.; Jeang, K.T.; Grassmann, R. Physical interaction of human T-cell leukemia virus type 1 Tax with cyclin-dependent kinase 4 stimulates the phosphorylation of retinoblastoma protein. Mol. Cell. Biol. 2002, 22, 3327–3338. [Google Scholar] [CrossRef]
- Huang, Y.; Ohtani, K.; Iwanaga, R.; Matsumura, Y.; Nakamura, M. Direct trans-activation of the human cyclin D2 gene by the oncogene product Tax of human T-cell leukemia virus type I. Oncogene 2001, 20, 1094–1102. [Google Scholar] [CrossRef] [PubMed]
- Sivasudhan, E.; Blake, N.; Lu, Z.; Meng, J.; Rong, R. Hepatitis B Viral Protein HBx and the Molecular Mechanisms Modulating the Hallmarks of Hepatocellular Carcinoma: A Comprehensive Review. Cells 2022, 11, 741. [Google Scholar] [CrossRef]
- Wei, F.; Gan, J.; Wang, C.; Zhu, C.; Cai, Q. Cell Cycle Regulatory Functions of the KSHV Oncoprotein LANA. Front. Microbiol. 2016, 7, 334. [Google Scholar] [CrossRef]
- Dyson, N.; Howley, P.M.; Munger, K.; Harlow, E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 1989, 243, 934–937. [Google Scholar] [CrossRef]
- Harris, K.F.; Christensen, J.B.; Imperiale, M.J. BK virus large T antigen: Interactions with the retinoblastoma family of tumor suppressor proteins and effects on cellular growth control. J. Virol. 1996, 70, 2378–2386. [Google Scholar] [CrossRef]
- Hesbacher, S.; Pfitzer, L.; Wiedorfer, K.; Angermeyer, S.; Borst, A.; Haferkamp, S.; Scholz, C.J.; Wobser, M.; Schrama, D.; Houben, R. RB1 is the crucial target of the Merkel cell polyomavirus Large T antigen in Merkel cell carcinoma cells. Oncotarget 2016, 7, 32956–32968. [Google Scholar] [CrossRef]
- Houben, R.; Adam, C.; Baeurle, A.; Hesbacher, S.; Grimm, J.; Angermeyer, S.; Henzel, K.; Hauser, S.; Elling, R.; Brocker, E.B.; et al. An intact retinoblastoma protein-binding site in Merkel cell polyomavirus large T antigen is required for promoting growth of Merkel cell carcinoma cells. Int. J. Cancer 2012, 130, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Munakata, T.; Liang, Y.; Kim, S.; McGivern, D.R.; Huibregtse, J.; Nomoto, A.; Lemon, S.M. Hepatitis C virus induces E6AP-dependent degradation of the retinoblastoma protein. PLoS Pathog. 2007, 3, 1335–1347. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Kim, D.H.; Lee, S.H.; Su, J.; Han, K.H. Structural investigation on the intrinsically disordered N-terminal region of HPV16 E7 protein. BMB Rep. 2016, 49, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Phelps, W.C.; Munger, K.; Yee, C.L.; Barnes, J.A.; Howley, P.M. Structure-function analysis of the human papillomavirus type 16 E7 oncoprotein. J. Virol. 1992, 66, 2418–2427. [Google Scholar] [CrossRef]
- Berlow, R.B.; Dyson, H.J.; Wright, P.E. Functional advantages of dynamic protein disorder. FEBS Lett. 2015, 589, 2433–2440. [Google Scholar] [CrossRef]
- Mishra, P.M.; Verma, N.C.; Rao, C.; Uversky, V.N.; Nandi, C.K. Intrinsically disordered proteins of viruses: Involvement in the mechanism of cell regulation and pathogenesis. Prog. Mol. Biol. Transl. Sci. 2020, 174, 1–78. [Google Scholar] [CrossRef]
- Mughal, F.; Caetano-Anolles, G. Evolution of intrinsic disorder in the structural domains of viral and cellular proteomes. Sci. Rep. 2025, 15, 2878. [Google Scholar] [CrossRef]
- Dyson, H.J. Making Sense of Intrinsically Disordered Proteins. Biophys. J. 2016, 110, 1013–1016. [Google Scholar] [CrossRef]
- Bah, A.; Forman-Kay, J.D. Modulation of Intrinsically Disordered Protein Function by Post-translational Modifications. J. Biol. Chem. 2016, 291, 6696–6705. [Google Scholar] [CrossRef]
- Kumar, N.; Kaushik, R.; Tennakoon, C.; Uversky, V.N.; Longhi, S.; Zhang, K.Y.J.; Bhatia, S. Comprehensive Intrinsic Disorder Analysis of 6108 Viral Proteomes: From the Extent of Intrinsic Disorder Penetrance to Functional Annotation of Disordered Viral Proteins. J. Proteome Res. 2021, 20, 2704–2713. [Google Scholar] [CrossRef]
- Genovese, N.J.; Banerjee, N.S.; Broker, T.R.; Chow, L.T. Casein kinase II motif-dependent phosphorylation of human papillomavirus E7 protein promotes p130 degradation and S-phase induction in differentiated human keratinocytes. J. Virol. 2008, 82, 4862–4873. [Google Scholar] [CrossRef] [PubMed]
- Lodewick, J.; Lamsoul, I.; Bex, F. Move or die: The fate of the Tax oncoprotein of HTLV-1. Viruses 2011, 3, 829–857. [Google Scholar] [CrossRef]
- Moens, U.; Passerini, S.; Falquet, M.; Sveinbjornsson, B.; Pietropaolo, V. Phosphorylation of Human Polyomavirus Large and Small T Antigens: An Ignored Research Field. Viruses 2023, 15, 2235. [Google Scholar] [CrossRef] [PubMed]
- Keating, J.A.; Striker, R. Phosphorylation events during viral infections provide potential therapeutic targets. Rev. Med. Virol. 2012, 22, 166–181. [Google Scholar] [CrossRef]
- Bex, F.; Murphy, K.; Wattiez, R.; Burny, A.; Gaynor, R.B. Phosphorylation of the human T-cell leukemia virus type 1 transactivator tax on adjacent serine residues is critical for tax activation. J. Virol. 1999, 73, 738–745. [Google Scholar] [CrossRef]
- Chemes, L.B.; Sanchez, I.E.; Smal, C.; de Prat-Gay, G. Targeting mechanism of the retinoblastoma tumor suppressor by a prototypical viral oncoprotein. FEBS J. 2010, 277, 973–988. [Google Scholar] [CrossRef]
- Prives, C. The replication functions of SV40 T antigen are regulated by phosphorylation. Cell 1990, 61, 735–738. [Google Scholar] [CrossRef]
- Schrama, D.; Hesbacher, S.; Angermeyer, S.; Schlosser, A.; Haferkamp, S.; Aue, A.; Adam, C.; Weber, A.; Schmidt, M.; Houben, R. Serine 220 phosphorylation of the Merkel cell polyomavirus large T antigen crucially supports growth of Merkel cell carcinoma cells. Int. J. Cancer 2016, 138, 1153–1162. [Google Scholar] [CrossRef] [PubMed]
- Meggio, F.; Pinna, L.A. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003, 17, 349–368. [Google Scholar] [CrossRef]
- St-Denis, N.A.; Litchfield, D.W. Protein kinase CK2 in health and disease: From birth to death: The role of protein kinase CK2 in the regulation of cell proliferation and survival. Cell. Mol. Life Sci. 2009, 66, 1817–1829. [Google Scholar] [CrossRef]
- Borgo, C.; D’Amore, C.; Sarno, S.; Salvi, M.; Ruzzene, M. Protein kinase CK2: A potential therapeutic target for diverse human diseases. Signal Transduct. Target. Ther. 2021, 6, 183. [Google Scholar] [CrossRef] [PubMed]
- Meggio, F.; Marin, O.; Pinna, L.A. Substrate specificity of protein kinase CK2. Cell. Mol. Biol. Res. 1994, 40, 401–409. [Google Scholar] [PubMed]
- Sabariegos, R.; Albentosa-Gonzalez, L.; Palmero, B.; Clemente-Casares, P.; Ramirez, E.; Garcia-Crespo, C.; Gallego, I.; de Avila, A.I.; Perales, C.; Domingo, E.; et al. Akt Phosphorylation of Hepatitis C Virus NS5B Regulates Polymerase Activity and Hepatitis C Virus Infection. Front. Microbiol. 2021, 12, 754664. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cohen, J.I. The role of PI3K/Akt in human herpesvirus infection: From the bench to the bedside. Virology 2015, 479–480, 568–577. [Google Scholar] [CrossRef]
- Graham, S.V. Human papillomavirus: Gene expression, regulation and prospects for novel diagnostic methods and antiviral therapies. Future Microbiol. 2010, 5, 1493–1506. [Google Scholar] [CrossRef]
- zur Hausen, H. Papillomaviruses in human cancers. Proc. Assoc. Am. Physicians 1999, 111, 581–587. [Google Scholar] [CrossRef]
- de Villiers, E.M.; Fauquet, C.; Broker, T.R.; Bernard, H.U.; zur Hausen, H. Classification of papillomaviruses. Virology 2004, 324, 17–27. [Google Scholar] [CrossRef]
- de Martel, C.; Ferlay, J.; Franceschi, S.; Vignat, J.; Bray, F.; Forman, D.; Plummer, M. Global burden of cancers attributable to infections in 2008: A review and synthetic analysis. Lancet Oncol. 2012, 13, 607–615. [Google Scholar] [CrossRef]
- Szymonowicz, K.A.; Chen, J. Biological and clinical aspects of HPV-related cancers. Cancer Biol. Med. 2020, 17, 864–878. [Google Scholar] [CrossRef]
- Yousaf, H.; Javed, A.; Sultana, N.; Farouk, K.; Usman, M.; Khan, G.R.; Gomez, M.K.; Asghar, S.; Justin, S. Unlocking the secrets: Exploring the connection between HPV and bladder cancer in Pakistan. Urol. Oncol. Semin. Orig. Investig. 2025. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Bruni, L.; Diaz, M.; Castellsague, X.; Ferrer, E.; Bosch, F.X.; de Sanjose, S. Cervical human papillomavirus prevalence in 5 continents: Meta-analysis of 1 million women with normal cytological findings. J. Infect. Dis. 2010, 202, 1789–1799. [Google Scholar] [CrossRef]
- Shapiro, G.K. HPV Vaccination: An Underused Strategy for the Prevention of Cancer. Curr. Oncol. 2022, 29, 3780–3792. [Google Scholar] [CrossRef] [PubMed]
- Munger, K.; White, E.A. What are the essential determinants of human papillomavirus carcinogenesis? mBio 2024, 15, e0046224. [Google Scholar] [CrossRef]
- Sund, D.T.; Brouwer, A.F.; Walline, H.M.; Carey, T.E.; Meza, R.; Jackson, T.; Eisenberg, M.C. Understanding the mechanisms of HPV-related carcinogenesis: Implications for cell cycle dynamics. J. Theor. Biol. 2022, 551–552, 111235. [Google Scholar] [CrossRef]
- Gbala, I.; Kavcic, N.; Banks, L. The retinoblastoma protein contributes to maintaining the stability of HPV E7 in cervical cancer cells. J. Virol. 2025, 99, e0220324. [Google Scholar] [CrossRef]
- Helt, A.M.; Galloway, D.A. Destabilization of the retinoblastoma tumor suppressor by human papillomavirus type 16 E7 is not sufficient to overcome cell cycle arrest in human keratinocytes. J. Virol. 2001, 75, 6737–6747. [Google Scholar] [CrossRef]
- Helt, A.M.; Galloway, D.A. Mechanisms by which DNA tumor virus oncoproteins target the Rb family of pocket proteins. Carcinogenesis 2003, 24, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Munger, K.; Jones, D.L. Human papillomavirus carcinogenesis: An identity crisis in the retinoblastoma tumor suppressor pathway. J. Virol. 2015, 89, 4708–4711. [Google Scholar] [CrossRef]
- Roman, A.; Munger, K. The papillomavirus E7 proteins. Virology 2013, 445, 138–168. [Google Scholar] [CrossRef]
- Antonio-Vejar, V.; Ortiz-Sanchez, E.; Rosendo-Chalma, P.; Patino-Morales, C.C.; Guido-Jimenez, M.C.; Alvarado-Ortiz, E.; Hernandez, G.; Garcia-Carranca, A. New insights into the interactions of HPV-16 E6*I and E6*II with p53 isoforms and induction of apoptosis in cancer-derived cell lines. Pathol. Res. Pract. 2022, 234, 153890. [Google Scholar] [CrossRef] [PubMed]
- Howie, H.L.; Katzenellenbogen, R.A.; Galloway, D.A. Papillomavirus E6 proteins. Virology 2009, 384, 324–334. [Google Scholar] [CrossRef]
- Rasi, F.; Zarredar, H.; Amini, M.; Onsori, H.; Dadashzadeh, K.; Khanmohammadi, M.; Vahedi, L.; Mokhtarzadeh, A.; Baradaran, B.; Bannazadeh Baghi, H. Suppression of E6 Oncogene Induces Apoptosis in CaSki Cervical Cancer Cells. Asian Pac. J. Cancer Prev. 2023, 24, 3389–3395. [Google Scholar] [CrossRef] [PubMed]
- Hoppe-Seyler, K.; Bossler, F.; Braun, J.A.; Herrmann, A.L.; Hoppe-Seyler, F. The HPV E6/E7 Oncogenes: Key Factors for Viral Carcinogenesis and Therapeutic Targets. Trends Microbiol. 2018, 26, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Clements, A.; Johnston, K.; Mazzarelli, J.M.; Ricciardi, R.P.; Marmorstein, R. Oligomerization properties of the viral oncoproteins adenovirus E1A and human papillomavirus E7 and their complexes with the retinoblastoma protein. Biochemistry 2000, 39, 16033–16045. [Google Scholar] [CrossRef]
- Liu, X.; Clements, A.; Zhao, K.; Marmorstein, R. Structure of the human Papillomavirus E7 oncoprotein and its mechanism for inactivation of the retinoblastoma tumor suppressor. J. Biol. Chem. 2006, 281, 578–586. [Google Scholar] [CrossRef]
- Ohlenschlager, O.; Seiboth, T.; Zengerling, H.; Briese, L.; Marchanka, A.; Ramachandran, R.; Baum, M.; Korbas, M.; Meyer-Klaucke, W.; Durst, M.; et al. Solution structure of the partially folded high-risk human papilloma virus 45 oncoprotein E7. Oncogene 2006, 25, 5953–5959. [Google Scholar] [CrossRef]
- Todorovic, B.; Massimi, P.; Hung, K.; Shaw, G.S.; Banks, L.; Mymryk, J.S. Systematic analysis of the amino acid residues of human papillomavirus type 16 E7 conserved region 3 involved in dimerization and transformation. J. Virol. 2011, 85, 10048–10057. [Google Scholar] [CrossRef]
- Bello-Rios, C.; Montano, S.; Garibay-Cerdenares, O.L.; Araujo-Arcos, L.E.; Leyva-Vazquez, M.A.; Illades-Aguiar, B. Modeling and Molecular Dynamics of the 3D Structure of the HPV16 E7 Protein and Its Variants. Int. J. Mol. Sci. 2021, 22, 1400. [Google Scholar] [CrossRef]
- Garcia-Alai, M.M.; Alonso, L.G.; de Prat-Gay, G. The N-terminal module of HPV16 E7 is an intrinsically disordered domain that confers conformational and recognition plasticity to the oncoprotein. Biochemistry 2007, 46, 10405–10412. [Google Scholar] [CrossRef]
- Jansma, A.L.; Martinez-Yamout, M.A.; Liao, R.; Sun, P.; Dyson, H.J.; Wright, P.E. The high-risk HPV16 E7 oncoprotein mediates interaction between the transcriptional coactivator CBP and the retinoblastoma protein pRb. J. Mol. Biol. 2014, 426, 4030–4048. [Google Scholar] [CrossRef] [PubMed]
- Dick, F.A.; Dyson, N.J. Three regions of the pRB pocket domain affect its inactivation by human papillomavirus E7 proteins. J. Virol. 2002, 76, 6224–6234. [Google Scholar] [CrossRef]
- Lee, J.O.; Russo, A.A.; Pavletich, N.P. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature 1998, 391, 859–865. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.S.; Patrick, D.R.; Edwards, G.; Goodhart, P.J.; Huber, H.E.; Miles, L.; Garsky, V.M.; Oliff, A.; Heimbrook, D.C. Protein domains governing interactions between E2F, the retinoblastoma gene product, and human papillomavirus type 16 E7 protein. Mol. Cell. Biol. 1993, 13, 953–960. [Google Scholar] [CrossRef]
- Blakely, W.J.; Hatterschide, J.; White, E.A. HPV18 E7 inhibits LATS1 kinase and activates YAP1 by degrading PTPN14. bioRxiv 2024, 15, e0181124. [Google Scholar] [CrossRef]
- Gelbard, M.K.; Munger, K. Human papillomaviruses: Knowns, mysteries, and unchartered territories. J. Med. Virol. 2023, 95, e29191. [Google Scholar] [CrossRef] [PubMed]
- Risor, M.W.; Jansma, A.L.; Medici, N.; Thomas, B.; Dyson, H.J.; Wright, P.E. Characterization of the High-Affinity Fuzzy Complex between the Disordered Domain of the E7 Oncoprotein from High-Risk HPV and the TAZ2 Domain of CBP. Biochemistry 2021, 60, 3887–3898. [Google Scholar] [CrossRef]
- Basukala, O.; Trejo-Cerro, O.; Myers, M.P.; Pim, D.; Massimi, P.; Thomas, M.; Guarnaccia, C.; Owen, D.; Banks, L. HPV-16 E7 Interacts with the Endocytic Machinery via the AP2 Adaptor mu2 Subunit. mBio 2022, 13, e0230222. [Google Scholar] [CrossRef]
- Barbosa, M.S.; Edmonds, C.; Fisher, C.; Schiller, J.T.; Lowy, D.R.; Vousden, K.H. The region of the HPV E7 oncoprotein homologous to adenovirus E1a and Sv40 large T antigen contains separate domains for Rb binding and casein kinase II phosphorylation. EMBO J. 1990, 9, 153–160. [Google Scholar] [CrossRef]
- Nogueira, M.O.; Hosek, T.; Calcada, E.O.; Castiglia, F.; Massimi, P.; Banks, L.; Felli, I.C.; Pierattelli, R. Monitoring HPV-16 E7 phosphorylation events. Virology 2017, 503, 70–75. [Google Scholar] [CrossRef]
- Zine El Abidine, A.; Tomaic, V.; Bel Haj Rhouma, R.; Massimi, P.; Guizani, I.; Boubaker, S.; Ennaifer, E.; Banks, L. A naturally occurring variant of HPV-16 E7 exerts increased transforming activity through acquisition of an additional phospho-acceptor site. Virology 2017, 500, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Noval, M.G.; Gallo, M.; Perrone, S.; Salvay, A.G.; Chemes, L.B.; de Prat-Gay, G. Conformational dissection of a viral intrinsically disordered domain involved in cellular transformation. PLoS ONE 2013, 8, e72760. [Google Scholar] [CrossRef] [PubMed]
- Calcada, E.O.; Felli, I.C.; Hosek, T.; Pierattelli, R. The heterogeneous structural behavior of E7 from HPV16 revealed by NMR spectroscopy. Chembiochem 2013, 14, 1876–1882. [Google Scholar] [CrossRef]
- Kukic, P.; Lo Piccolo, G.M.; Nogueira, M.O.; Svergun, D.I.; Vendruscolo, M.; Felli, I.C.; Pierattelli, R. The free energy landscape of the oncogene protein E7 of human papillomavirus type 16 reveals a complex interplay between ordered and disordered regions. Sci. Rep. 2019, 9, 5822. [Google Scholar] [CrossRef]
- Malone, M.; Maeyama, A.; Ogden, N.; Perry, K.N.; Kramer, A.; Bates, C.; Marble, C.; Orlando, R.; Rausch, A.; Smeraldi, C.; et al. The effect of phosphorylation efficiency on the oncogenic properties of the protein E7 from high-risk HPV. Virus Res. 2024, 348, 199446. [Google Scholar] [CrossRef] [PubMed]
- Czech-Sioli, M.; Gunther, T.; Therre, M.; Spohn, M.; Indenbirken, D.; Theiss, J.; Riethdorf, S.; Qi, M.; Alawi, M.; Wulbeck, C.; et al. High-resolution analysis of Merkel Cell Polyomavirus in Merkel Cell Carcinoma reveals distinct integration patterns and suggests NHEJ and MMBIR as underlying mechanisms. PLoS Pathog. 2020, 16, e1008562. [Google Scholar] [CrossRef]
- Myrda, J.; Bremm, F.; Schaft, N.; Dorrie, J. The Role of the Large T Antigen in the Molecular Pathogenesis of Merkel Cell Carcinoma. Genes 2024, 15, 1127. [Google Scholar] [CrossRef]
- Pietropaolo, V.; Prezioso, C.; Moens, U. Merkel Cell Polyomavirus and Merkel Cell Carcinoma. Cancers 2020, 12, 1774. [Google Scholar] [CrossRef]
- Feng, H.; Shuda, M.; Chang, Y.; Moore, P.S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 2008, 319, 1096–1100. [Google Scholar] [CrossRef]
- Li, J.; Wang, X.; Diaz, J.; Tsang, S.H.; Buck, C.B.; You, J. Merkel cell polyomavirus large T antigen disrupts host genomic integrity and inhibits cellular proliferation. J. Virol. 2013, 87, 9173–9188. [Google Scholar] [CrossRef]
- Tolstov, Y.L.; Pastrana, D.V.; Feng, H.; Becker, J.C.; Jenkins, F.J.; Moschos, S.; Chang, Y.; Buck, C.B.; Moore, P.S. Human Merkel cell polyomavirus infection II. MCV is a common human infection that can be detected by conformational capsid epitope immunoassays. Int. J. Cancer 2009, 125, 1250–1256. [Google Scholar] [CrossRef] [PubMed]
- Harms, K.L.; Healy, M.A.; Nghiem, P.; Sober, A.J.; Johnson, T.M.; Bichakjian, C.K.; Wong, S.L. Analysis of Prognostic Factors from 9387 Merkel Cell Carcinoma Cases Forms the Basis for the New 8th Edition AJCC Staging System. Ann. Surg. Oncol. 2016, 23, 3564–3571. [Google Scholar] [CrossRef] [PubMed]
- Paulson, K.G.; Park, S.Y.; Vandeven, N.A.; Lachance, K.; Thomas, H.; Chapuis, A.G.; Harms, K.L.; Thompson, J.A.; Bhatia, S.; Stang, A.; et al. Merkel cell carcinoma: Current US incidence and projected increases based on changing demographics. J. Am. Acad. Dermatol. 2018, 78, 457–463.e2. [Google Scholar] [CrossRef]
- Gambichler, T.; Schrama, D.; Kapynen, R.; Weyer-Fahlbusch, S.S.; Becker, J.C.; Susok, L.; Kreppel, F.; Abu Rached, N. Current Progress in Vaccines against Merkel Cell Carcinoma: A Narrative Review and Update. Vaccines 2024, 12, 533. [Google Scholar] [CrossRef]
- Becker, J.C.; Stang, A.; DeCaprio, J.A.; Cerroni, L.; Lebbe, C.; Veness, M.; Nghiem, P. Merkel cell carcinoma. Nat. Rev. Dis. Primers 2017, 3, 17077. [Google Scholar] [CrossRef]
- Verhaegen, M.E.; Mangelberger, D.; Harms, P.W.; Vozheiko, T.D.; Weick, J.W.; Wilbert, D.M.; Saunders, T.L.; Ermilov, A.N.; Bichakjian, C.K.; Johnson, T.M.; et al. Merkel cell polyomavirus small T antigen is oncogenic in transgenic mice. J. Investig. Dermatol. 2015, 135, 1415–1424. [Google Scholar] [CrossRef] [PubMed]
- Berrios, C.; Padi, M.; Keibler, M.A.; Park, D.E.; Molla, V.; Cheng, J.; Lee, S.M.; Stephanopoulos, G.; Quackenbush, J.; DeCaprio, J.A. Merkel Cell Polyomavirus Small T Antigen Promotes Pro-Glycolytic Metabolic Perturbations Required for Transformation. PLoS Pathog. 2016, 12, e1006020. [Google Scholar] [CrossRef]
- Nwogu, N.; Ortiz, L.E.; Kwun, H.J. Merkel Cell Polyomavirus Large T Antigen Unique Domain Regulates Its Own Protein Stability and Cell Growth. Viruses 2020, 12, 1043. [Google Scholar] [CrossRef]
- Shuda, M.; Feng, H.; Kwun, H.J.; Rosen, S.T.; Gjoerup, O.; Moore, P.S.; Chang, Y. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc. Natl. Acad. Sci. USA 2008, 105, 16272–16277. [Google Scholar] [CrossRef]
- Ortiz, L.E.; Pham, A.M.; Kwun, H.J. Identification of the Merkel Cell Polyomavirus Large Tumor Antigen Ubiquitin Conjugation Residue. Int. J. Mol. Sci. 2021, 22, 7169. [Google Scholar] [CrossRef]
- Houben, R.; Celikdemir, B.; Kervarrec, T.; Schrama, D. Merkel Cell Polyomavirus: Infection, Genome, Transcripts and Its Role in Development of Merkel Cell Carcinoma. Cancers 2023, 15, 444. [Google Scholar] [CrossRef] [PubMed]
- Borchert, S.; Czech-Sioli, M.; Neumann, F.; Schmidt, C.; Wimmer, P.; Dobner, T.; Grundhoff, A.; Fischer, N. High-affinity Rb binding, p53 inhibition, subcellular localization, and transformation by wild-type or tumor-derived shortened Merkel cell polyomavirus large T antigens. J. Virol. 2014, 88, 3144–3160. [Google Scholar] [CrossRef] [PubMed]
- DeCaprio, J.A.; Garcea, R.L. A cornucopia of human polyomaviruses. Nat. Rev. Microbiol. 2013, 11, 264–276. [Google Scholar] [CrossRef]
- Wendzicki, J.A.; Moore, P.S.; Chang, Y. Large T and small T antigens of Merkel cell polyomavirus. Curr. Opin. Virol. 2015, 11, 38–43. [Google Scholar] [CrossRef]
- Dyson, H.J.; Wright, P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005, 6, 197–208. [Google Scholar] [CrossRef]
- Li, J.; Diaz, J.; Wang, X.; Tsang, S.H.; You, J. Phosphorylation of Merkel cell polyomavirus large tumor antigen at serine 816 by ATM kinase induces apoptosis in host cells. J. Biol. Chem. 2015, 290, 1874–1884. [Google Scholar] [CrossRef] [PubMed]
- McVey, D.; Brizuela, L.; Mohr, I.; Marshak, D.R.; Gluzman, Y.; Beach, D. Phosphorylation of large tumour antigen by cdc2 stimulates SV40 DNA replication. Nature 1989, 341, 503–507. [Google Scholar] [CrossRef]
- Hrubey, T.W.; Roach, P.J. Phosphoserine in peptide substrates can specify casein kinase II action. Biochem. Biophys. Res. Commun. 1990, 172, 190–196. [Google Scholar] [CrossRef] [PubMed]
- St-Denis, N.; Gabriel, M.; Turowec, J.P.; Gloor, G.B.; Li, S.S.C.; Gingras, A.C.; Litchfield, D.W. Litchfield, Systematic investigation of hierarchical phosphorylation by protein kinase CK2. J. Proteom. 2015, 118, 49–62. [Google Scholar] [CrossRef]
- Gardner, S.D.; Field, A.M.; Coleman, D.V.; Hulme, B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet 1971, 1, 1253–1257. [Google Scholar] [CrossRef]
- Hirsch, H.H.; Steiger, J. Polyomavirus BK. Lancet Infect. Dis. 2003, 3, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Viscidi, R.P.; Rollison, D.E.; Sondak, V.K.; Silver, B.; Messina, J.L.; Giuliano, A.R.; Fulp, W.; Ajidahun, A.; Rivanera, D. Age-specific seroprevalence of Merkel cell polyomavirus, BK virus, and JC virus. Clin. Vaccine Immunol. 2011, 18, 1737–1743. [Google Scholar] [CrossRef]
- Furmaga, J.; Kowalczyk, M.; Zapolski, T.; Furmaga, O.; Krakowski, L.; Rudzki, G.; Jaroszynski, A.; Jakubczak, A. BK Polyomavirus-Biology, Genomic Variation and Diagnosis. Viruses 2021, 13, 1502. [Google Scholar] [CrossRef]
- Zhou, X.; Zhu, C.; Li, H. BK polyomavirus: Latency, reactivation, diseases and tumorigenesis. Front. Cell. Infect. Microbiol. 2023, 13, 1263983. [Google Scholar] [CrossRef]
- Kotton, C.N.; Kamar, N.; Wojciechowski, D.; Eder, M.; Hopfer, H.; Randhawa, P.; Sester, M.; Comoli, P.; Tedesco Silva, H.; Knoll, G.; et al. The Second International Consensus Guidelines on the Management of BK Polyomavirus in Kidney Transplantation. Transplantation 2024, 108, 1834–1866. [Google Scholar] [CrossRef]
- Yamaguchi, K.; Yamamoto, H.; Izutsu, K.; Yuasa, M.; Kaji, D.; Nishida, A.; Ishiwata, K.; Takagi, S.; Yamamoto, G.; Asano-Mori, Y.; et al. Fatal outcome of BK virus encephalitis in an allogeneic stem cell transplantation recipient. J. Infect. Chemother. 2024, 30, 1166–1169. [Google Scholar] [CrossRef] [PubMed]
- Borriello, M.; Ingrosso, D.; Perna, A.F.; Lombardi, A.; Maggi, P.; Altucci, L.; Caraglia, M. BK Virus Infection and BK-Virus-Associated Nephropathy in Renal Transplant Recipients. Genes 2022, 13, 1290. [Google Scholar] [CrossRef] [PubMed]
- Dadwal, S.S.; Papanicolaou, G.A.; Boeckh, M. How I prevent viral reactivation in high-risk patients. Blood 2023, 141, 2062–2074. [Google Scholar] [CrossRef]
- Chen, C.H.; Wen, M.C.; Wang, M.; Lian, J.D.; Cheng, C.H.; Wu, M.J.; Yu, T.M.; Chuang, Y.W.; Chang, D.; Shu, K.H. High incidence of malignancy in polyomavirus-associated nephropathy in renal transplant recipients. Transplant. Proc. 2010, 42, 817–818. [Google Scholar] [CrossRef]
- Papadimitriou, J.C.; Randhawa, P.; Rinaldo, C.H.; Drachenberg, C.B.; Alexiev, B.; Hirsch, H.H. BK Polyomavirus Infection and Renourinary Tumorigenesis. Am. J. Transplant. 2016, 16, 398–406. [Google Scholar] [CrossRef]
- Kenan, D.J.; Mieczkowski, P.A.; Burger-Calderon, R.; Singh, H.K.; Nickeleit, V. The oncogenic potential of BK-polyomavirus is linked to viral integration into the human genome. J. Pathol. 2015, 237, 379–389. [Google Scholar] [CrossRef]
- Kenan, D.J.; Mieczkowski, P.A.; Latulippe, E.; Cote, I.; Singh, H.K.; Nickeleit, V. BK Polyomavirus Genomic Integration and Large T Antigen Expression: Evolving Paradigms in Human Oncogenesis. Am. J. Transplant. 2017, 17, 1674–1680. [Google Scholar] [CrossRef]
- Monini, P.; de Lellis, L.; Rotola, A.; Di Luca, D.; Ravaioli, T.; Bigoni, B.; Cassai, E. Chimeric BK virus DNA episomes in a papillary urothelial bladder carcinoma. Intervirology 1995, 38, 304–308. [Google Scholar] [CrossRef] [PubMed]
- Sirohi, D.; Vaske, C.; Sanborn, Z.; Smith, S.C.; Don, M.D.; Lindsey, K.G.; Federman, S.; Vankalakunti, M.; Koo, J.; Bose, S.; et al. Polyoma virus-associated carcinomas of the urologic tract: A clinicopathologic and molecular study. Mod. Pathol. 2018, 31, 1429–1441. [Google Scholar] [CrossRef]
- Baez, C.F.; Brandao Varella, R.; Villani, S.; Delbue, S. Human Polyomaviruses: The Battle of Large and Small Tumor Antigens. Virology 2017, 8, 1178122X17744785. [Google Scholar] [CrossRef] [PubMed]
- Durairaj, J.; Follonier, O.M.; Leuzinger, K.; Alexander, L.T.; Wilhelm, M.; Pereira, J.; Hillenbrand, C.A.; Weissbach, F.H.; Schwede, T.; Hirsch, H.H. Structural implications of BK polyomavirus sequence variations in the major viral capsid protein Vp1 and large T-antigen: A computational study. mSphere 2024, 9, e0079923. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.K.; Vaithiyalingam, S.; Hammel, M.; Pipas, J.; Chazin, W.J. Binding to retinoblastoma pocket domain does not alter the inter-domain flexibility of the J domain of SV40 large T antigen. Arch. Biochem. Biophys. 2012, 518, 111–118. [Google Scholar] [CrossRef][Green Version]
- Helle, F.; Brochot, E.; Handala, L.; Martin, E.; Castelain, S.; Francois, C.; Duverlie, G. Biology of the BKPyV: An Update. Viruses 2017, 9, 327. [Google Scholar] [CrossRef]
- Seguin, S.P.; Ireland, A.W.; Gupta, T.; Wright, C.M.; Miyata, Y.; Wipf, P.; Pipas, J.M.; Gestwicki, J.E.; Brodsky, J.L. A screen for modulators of large T antigen’s ATPase activity uncovers novel inhibitors of Simian Virus 40 and BK virus replication. Antivir. Res. 2012, 96, 70–81. [Google Scholar] [CrossRef][Green Version]
- Poiesz, B.J.; Ruscetti, F.W.; Gazdar, A.F.; Bunn, P.A.; Minna, J.D.; Gallo, R.C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA 1980, 77, 7415–7419. [Google Scholar] [CrossRef]
- Bryan, E.S.; Tadi, P. Human T-Cell Lymphotropic Virus. In StatPearls; StatPearls: Treasure Island, FL, USA, 2025. [Google Scholar]
- Mazurov, D.; Ilinskaya, A.; Heidecker, G.; Lloyd, P.; Derse, D. Quantitative comparison of HTLV-1 and HIV-1 cell-to-cell infection with new replication dependent vectors. PLoS Pathog. 2010, 6, e1000788. [Google Scholar] [CrossRef] [PubMed]
- Bittner, J.J. Some Possible Effects of Nursing on the Mammary Gland Tumor Incidence in Mice. Science 1936, 84, 162. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Geographical Distribution of Areas with a High Prevalence of HTLV-1 Infection; ECDC: Stockholm, Sweden, 2015.
- Myers, C.S.; Williams, E.; Cornejo, C.B.; Pongas, G.; Toomey, N.L.; Sanches, J.A.; Battistella, M.; Mo, S.; Pulitzer, M.; Moskaluk, C.A.; et al. Distinctive genomic features of human T-lymphotropic virus type 1-related adult T-cell leukemia-lymphoma in Western populations. Haematologica 2024, 109, 4021–4039. [Google Scholar] [CrossRef] [PubMed]
- Einsiedel, L.J.; Pham, H.; Woodman, R.J.; Pepperill, C.; Taylor, K.A. The prevalence and clinical associations of HTLV-1 infection in a remote Indigenous community. Med. J. Aust. 2016, 205, 305–309. [Google Scholar] [CrossRef]
- Hirons, A.; Khoury, G.; Purcell, D.F.J. Human T-cell lymphotropic virus type-1: A lifelong persistent infection, yet never truly silent. Lancet Infect. Dis. 2021, 21, e2–e10. [Google Scholar] [CrossRef]
- Nakahata, S.; Enriquez-Vera, D.; Jahan, M.I.; Sugata, K.; Satou, Y. Understanding the Immunopathology of HTLV-1-Associated Adult T-Cell Leukemia/Lymphoma: A Comprehensive Review. Biomolecules 2023, 13, 1543. [Google Scholar] [CrossRef]
- Suzumiya, J.; Ohshima, K.; Tamura, K.; Karube, K.; Uike, N.; Tobinai, K.; Gascoyne, R.D.; Vose, J.M.; Armitage, J.O.; Weisenburger, D.D.; et al. The International Prognostic Index predicts outcome in aggressive adult T-cell leukemia/lymphoma: Analysis of 126 patients from the International Peripheral T-Cell Lymphoma Project. Ann. Oncol. 2009, 20, 715–721. [Google Scholar] [CrossRef]
- Weterings, D.A.; Rowan, A.G.; Cook, L.B. Immunological aspects of HTLV-1 persistence; for the prevention and treatment of Adult T-cell leukaemia-lymphoma (ATL). Leuk. Res. 2025, 148, 107635. [Google Scholar] [CrossRef]
- Forlani, G.; Shallak, M.; Accolla, R.S.; Romanelli, M.G. HTLV-1 Infection and Pathogenesis: New Insights from Cellular and Animal Models. Int. J. Mol. Sci. 2021, 22, 8001. [Google Scholar] [CrossRef]
- Giam, C.Z. HTLV-1 Replication and Adult T Cell Leukemia Development. Recent Results Cancer Res. 2021, 217, 209–243. [Google Scholar] [CrossRef]
- Hong, W.; Cheng, W.; Zheng, T.; Jiang, N.; Xu, R. AHR is a tunable knob that controls HTLV-1 latency-reactivation switching. PLoS Pathog. 2020, 16, e1008664. [Google Scholar] [CrossRef] [PubMed]
- Melamed, A.; Laydon, D.J.; Gillet, N.A.; Tanaka, Y.; Taylor, G.P.; Bangham, C.R. Genome-wide determinants of proviral targeting, clonal abundance and expression in natural HTLV-1 infection. PLoS Pathog. 2013, 9, e1003271. [Google Scholar] [CrossRef]
- Rowan, A.G.; Dillon, R.; Witkover, A.; Melamed, A.; Demontis, M.A.; Gillet, N.A.; Mun, L.J.; Bangham, C.R.M.; Cook, L.B.; Fields, P.A.; et al. Evolution of retrovirus-infected premalignant T-cell clones prior to adult T-cell leukemia/lymphoma diagnosis. Blood 2020, 135, 2023–2032. [Google Scholar] [CrossRef]
- Bazarbachi, A.; Suarez, F.; Fields, P.; Hermine, O. How I treat adult T-cell leukemia/lymphoma. Blood 2011, 118, 1736–1745. [Google Scholar] [CrossRef] [PubMed]
- Durer, C.; Babiker, H.M. Adult T-Cell Leukemia. In StatPearls; StatPearls: Treasure Island, FL, USA, 2025. [Google Scholar]
- Grassmann, R.; Aboud, M.; Jeang, K.T. Molecular mechanisms of cellular transformation by HTLV-1 Tax. Oncogene 2005, 24, 5976–5985. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, M.; Jeang, K.T. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat. Rev. Cancer 2007, 7, 270–280. [Google Scholar] [CrossRef]
- Zhao, T.; Wang, Z.; Fang, J.; Cheng, W.; Zhang, Y.; Huang, J.; Xu, L.; Gou, H.; Zeng, L.; Jin, Z.; et al. HTLV-1 activates YAP via NF-kappaB/p65 to promote oncogenesis. Proc. Natl. Acad. Sci. USA 2022, 119, e2115316119. [Google Scholar] [CrossRef]
- Boxus, M.; Twizere, J.C.; Legros, S.; Dewulf, J.F.; Kettmann, R.; Willems, L. The HTLV-1 Tax interactome. Retrovirology 2008, 5, 76. [Google Scholar] [CrossRef]
- Schmitt, I.; Rosin, O.; Rohwer, P.; Gossen, M.; Grassmann, R. Stimulation of cyclin-dependent kinase activity and G1- to S-phase transition in human lymphocytes by the human T-cell leukemia/lymphotropic virus type 1 Tax protein. J. Virol. 1998, 72, 633–640. [Google Scholar] [CrossRef]
- Ohtani, K.; Iwanaga, R.; Arai, M.; Huang, Y.; Matsumura, Y.; Nakamura, M. Cell type-specific E2F activation and cell cycle progression induced by the oncogene product Tax of human T-cell leukemia virus type I. J. Biol. Chem. 2000, 275, 11154–11163. [Google Scholar] [CrossRef]
- Lemoine, F.J.; Marriott, S.J. Accelerated G(1) phase progression induced by the human T cell leukemia virus type I (HTLV-I) Tax oncoprotein. J. Biol. Chem. 2001, 276, 31851–31857. [Google Scholar] [CrossRef] [PubMed]
- Kehn, K.; Deng, L.; de la Fuente, C.; Strouss, K.; Wu, K.; Maddukuri, A.; Baylor, S.; Rufner, R.; Pumfery, A.; Bottazzi, M.E.; et al. The role of cyclin D2 and p21/waf1 in human T-cell leukemia virus type 1 infected cells. Retrovirology 2004, 1, 6. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Santiago, F.; Clark, E.; Chong, S.; Molina, C.; Mozafari, F.; Mahieux, R.; Fujii, M.; Azimi, N.; Kashanchi, F. Transcriptional up-regulation of the cyclin D2 gene and acquisition of new cyclin-dependent kinase partners in human T-cell leukemia virus type 1-infected cells. J. Virol. 1999, 73, 9917–9927. [Google Scholar] [CrossRef]
- Ferreon, J.C.; Martinez-Yamout, M.A.; Dyson, H.J.; Wright, P.E. Structural basis for subversion of cellular control mechanisms by the adenoviral E1A oncoprotein. Proc. Natl. Acad. Sci. USA 2009, 106, 13260–13265. [Google Scholar] [CrossRef]
- Bertazzoni, U.; Turci, M.; Avesani, F.; Di Gennaro, G.; Bidoia, C.; Romanelli, M.G. Intracellular localization and cellular factors interaction of HTLV-1 and HTLV-2 Tax proteins: Similarities and functional differences. Viruses 2011, 3, 541–560. [Google Scholar] [CrossRef]
- Radkov, S.A.; Kellam, P.; Boshoff, C. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 2000, 6, 1121–1127. [Google Scholar] [CrossRef]
- World Health Organization. Hepatitis C. Available online: https://www.who.int/news-room/fact-sheets/detail/hepatitis-c (accessed on 5 April 2025).
- Guntipalli, P.; Pakala, R.; Kumari Gara, S.; Ahmed, F.; Bhatnagar, A.; Endaya Coronel, M.K.; Razzack, A.A.; Solimando, A.G.; Thompson, A.; Andrews, K.; et al. Cozzolongo R, Shahini E. Worldwide prevalence, genotype distribution and management of hepatitis C. Acta Gastroenterol Belg. 2021, 84, 637–656. [Google Scholar] [CrossRef]
- Laugi, H. Discovery of Hepatitis C Virus: 2020 Nobel Prize in Medicine. Euroasian J. Hepatogastroenterol. 2020, 10, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Axley, P.; Ahmed, Z.; Ravi, S.; Singal, A.K. Hepatitis C Virus and Hepatocellular Carcinoma: A Narrative Review. J. Clin. Transl. Hepatol. 2018, 6, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Germer, J.J.; Mandrekar, J.N.; Bendel, J.L.; Mitchell, P.S.; Yao, J.D. Hepatitis C virus genotypes in clinical specimens tested at a national reference testing laboratory in the United States. J. Clin. Microbiol. 2011, 49, 3040–3043. [Google Scholar] [CrossRef]
- Zein, N.N. Clinical significance of hepatitis C virus genotypes. Clin. Microbiol. Rev. 2000, 13, 223–235. [Google Scholar] [CrossRef]
- Mittal, G.; Gupta, P.; Gupta, R.; Ahuja, V.; Mittal, M.; Dhar, M. Seroprevalence and risk factors of hepatitis B and hepatitis C virus infections in uttarakhand, India. J. Clin. Exp. Hepatol. 2013, 3, 296–300. [Google Scholar] [CrossRef] [PubMed]
- Lemon, S.M.; McGivern, D.R. Is hepatitis C virus carcinogenic? Gastroenterology 2012, 142, 1274–1278. [Google Scholar] [CrossRef] [PubMed]
- Boyce, S.E.; Tirunagari, N.; Niedziela-Majka, A.; Perry, J.; Wong, M.; Kan, E.; Lagpacan, L.; Barauskas, O.; Hung, M.; Fenaux, M.; et al. Structural and regulatory elements of HCV NS5B polymerase—beta-loop and C-terminal tail—are required for activity of allosteric thumb site II inhibitors. PLoS ONE 2014, 9, e84808. [Google Scholar] [CrossRef] [PubMed]
- Bressanelli, S.; Tomei, L.; Roussel, A.; Incitti, I.; Vitale, R.L.; Mathieu, M.; De Francesco, R.; Rey, F.A. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA 1999, 96, 13034–13039. [Google Scholar] [CrossRef]
- Lesburg, C.A.; Cable, M.B.; Ferrari, E.; Hong, Z.; Mannarino, A.F.; Weber, P.C. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Mol. Biol. 1999, 6, 937–943. [Google Scholar] [CrossRef]
- LaPlante, S.R.; Gillard, J.R.; Jakalian, A.; Aubry, N.; Coulombe, R.; Brochu, C.; Tsantrizos, Y.S.; Poirier, M.; Kukolj, G.; Beaulieu, P.L. Importance of ligand bioactive conformation in the discovery of potent indole-diamide inhibitors of the hepatitis C virus NS5B. J. Am. Chem. Soc. 2010, 132, 15204–15212. [Google Scholar] [CrossRef]
- Li, H.C.; Yang, C.H.; Lo, S.Y. Hepatitis C Viral Replication Complex. Viruses 2021, 13, 520. [Google Scholar] [CrossRef]
- Amon, W.; Farrell, P.J. Reactivation of Epstein-Barr virus from latency. Rev. Med. Virol. 2005, 15, 149–156. [Google Scholar] [CrossRef]
- Davison, A.J.; Eberle, R.; Ehlers, B.; Hayward, G.S.; McGeoch, D.J.; Minson, A.C.; Pellett, P.E.; Roizman, B.; Studdert, M.J.; Thiry, E. The order Herpesvirales. Arch. Virol. 2009, 154, 171–177. [Google Scholar] [CrossRef]
- Epstein, M.A.; Achong, B.G.; Barr, Y.M. Virus Particles in Cultured Lymphoblasts from Burkitt’s Lymphoma. Lancet 1964, 1, 702–703. [Google Scholar] [CrossRef] [PubMed]
- Dojcinov, S.D.; Fend, F.; Quintanilla-Martinez, L. EBV-Positive Lymphoproliferations of B- T- and NK-Cell Derivation in Non-Immunocompromised Hosts. Pathogens 2018, 7, 28. [Google Scholar] [CrossRef] [PubMed]
- Taylor, G.S.; Long, H.M.; Brooks, J.M.; Rickinson, A.B.; Hislop, A.D. The immunology of Epstein-Barr virus-induced disease. Annu. Rev. Immunol. 2015, 33, 787–821. [Google Scholar] [CrossRef]
- Khan, G.; Fitzmaurice, C.; Naghavi, M.; Ahmed, L.A. Global and regional incidence, mortality and disability-adjusted life-years for Epstein-Barr virus-attributable malignancies, 1990-2017. BMJ Open 2020, 10, e037505. [Google Scholar] [CrossRef]
- Hoover, K.; Higginbotham, K. Epstein-Barr Virus. In StatPearls; StatPearls: Treasure Island, FL, USA, 2025. [Google Scholar]
- Jones, J.F.; Shurin, S.; Abramowsky, C.; Tubbs, R.R.; Sciotto, C.G.; Wahl, R.; Sands, J.; Gottman, D.; Katz, B.Z.; Sklar, J. T-cell lymphomas containing Epstein-Barr viral DNA in patients with chronic Epstein-Barr virus infections. N. Engl. J. Med. 1988, 318, 733–741. [Google Scholar] [CrossRef]
- Malpica, L.; Marques-Piubelli, M.L.; Beltran, B.E.; Chavez, J.C.; Miranda, R.N.; Castillo, J.J. EBV-positive diffuse large B-cell lymphoma, not otherwise specified: 2024 update on the diagnosis, risk-stratification, and management. Am. J. Hematol. 2024, 99, 2002–2015. [Google Scholar] [CrossRef]
- Ross, A.M.; Leahy, C.I.; Neylon, F.; Steigerova, J.; Flodr, P.; Navratilova, M.; Urbankova, H.; Vrzalikova, K.; Mundo, L.; Lazzi, S.; et al. Epstein-Barr Virus and the Pathogenesis of Diffuse Large B-Cell Lymphoma. Life 2023, 13, 521. [Google Scholar] [CrossRef]
- de Lima, M.A.P.; Teodoro, I.P.P.; da Silva, C.G.L.; Lima, M.V.A. Role of Epstein-Barr Virus and Human Papillomavirus Coinfection in Oral and Anogenital Carcinogenesis: Potential Tumorigenic Pathways. Crit. Rev. Oncog. 2019, 24, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Nahand, J.S.; Khanaliha, K.; Mirzaei, H.; Moghoofei, M.; Baghi, H.B.; Esghaei, M.; Khatami, A.R.; Fatemipour, M.; Bokharaei-Salim, F. Possible role of HPV/EBV coinfection in anoikis resistance and development in prostate cancer. BMC Cancer 2021, 21, 926. [Google Scholar] [CrossRef]
- Thorley-Lawson, D.A. EBV Persistence—Introducing the Virus. Curr. Top. Microbiol. Immunol. 2015, 390, 151–209. [Google Scholar] [CrossRef]
- Kenney, S.C.; Mertz, J.E. Regulation of the latent-lytic switch in Epstein-Barr virus. Semin. Cancer Biol. 2014, 26, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Gewruz, B.E.; Longnecker, R.M.; Cohen, J.I. Epstein-Barr Virus (Chapter 11). In Fields Virology: DNA Viruses; Lippincott Williams and Wilkins: Philadelphia, PA, USA, 2021. [Google Scholar]
- Calderwood, M.A.; Venkatesan, K.; Xing, L.; Chase, M.R.; Vazquez, A.; Holthaus, A.M.; Ewence, A.E.; Li, N.; Hirozane-Kishikawa, T.; Hill, D.E.; et al. Epstein-Barr virus and virus human protein interaction maps. Proc. Natl. Acad. Sci. USA 2007, 104, 7606–7611. [Google Scholar] [CrossRef] [PubMed]
- Damania, B.; Kenney, S.C.; Raab-Traub, N. Epstein-Barr virus: Biology and clinical disease. Cell 2022, 185, 3652–3670. [Google Scholar] [CrossRef]
- Rowe, M.; Lear, A.L.; Croom-Carter, D.; Davies, A.H.; Rickinson, A.B. Three pathways of Epstein-Barr virus gene activation from EBNA1-positive latency in B lymphocytes. J. Virol. 1992, 66, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Kieff, E. Epstein-Barr Virus and its replication. In Fields Virology, 5th ed.; Lippincott Williams and Wilkins: Philadelphia, PA, USA, 2007. [Google Scholar]
- Bhattacharjee, S.; Ghosh Roy, S.; Bose, P.; Saha, A. Role of EBNA-3 Family Proteins in EBV Associated B-cell Lymphomagenesis. Front. Microbiol. 2016, 7, 457. [Google Scholar] [CrossRef]
- Wang, D.; Liebowitz, D.; Kieff, E. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 1985, 43, 831–840. [Google Scholar] [CrossRef]
- Yu, H.; Robertson, E.S. Epstein-Barr Virus History and Pathogenesis. Viruses 2023, 15, 714. [Google Scholar] [CrossRef]
- Saha, A.; Robertson, E.S. Impact of EBV essential nuclear protein EBNA-3C on B-cell proliferation and apoptosis. Future Microbiol. 2013, 8, 323–352. [Google Scholar] [CrossRef]
- Jha, H.C.; Lu, J.; Saha, A.; Cai, Q.; Banerjee, S.; Prasad, M.A.; Robertson, E.S. EBNA3C-mediated regulation of aurora kinase B contributes to Epstein-Barr virus-induced B-cell proliferation through modulation of the activities of the retinoblastoma protein and apoptotic caspases. J. Virol. 2013, 87, 12121–12138. [Google Scholar] [CrossRef]
- Knight, J.S.; Sharma, N.; Kalman, D.E.; Robertson, E.S. A cyclin-binding motif within the amino-terminal homology domain of EBNA3C binds cyclin A and modulates cyclin A-dependent kinase activity in Epstein-Barr virus-infected cells. J. Virol. 2004, 78, 12857–12867. [Google Scholar] [CrossRef]
- Knight, J.S.; Sharma, N.; Robertson, E.S. Epstein-Barr virus latent antigen 3C can mediate the degradation of the retinoblastoma protein through an SCF cellular ubiquitin ligase. Proc. Natl. Acad. Sci. USA 2005, 102, 18562–18566. [Google Scholar] [CrossRef] [PubMed]
- Parker, G.A.; Touitou, R.; Allday, M.J. Epstein-Barr virus EBNA3C can disrupt multiple cell cycle checkpoints and induce nuclear division divorced from cytokinesis. Oncogene 2000, 19, 700–709. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Cesarman, E.; Pessin, M.S.; Lee, F.; Culpepper, J.; Knowles, D.M.; Moore, P.S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 1994, 266, 1865–1869. [Google Scholar] [CrossRef]
- Russo, J.J.; Bohenzky, R.A.; Chien, M.C.; Chen, J.; Yan, M.; Maddalena, D.; Parry, J.P.; Peruzzi, D.; Edelman, I.S.; Chang, Y.; et al. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 1996, 93, 14862–14867. [Google Scholar] [CrossRef]
- Yan, L.; Majerciak, V.; Zheng, Z.M.; Lan, K. Towards Better Understanding of KSHV Life Cycle: From Transcription and Posttranscriptional Regulations to Pathogenesis. Virol. Sin. 2019, 34, 135–161. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.W.; Dittmer, D.P. Diagnosis and Treatment of Kaposi Sarcoma. Am. J. Clin. Dermatol. 2017, 18, 529–539. [Google Scholar] [CrossRef]
- Dupin, N.; Fisher, C.; Kellam, P.; Ariad, S.; Tulliez, M.; Franck, N.; van Marck, E.; Salmon, D.; Gorin, I.; Escande, J.P.; et al. Distribution of human herpesvirus-8 latently infected cells in Kaposi’s sarcoma, multicentric Castleman’s disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. USA 1999, 96, 4546–4551. [Google Scholar] [CrossRef]
- Dittmer, D.P. Restricted Kaposi’s sarcoma (KS) herpesvirus transcription in KS lesions from patients on successful antiretroviral therapy. mBio 2011, 2, e00138-11. [Google Scholar] [CrossRef]
- Radu, O.; Pantanowitz, L. Kaposi sarcoma. Arch. Pathol. Lab. Med. 2013, 137, 289–294. [Google Scholar] [CrossRef]
- Gathers, D.A.; Galloway, E.; Kelemen, K.; Rosenthal, A.; Gibson, S.E.; Munoz, J. Primary Effusion Lymphoma: A Clinicopathologic Perspective. Cancers 2022, 14, 722. [Google Scholar] [CrossRef]
- Rainbow, L.; Platt, G.M.; Simpson, G.R.; Sarid, R.; Gao, S.J.; Stoiber, H.; Herrington, C.S.; Moore, P.S.; Schulz, T.F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 1997, 71, 5915–5921. [Google Scholar] [CrossRef] [PubMed]
- Cesarman, E.; Nador, R.G.; Bai, F.; Bohenzky, R.A.; Russo, J.J.; Moore, P.S.; Chang, Y.; Knowles, D.M. Kaposi’s sarcoma-associated herpesvirus contains G protein-coupled receptor and cyclin D homologs which are expressed in Kaposi’s sarcoma and malignant lymphoma. J. Virol. 1996, 70, 8218–8223. [Google Scholar] [CrossRef] [PubMed]
- Horenstein, M.G.; Cesarman, E.; Wang, X.; Linkov, I.; Prieto, V.G.; Louie, D.C. Cyclin D1 and retinoblastoma protein expression in Kaposi’s sarcoma. J. Cutan. Pathol. 1997, 24, 585–589. [Google Scholar] [CrossRef]
- Li, M.; Lee, H.; Yoon, D.W.; Albrecht, J.C.; Fleckenstein, B.; Neipel, F.; Jung, J.U. Kaposi’s sarcoma-associated herpesvirus encodes a functional cyclin. J. Virol. 1997, 71, 1984–1991. [Google Scholar] [CrossRef]
- Cotter, M.A., 2nd; Robertson, E.S. The latency-associated nuclear antigen tethers the Kaposi’s sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 1999, 264, 254–264. [Google Scholar] [CrossRef]
- Le Cam, L.; Polanowska, J.; Fabbrizio, E.; Olivier, M.; Philips, A.; Ng Eaton, E.; Classon, M.; Geng, Y.; Sardet, C. Timing of cyclin E gene expression depends on the regulated association of a bipartite repressor element with a novel E2F complex. EMBO J. 1999, 18, 1878–1890. [Google Scholar] [CrossRef]
- Borah, S.; Verma, S.C.; Robertson, E.S. ORF73 of herpesvirus saimiri, a viral homolog of Kaposi’s sarcoma-associated herpesvirus, modulates the two cellular tumor suppressor proteins p53 and pRb. J. Virol. 2004, 78, 10336–10347. [Google Scholar] [CrossRef] [PubMed]
- Hinds, P.W.; Mittnacht, S.; Dulic, V.; Arnold, A.; Reed, S.I.; Weinberg, R.A. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell 1992, 70, 993–1006. [Google Scholar] [CrossRef]
- Ponnusamy, R.; Petoukhov, M.V.; Correia, B.; Custodio, T.F.; Juillard, F.; Tan, M.; Pires de Miranda, M.; Carrondo, M.A.; Simas, J.P.; Kaye, K.M.; et al. KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA. Nucleic Acids Res. 2015, 43, 10039–10054. [Google Scholar] [CrossRef]
- Weidner-Glunde, M.; Mariggio, G.; Schulz, T.F. Kaposi’s Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen: Replicating and Shielding Viral DNA during Viral Persistence. J. Virol. 2017, 91, e01083-16. [Google Scholar] [CrossRef]
- Razavi-Shearer, D.; Razavi, H. Global prevalence of hepatitis B virus infection and prevention of mother-to-child transmission—Authors’ reply. Lancet Gastroenterol. Hepatol. 2018, 3, 599. [Google Scholar] [CrossRef] [PubMed]
- Schweitzer, A.; Horn, J.; Mikolajczyk, R.T.; Krause, G.; Ott, J.J. Estimations of worldwide prevalence of chronic hepatitis B virus infection: A systematic review of data published between 1965 and 2013. Lancet 2015, 386, 1546–1555. [Google Scholar] [CrossRef]
- Al-Busafi, S.A.; Alwassief, A. Global Perspectives on the Hepatitis B Vaccination: Challenges, Achievements, and the Road to Elimination by 2030. Vaccines 2024, 12, 288. [Google Scholar] [CrossRef]
- The Nobel Prize in Physiology or Medicine 1976. NobelPrize.org. Nobel Prize Outreach 2025. Mon. 9 Jun 2025. Available online: https://www.nobelprize.org/prizes/medicine/1976/summary/ (accessed on 5 April 2025).
- Campbell, C.; Wang, T.; McNaughton, A.L.; Barnes, E.; Matthews, P.C. Risk factors for the development of hepatocellular carcinoma (HCC) in chronic hepatitis B virus (HBV) infection: A systematic review and meta-analysis. J. Viral. Hepat. 2021, 28, 493–507. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.H.; Lucko, A.; Vachon, A.; Doucette, K.E.; Ramji, A.; Sycuro, L.; Patel, T.R.; Chadee, K.; Raman, M.; van Marle, G.; et al. Assessment of HBV variants and novel viral and immune biomarkers in chronic hepatitis B patients with metabolic dysfunction associated steatotic liver disease. J. Viral. Hepat. 2024, 31, 582–591. [Google Scholar] [CrossRef]
- Xu, P.; Liu, M.; Liu, M.; Shen, A. Management of non-alcoholic fatty liver disease-associated hepatocellular carcinoma. Biosci. Trends 2024, 18, 431–443. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.L.; Kao, J.H. The clinical implications of hepatitis B virus genotype: Recent advances. J. Gastroenterol. Hepatol. 2011, 26 (Suppl. 1), 123–130. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, C.; Mao, X.; Guo, C.; Suo, C.; Zhu, D.; Jiang, W.; Li, Y.; Fan, J.; Song, C.; et al. Changing prevalence of chronic hepatitis B virus infection in China between 1973 and 2021: A systematic literature review and meta-analysis of 3740 studies and 231 million people. Gut 2023, 72, 2354–2363. [Google Scholar] [CrossRef]
- Tu, T.; Budzinska, M.A.; Shackel, N.A.; Urban, S. HBV DNA Integration: Molecular Mechanisms and Clinical Implications. Viruses 2017, 9, 75. [Google Scholar] [CrossRef]
- Sekiba, K.; Otsuka, M.; Koike, K. Identifying Inhibitors of the HBx-DDB1 Interaction Using a Split Luciferase Assay System. J. Vis. Exp. 2019, 154, e60652. [Google Scholar] [CrossRef]
- Hodgson, A.J.; Hyser, J.M.; Keasler, V.V.; Cang, Y.; Slagle, B.L. Hepatitis B virus regulatory HBx protein binding to DDB1 is required but is not sufficient for maximal HBV replication. Virology 2012, 426, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Pandey, V.; Kumar, V. HBx protein of hepatitis B virus promotes reinitiation of DNA replication by regulating expression and intracellular stability of replication licensing factor CDC6. J. Biol. Chem. 2012, 287, 20545–20554. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Cha, E.J.; Lim, J.E.; Kwon, S.H.; Kim, D.H.; Cho, H.; Han, K.H. Structural characterization of an intrinsically unfolded mini-HBX protein from hepatitis B virus. Mol. Cells 2012, 34, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Prescott, N.A.; Bram, Y.; Schwartz, R.E.; David, Y. Targeting Hepatitis B Virus Covalently Closed Circular DNA and Hepatitis B Virus X Protein: Recent Advances and New Approaches. ACS Infect. Dis. 2019, 5, 1657–1667. [Google Scholar] [CrossRef]
- Lin, M.H.; Lo, S.C. Dimerization of hepatitis B viral X protein synthesized in a cell-free system. Biochem. Biophys. Res. Commun. 1989, 164, 14–21. [Google Scholar] [CrossRef]
- Farshid, M.; Nedjar, S.; Mitchell, F.; Biswas, R. Effect of hepatitis B virus X protein on the expression of retinoblastoma gene product. Acta. Virol. 1997, 41, 125–129. [Google Scholar]
- Hernandez, S.; Venegas, M.; Brahm, J.; Villanueva, R.A. The viral transactivator HBx protein exhibits a high potential for regulation via phosphorylation through an evolutionarily conserved mechanism. Infect. Agent. Cancer 2012, 7, 27. [Google Scholar] [CrossRef]
- Schillie, S.; Wester, C.; Osborne, M.; Wesolowski, L.; Ryerson, A.B. CDC Recommendations for Hepatitis C Screening Among Adults—United States, 2020. MMWR Recomm. Rep. 2020, 69, 1–17. [Google Scholar] [CrossRef]
- Mast, E.E.; Weinbaum, C.M.; Fiore, A.E.; Alter, M.J.; Bell, B.P.; Finelli, L.; Rodewald, L.E.; Douglas, J.M., Jr.; Janssen, R.S.; Ward, J.W.; et al. A comprehensive immunization strategy to eliminate transmission of hepatitis B virus infection in the United States: Recommendations of the Advisory Committee on Immunization Practices (ACIP) Part II: Immunization of adults. MMWR Recomm. Rep. 2006, 55, 1–33, quiz CE31-34. [Google Scholar]
- Meites, E.; Szilagyi, P.G.; Chesson, H.W.; Unger, E.R.; Romero, J.R.; Markowitz, L.E. Human Papillomavirus Vaccination for Adults: Updated Recommendations of the Advisory Committee on Immunization Practices. MMWR Morb. Mortal Wkly. Rep. 2019, 68, 698–702. [Google Scholar] [CrossRef]
- Cheng, L.; Wang, Y.; Du, J. Human Papillomavirus Vaccines: An Updated Review. Vaccines 2020, 8, 391. [Google Scholar] [CrossRef]
- Lei, J.; Ploner, A.; Elfstrom, K.M.; Wang, J.; Roth, A.; Fang, F.; Sundstrom, K.; Dillner, J.; Sparen, P. HPV Vaccination and the Risk of Invasive Cervical Cancer. N. Engl. J. Med. 2020, 383, 1340–1348. [Google Scholar] [CrossRef]
- Dorali, P.; Damgacioglu, H.; Clarke, M.A.; Wentzensen, N.; Orr, B.C.; Sonawane, K.; Deshmukh, A.A. Cervical Cancer Mortality Among US Women Younger Than 25 Years, 1992–2021. JAMA 2025, 333, 165–166. [Google Scholar] [CrossRef]
- Madeira, F.; Madhusoodanan, N.; Lee, J.; Eusebi, A.; Niewielska, A.; Tivey, A.R.N.; Lopez, R.; Butcher, S. The EMBL-EBI Job Dispatcher sequence analysis tools framework in 2024. Nucleic Acids Res. 2024, 52, W521–W525. [Google Scholar] [CrossRef]



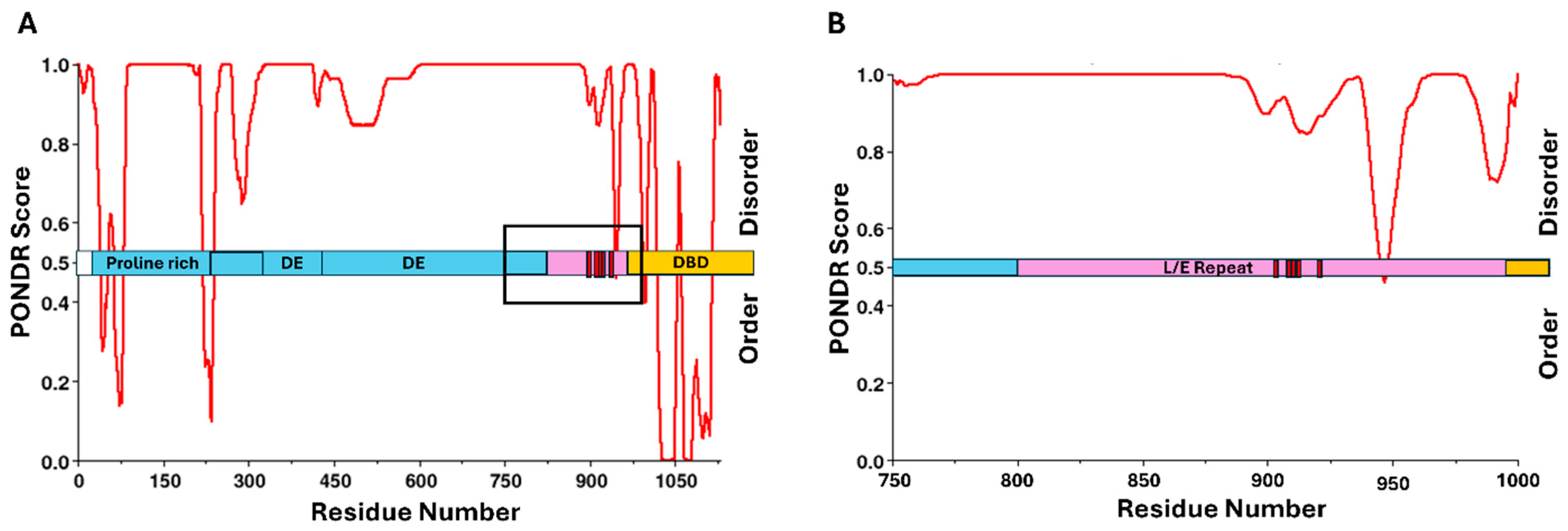
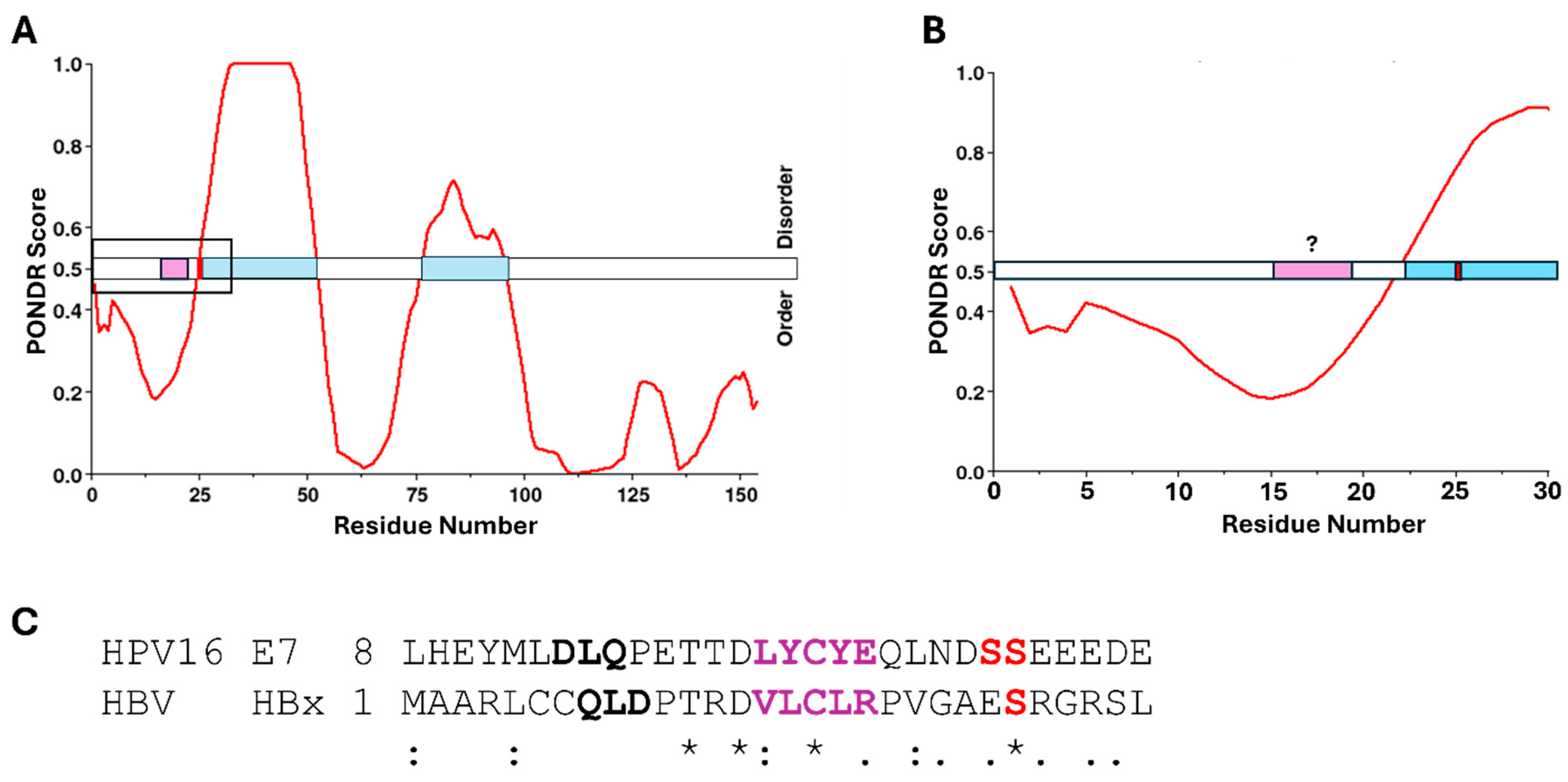
| Virus | Abbreviation | Virus Family | Oncoprotein Causing pRb Dysregulation | Mode of pRb Interaction | Phorphorylation Site(s) Proximal to pRb Binding | Kinase |
|---|---|---|---|---|---|---|
| Human Papillomavirus | HPV | Papillomaviridae | E7 | LxCxE | S31/S32 | CK2 |
| Merkel Cell Polyomavirus | MCPyV | Polyomaviridae | LTAg | LxCxE | S220 | Unknown |
| BK Polyomavirus | BKPyV | Polyomaviridae | LTAg | LxCxE | S114 | CK2 * |
| Human T-cell Lymphotropic Virus-1 | HTLV-1 | Retroviridae | Tax | LxCxE-mimic | S301/S302 | Unknown |
| Hepatitis C Virus | HCV | Hepaviridae | NS5B | LxCxE-mimic | S326 | Akt kinase |
| Epstein-Barr Virus | EBV (HHV-4) | Herpesviridae | EBNA-3C | LxCxE-mimic | S134 * | CK2 * |
| Kaposi Sarcoma-associated Herpesvirus | KSHV (HHV-8) | Herpesviridae | LANA | LxCxE-Independent | Unknown | NA |
| Hepatitis B Virus | HBV | Hepaviridae | HBx ? | Unknown | S25 ? | Unknown |
| Virus | Abbreviation | Virus Family | Viral Oncoprotein | NCBI Accession | % Predicted Disorder |
|---|---|---|---|---|---|
| Human Papillomavirus | HPV | Papillomaviridae | E7 (HPV16) | NP_041326.1 | 27.6 |
| Merkel Cell Polyomavirus | MCPyV | Polyomaviridae | LTAg | YP_009111421.1 | 32.7 |
| BK Polyomavirus | BKPyV | Polyomaviridae | LTAg | PY_717940.1 | 24.5 |
| Human T-cell Lyphotropic Virus-1 | HTLV-1 | Retrovridae | Tax | NP_957864.1 | 13 |
| Hepatisis C Virus | HCV | Hepaviridae | NS5B RNA dependent RNA polymerase | CAB46677.1 NS5B (2420-3010 of the polypeptide) | 18.6 |
| Epstein-Barr Virus | EBV (HHV-4) | Herpesviridae | EBNA-3C | YP_401671.1 | 66.8 |
| Kaposi Sarcoma-associated Herpesvirus | KSHV (HHV-8) | Herpesviridae | LANA | YP_001129431.1 | 86.7 |
| Hepatisis B Virus | HBV | Hepaviridae | HBx | ABG23459.1 | 30.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kast-Woelbern, H.; Martinho, S.K.; Julio, K.T.; Vazzana, A.M.; Mandagie, A.E.; Jansma, A.L. The Use of Intrinsic Disorder and Phosphorylation by Oncogenic Viral Proteins to Dysregulate the Host Cell Cycle Through Interaction with pRb. Viruses 2025, 17, 835. https://doi.org/10.3390/v17060835
Kast-Woelbern H, Martinho SK, Julio KT, Vazzana AM, Mandagie AE, Jansma AL. The Use of Intrinsic Disorder and Phosphorylation by Oncogenic Viral Proteins to Dysregulate the Host Cell Cycle Through Interaction with pRb. Viruses. 2025; 17(6):835. https://doi.org/10.3390/v17060835
Chicago/Turabian StyleKast-Woelbern, Heidi, Sarah K. Martinho, Kayla T. Julio, Audrey M. Vazzana, Abbey E. Mandagie, and Ariane L. Jansma. 2025. "The Use of Intrinsic Disorder and Phosphorylation by Oncogenic Viral Proteins to Dysregulate the Host Cell Cycle Through Interaction with pRb" Viruses 17, no. 6: 835. https://doi.org/10.3390/v17060835
APA StyleKast-Woelbern, H., Martinho, S. K., Julio, K. T., Vazzana, A. M., Mandagie, A. E., & Jansma, A. L. (2025). The Use of Intrinsic Disorder and Phosphorylation by Oncogenic Viral Proteins to Dysregulate the Host Cell Cycle Through Interaction with pRb. Viruses, 17(6), 835. https://doi.org/10.3390/v17060835







