Assessment and Performance of Pooled Serum Samples for Monitoring Farm-Level Immunity in Tilapia Infected with Tilapia Lake Virus
Abstract
1. Introduction
2. Materials and Methods
2.1. Serum Samples and Ethical Approval
2.2. Preparation of Recombinant TiLV Segment 4 Protein
2.2.1. Construction of Recombinant TiLV Segment 4 Plasmid
2.2.2. Expression and Purification of Recombinant Protein
2.3. Optimization of TiLV-S4 ELISA
2.4. Cut-Off Value for Classification of an ELISA Result as Positive or Negative
2.5. Repeatability and Reproducibility of TiLV-S4 ELISA
2.6. Sensitivity and Specificity of TiLV-S4 ELISA
2.7. Viral Antigen Preparation and TiLV-WV ELISA Protocol
2.8. Design of Pooling Experiments
2.9. Data Management and Statistical Analysis
3. Results
3.1. Production of TiLV-S4 Protein and Its Specificity
3.2. Development and Optimization of a TiLV-S4 ELISA
3.2.1. Validation of TiLV-S4 Recombinant Protein and Primary Antibody Concentrations
3.2.2. Repeatability and Reproducibility of the TiLV-S4 ELISA
3.2.3. Determination of the Cut-Off Value for TiLV-S4 ELISA for Classification of Results as Positive or Negative
3.2.4. Efficiency of TiLV-S4 ELISA Compared with the Reference Method
3.3. Pooling Efficiency
3.3.1. Pooling Scenarios
3.3.2. Efficiency of Pooling Technique to Detect Antibodies Against TiLV in Fish Populations
3.4. Pooled Compared with Individual Test Results in Farms A, B, C, and D
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. The State of World Fisheries and Aquaculture (SOFIA). In Towards Blue Transformation [Internet]; FAO: Rome, Italy, 2022; Available online: https://openknowledge.fao.org/handle/20.500.14283/cc0461en (accessed on 26 July 2024).
- WOAH. Infection with Tilapia Lake Virus (TiLV)—A Novel Orthomyxo-like Virus. In WOAH Disease Card [Internet]; WOAH: Paris, France, 2022; Available online: https://www.woah.org/en/document/infection-with-tilapia-lake-virus-tilv/ (accessed on 26 July 2024).
- Surachetpong, W.; Roy, S.R.K.; Nicholson, P. Tilapia lake virus: The story so far. J. Fish Dis. 2020, 43, 1115–1132. [Google Scholar] [CrossRef] [PubMed]
- Jaemwimol, P.; Rawiwan, P.; Tattiyapong, P.; Kamlangdee, A.; Surachetpong, W. Susceptibility of important warm water fish species to tilapia lake virus (TiLV) infection. Aquaculture 2018, 497, 462–468. [Google Scholar] [CrossRef]
- Phusantisampan, T.; Tattiyapong, P.; Mutrakulcharoen, P.; Sriariyanun, M.; Surachetpong, W. Rapid detection of tilapia lake virus using a one-step reverse transcription loop-mediated isothermal amplification assay. Aquaculture 2019, 507, 35–39. [Google Scholar] [CrossRef]
- Piewbang, C.; Tattiyapong, P.; Techangamsuwan, S.; Surachetpong, W. Tilapia lake virus immunoglobulin G (TiLV IgG) antibody: Immunohistochemistry application reveals cellular tropism of TiLV infection. Fish Shellfish Immunol. 2021, 116, 115–123. [Google Scholar] [CrossRef]
- Tattiyapong, P.; Sirikanchana, K.; Surachetpong, W. Development and validation of a reverse transcription quantitative polymerase chain reaction for tilapia lake virus detection in clinical samples and experimentally challenged fish. J. Fish Dis. 2018, 41, 255–261. [Google Scholar] [CrossRef]
- Tattiyapong, P.; Dechavichitlead, W.; Waltzek, T.B.; Surachetpong, W. Tilapia develop protective immunity including a humoral response following exposure to tilapia lake virus. Fish Shellfish Immunol. 2020, 106, 666–674. [Google Scholar] [CrossRef]
- Hu, H.; Zeng, W.; Wang, Y.; Bergmann, S.M.; Yin, J. Development and application of a recombinant protein-based indirect ELISA for detection of anti-tilapia lake virus IgM in sera from tilapia. Aquaculture 2020, 520, 734756. [Google Scholar] [CrossRef]
- Lalruatfela; Bedekar, M.K.; Godavarikar, A.; Valsalam, A.; Gireesh Babu, P.; Rajendran, K.V. Molecular cloning and expression of codon-optimized segment 4 hypothetical protein (35 kDa) of tilapia lake virus (TiLV) in pET-28a( +) expression vector and development of indirect ELISA test. Aquac. Int. 2024, 32, 5997–6015. [Google Scholar] [CrossRef]
- Bergmann, S.M.; Wang, Q.; Zeng, W.; Li, Y.; Wang, Y.; Matras, M. Validation of a KHV antibody enzyme-linked immunosorbent assay (ELISA). J. Fish Dis. 2017, 40, 1511–1527. [Google Scholar] [CrossRef]
- Kim, H.J.; Park, J.S.; Kwon, S.R. Development of a stringent ELISA protocol to evaluate anti-viral hemorrhagic septicemia virus-specific antibodies in olive flounder Paralichthys olivaceus with improved specificity. J. Microbiol. 2015, 53, 481–485. [Google Scholar] [CrossRef]
- Zeng, W.; Wang, Y.; Guo, Y.; Bergmann, S.M.; Yin, Y.; Li, Y. Development of a VP38 recombinant protein-based indirect ELISA for detection of antibodies against grass carp reovirus genotype II (iELISA for detection of antibodies against GCRV II). J. Fish Dis. 2018, 41, 1811–1819. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Oseka, N.; Nishizawa, T.; Yoshimizu, M. Protection of rainbow trout from infectious hematopoietic necrosis (IHN) by injection of infectious pancreatic necrosis virus (IPNV) or poly(I:C). Dis. Aquat. Organ. 2009, 83, 105–113. [Google Scholar] [CrossRef]
- Nishizawa, T.; Takami, I.; Kokawa, Y.; Yoshimizu, M. Fish immunization using a synthetic double-stranded RNA Poly(I:C), an interferon inducer, offers protection against RGNNV, a fish nodavirus. Dis. Aquat. Organ. 2009, 83, 115–122. [Google Scholar] [CrossRef]
- Takami, I.; Kwon, S.R.; Nishizawa, T.; Yoshimizu, M. Protection of Japanese flounder Paralichthys olivaceus from viral hemorrhagic septicemia (VHS) by Poly(I:C) immunization. Dis. Aquat. Organ. 2010, 89, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Fereidouni, S.R.; Harder, T.C.; Gaidet, N.; Ziller, M.; Hoffmann, B.; Hammoumi, S. Saving resources: Avian influenza surveillance using pooled swab samples and reduced reaction volumes in real-time RT-PCR. J. Virol. Methods 2012, 186, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.J.; Hick, P.M.; Robinson, A.P.; Rimmer, A.E.; Tweedie, A.; Becker, J.A. The impact of pooling samples on surveillance sensitivity for the megalocytivirus Infectious spleen and kidney necrosis virus. Transbound. Emerg. Dis. 2019, 66, 2318–2328. [Google Scholar] [CrossRef]
- Nguyen, N.T.; Bish, E.K.; Bish, D.R. Optimal pooled testing design for prevalence estimation under resource constraints. Omega 2021, 105, 102504. [Google Scholar] [CrossRef]
- Yamkasem, J.; Roy, S.R.K.; Khemthong, M.; Gardner, I.A.; Syrachetpong, W. Diagnostic sensitivity of pooled samples for the detection of tilapia lake virus and application to the estimation of within-farm prevalence. Transbound. Emerg. Dis. 2021, 68, 3519–3528. [Google Scholar] [CrossRef]
- Kembou-Ringert, J.E.; Steinhagen, D.; Readman, J.; Daly, J.M.; Adamek, M. Tilapia Lake Virus Vaccine Development: A Review on the Recent Advances. Vaccines 2023, 11, 251. [Google Scholar] [CrossRef]
- Tran, T.H.; Nguyen, V.T.H.; Bui, H.C.N.; Tran, Y.B.T.; Tran, H.T.T.; Le, T.T.T. Tilapia Lake Virus (TiLV) from Vietnam is genetically distantly related to TiLV strains from other countries. J. Fish Dis. 2022, 45, 1389–1401. [Google Scholar] [CrossRef]
- Dong, H.; Siriroob, S.; Meemetta, W.; Santimanawong, W.; Gangnonngiw, W.; Pirarat, N. Emergence of tilapia lake virus in Thailand and an alternative semi-nested RT-PCR for detection. Aquaculture 2017, 476, 111–118. [Google Scholar] [CrossRef]
- Eyngor, M.; Zamostiano, R.; Kembou Tsofack, J.E.; Berkowitz, A.; Bercovier, H.; Tinman, S. Identification of a novel RNA virus lethal to tilapia. J. Clin. Microbiol. 2014, 52, 4137–4146. [Google Scholar] [CrossRef]
- Jaramillo, D.; Peeler, E.J.; Laurin, E.; Gardner, I.A.; Whittington, R.J. Serology in Finfish for Diagnosis, Surveillance, and Research: A Systematic Review. J. Aquat. Anim. Health 2017, 29, 1–14. [Google Scholar] [CrossRef]
- Matsuyama, T.; Sano, N.; Takano, T.; Sakai, T.; Yasuike, M.; Fujiwara, A. Antibody profiling using a recombinant protein-based multiplex ELISA array accelerates recombinant vaccine development: Case study on red sea bream iridovirus as a reverse vaccinology model. Vaccine 2018, 36, 2643–2649. [Google Scholar] [CrossRef]
- Rahnama, R.; Peyghan, R.; Reza Seyfi Abad Shapouri, M.; Rezaie, A.; Shahbazian, N. Designing an in-house ELISA to detect antibody against viral haemorrhagic septicaemia virus using recombinant N protein in Iranian farmed rainbow trout (Oncorhynchus mykiss). Aquac. Res. 2019, 50, 474–482. [Google Scholar] [CrossRef]
- Watanabe, K.I.; Nishizawa, T.; Yoshimizu, M. Selection of brood stock candidates of barfin flounder using an ELISA system with recombinant protein of barfin flounder nervous necrosis virus. Dis. Aquat. Organ. 2000, 41, 219–223. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.; Zhang, G.; Zhang, X.; Chen, X.; Zhang, M.; Li, L. Development of an Indirect ELISA Based on Spike Protein to Detect Antibodies against Feline Coronavirus. Viruses 2021, 13, 2496. [Google Scholar] [CrossRef]
- Lu, M.; Liu, Q.; Wang, X.; Zhang, J.; Zhang, X.; Shi, D. Development of an indirect ELISA for detecting porcine deltacoronavirus IgA antibodies. Arch. Virol. 2020, 165, 845–851. [Google Scholar] [CrossRef]
- Jacobson, R.H. Validation of serological assays for diagnosis of infectious diseases. Rev. Sci. Tech. 1998, 17, 469–526. [Google Scholar] [CrossRef]
- Baruch, J.; Suanes, A.; Piaggio, J.M.; Gil, A.D. Analytic Sensitivity of an ELISA Test on Pooled Sera Samples for Detection of Bovine Brucellosis in Eradication Stages in Uruguay. Front. Vet. Sci. 2020, 7, 178. [Google Scholar] [CrossRef]
- Hernandez-Medrano, J.H.; Espinosa-Castillo, L.F.; Rodriguez, A.D.; Gutierrez, C.G.; Wapenaar, W. Use of pooled serum samples to assess herd disease status using commercially available ELISAs. Trop. Anim. Health Prod. 2021, 53, 507. [Google Scholar] [CrossRef]
- Nezlin, R. The Immunoglobulins: Structure and Function; Academic Press: Cambridge, MA, USA, 1998; Volume 10. [Google Scholar]
- Wang, W.; Nema, S.; Teagarden, D. Protein aggregation-pathways and influencing factors. Int. J. Pharm. 2010, 390, 89–99. [Google Scholar] [CrossRef]

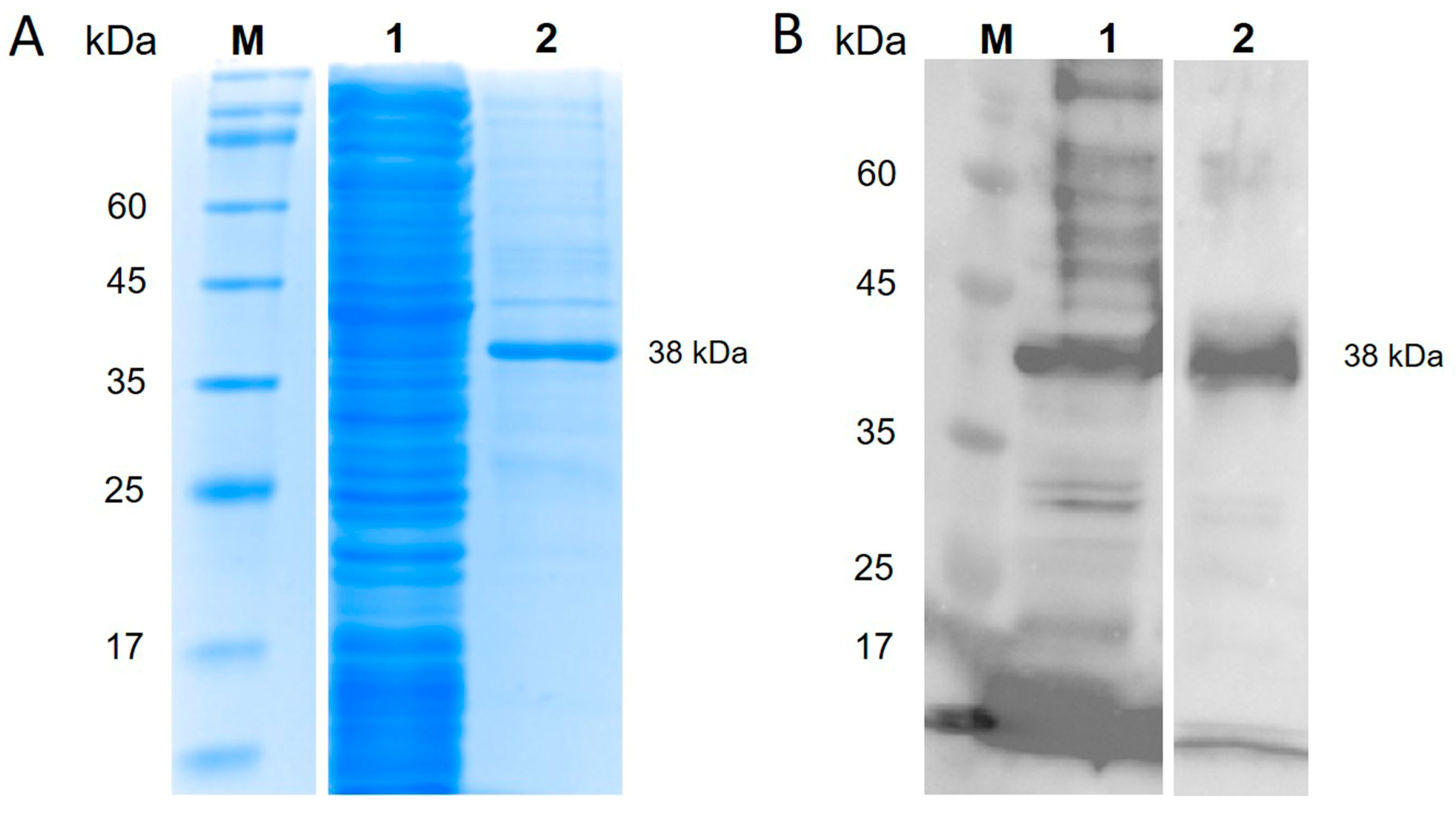
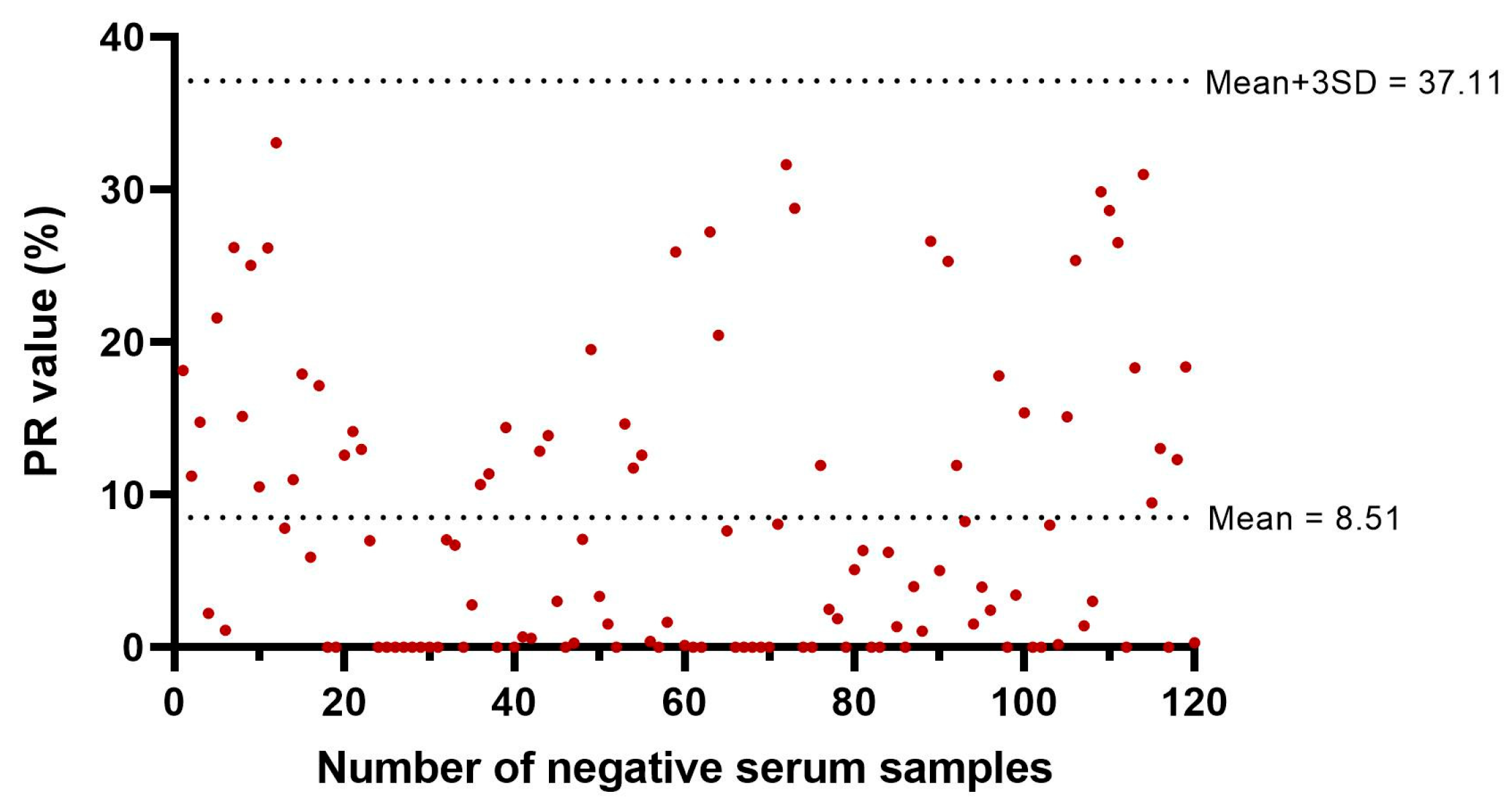

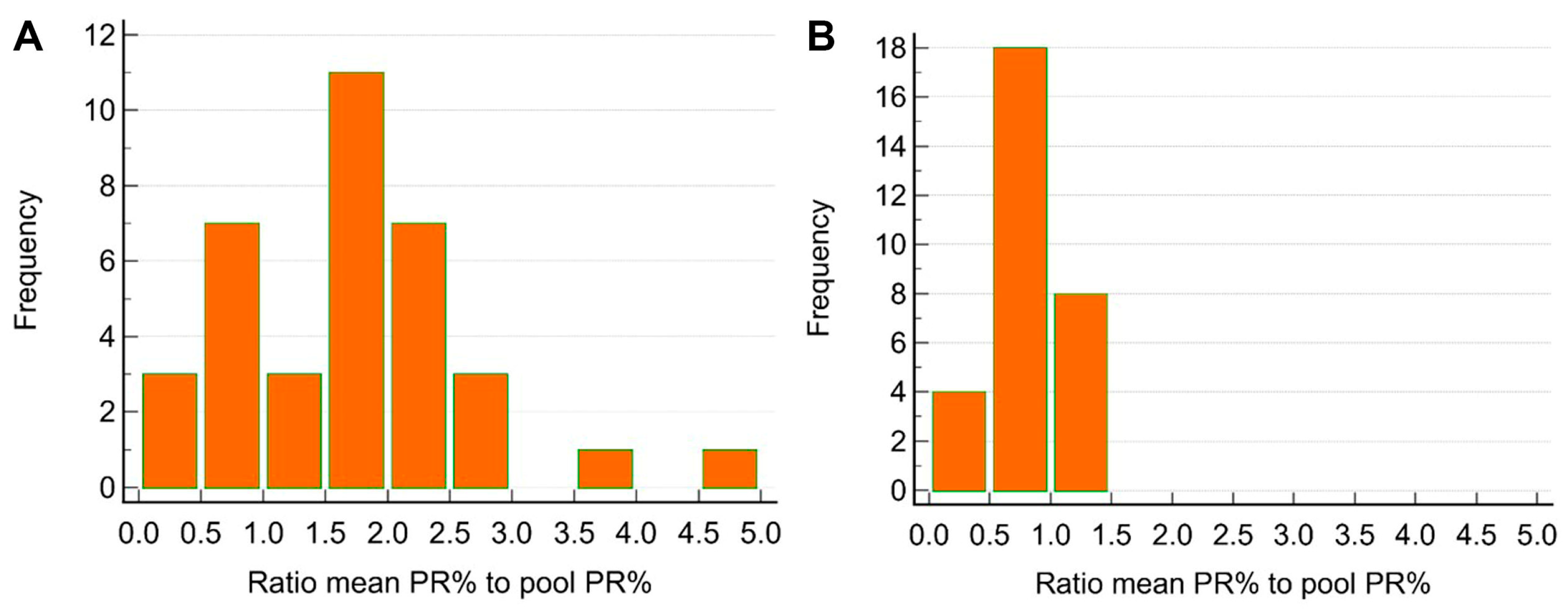
| Farms | Total Sample | Cumulative Mortality (%) During Outbreak | Day Post Infection (dpi) | Outbreak Start Date | Blood Collection Date | Average Fish Weight (g) |
|---|---|---|---|---|---|---|
| A | 40 | 33 | 28 | 10 September 2022 | 7 October 2022 | 150 |
| B | 38 | 23 | 39 | 28 August 2022 | 7 October 2022 | 30 |
| C | 42 | 40 | 60 | 6 July 2022 | 9 September 2022 | 200 |
| D | 40 | 40 | 90 | 6 July 2022 | 7 October 2022 | 400 |
| Samples | Mean OD Value | SD | CV% | |
|---|---|---|---|---|
| Repeatability (Intra-assay) | 1 | 1.06 | 0.04 | 3.6 |
| 2 | 0.91 | 0.01 | 1.1 | |
| 3 | 1.47 | 0.13 | 8.7 | |
| 4 | 0.08 | 0.00 | 2.7 | |
| 5 | 0.17 | 0.00 | 2.7 | |
| 6 | 0.21 | 0.02 | 7.7 | |
| Reproducibility (Inter-assay) | 1 | 1.03 | 0.08 | 7.7 |
| 2 | 0.91 | 0.03 | 3.1 | |
| 3 | 1.45 | 0.11 | 7.6 | |
| 4 | 0.08 | 0.01 | 6.8 | |
| 5 | 0.18 | 0.01 | 5.0 | |
| 6 | 0.20 | 0.00 | 1.7 |
| Sample No. | PR Value (%) of Pooled Sample | PR Value (%) of Individual Sample | ||||
|---|---|---|---|---|---|---|
| 1 | 14.2 | 223.6 | 26.4 | 21.9 | 26.5 | 20.8 |
| 2 | 20.8 | 94.0 | 21.9 | 26.5 | 20.8 | 20.1 |
| 3 | 14.2 | 55.4 | 15.8 | 26.4 | 21.9 | 26.5 |
| 4 | 16.5 | 49.2 | 26.4 | 21.9 | 26.5 | 20.8 |
| 5 | 12.3 | 44.0 | 21.9 | 26.5 | 20.8 | 20.1 |
| 6 | 14.9 | 40.1 | 39.7 | 15.8 | 26.4 | 24.6 |
| 7 | 29.4 | 176.5 | 44.0 | 20.1 | 31.4 | 24.6 |
| 8 | 16.2 | 70.9 | 40.1 | 15.8 | 26.4 | 21.9 |
| 9 | 26.8 | 67.0 | 44.0 | 20.8 | 20.1 | 26.5 |
| 10 | 21.1 | 53.4 | 44.0 | 20.1 | 20.8 | 31.4 |
| 11 | 26.0 | 45.2 | 44.0 | 20.1 | 31.4 | 24.6 |
| 12 | 131.2 | 248.9 | 223.6 | 21.9 | 26.5 | 20.8 |
| 13 | 35.1 | 153.5 | 67.0 | 15.8 | 26.4 | 21.9 |
| 14 | 16.2 | 176.5 | 58.5 | 26.5 | 20.8 | 20.1 |
| 15 | 20.4 | 70.9 | 63.2 | 26.4 | 21.9 | 26.5 |
| 16 | 17.1 | 45.2 | 44.5 | 21.9 | 26.5 | 20.8 |
| 17 | 16.9 | 44.5 | 39.7 | 40.1 | 15.8 | 26.4 |
| 18 | 25.7 | 59.7 | 39.7 | 40.1 | 31.4 | 24.6 |
| 19 | 13.4 | 52.8 | 39.7 | 40.1 | 24.6 | 15.8 |
| 20 | 37.7 | 58.5 | 55.4 | 45.2 | 21.9 | 26.5 |
| 21 | 15.5 | 59.4 | 49.2 | 39.7 | 31.4 | 24.6 |
| 22 | 95.8 | 59.7 | 53.4 | 44.5 | 26.4 | 20.8 |
| 23 | 22.9 | 63.5 | 45.2 | 40.1 | 15.8 | 26.4 |
| 24 | 23.9 | 72.0 | 55.4 | 44.0 | 20.8 | 20.1 |
| 25 | 54.5 | 76.0 | 58.5 | 45.2 | 24.6 | 15.8 |
| 26 | 17.5 | 76.0 | 44.0 | 39.7 | 24.6 | 31.4 |
| 27 | 72.5 | 89.6 | 68.5 | 55.4 | 20.8 | 20.1 |
| 28 | 137.5 | 94.0 | 70.9 | 63.5 | 15.8 | 26.4 |
| 29 | 60.1 | 92.7 | 70.9 | 63.2 | 21.9 | 26.5 |
| 30 | 24.6 | 112.0 | 39.7 | 40.1 | 15.8 | 26.4 |
| 31 | 133.3 | 153.5 | 58.5 | 49.2 | 31.4 | 24.6 |
| 32 | 27.4 | 153.5 | 44.5 | 44.0 | 31.4 | 24.6 |
| 33 | 80.4 | 176.5 | 89.6 | 72.0 | 26.4 | 21.9 |
| 34 | 39.0 | 223.6 | 176.5 | 44.5 | 26.5 | 20.8 |
| 35 | 87.3 | 248.9 | 39.7 | 40.1 | 24.6 | 15.8 |
| 36 | 157.5 | 248.9 | 94.0 | 44.0 | 20.1 | 31.4 |
| Sample No. | Farm | PR Value (%) of Pooled Sample | PR Value (%) of Individual Sample | ||||
|---|---|---|---|---|---|---|---|
| 1 | B | 41.5 | 60.2 | 63.5 | 43.9 | 11.5 | 13.6 |
| 2 | 49.2 | 71.6 | 79.7 | 12.5 | 18.1 | 25.2 | |
| 3 | 26.4 | 60.1 | 29.1 | 15.1 | 29.0 | 8.0 | |
| 4 | 47.5 | 104.2 | 77.0 | 30.5 | 25.5 | 18.7 | |
| 5 | 19.2 | 40.0 | 24.7 | 13.0 | 31.4 | 7.9 | |
| 6 | 26.2 | 48.6 | 70.4 | 31.9 | 3.6 | 12.6 | |
| 7 | 84.0 | 57.6 | 75.4 | 47.4 | 41.2 | 31.5 | |
| 8 | 30.2 | 71.6 | 43.9 | 15.1 | 14.3 | 12.9 | |
| 9 | 40.9 | 104.2 | 48.6 | 57.6 | 29.0 | 7.9 | |
| 10 | 38.1 | 79.7 | 11.5 | 12.9 | 29.1 | 30.5 | |
| 11 | C | 83.5 | 101.1 | 48.4 | 19.2 | 27.7 | 34.7 |
| 12 | 146.5 | 63.7 | 160.8 | 128.4 | 27.0 | 27.2 | |
| 13 | 114.5 | 162.9 | 46.7 | 37.7 | 32.7 | 13.5 | |
| 14 | 16.3 | 18.1 | 2.9 | 12.1 | 16.1 | 24.1 | |
| 15 | 103.9 | 171.9 | 41.9 | 29.4 | 17.4 | 32.6 | |
| 16 | 192.0 | 60.3 | 68.5 | 142.8 | 27.1 | 13.8 | |
| 17 | 94.9 | 113.1 | 82.2 | 97.5 | 99.7 | 23.7 | |
| 18 | 66.5 | 101.6 | 15.5 | 24.7 | 35.9 | 23.8 | |
| 19 | 166.7 | 91.3 | 63.7 | 162.9 | 101.1 | 11.5 | |
| 20 | 89.0 | 113.1 | 60.3 | 18.1 | 32.6 | 23.8 | |
| 21 | D | 14.0 | 8.5 | 8.6 | 19.5 | 24.5 | 9.7 |
| 22 | 22.3 | 64.8 | 19.5 | 17.4 | 22.5 | 3.7 | |
| 23 | 27.9 | 10.1 | 16.6 | 18.6 | 29.0 | 35.7 | |
| 24 | 32.7 | 14.1 | 8.0 | 18.4 | 29.9 | 27.5 | |
| 25 | 12.6 | 6.4 | 2.3 | 1.7 | 7.7 | 7.3 | |
| 26 | 21.1 | 11.5 | 7.4 | 8.4 | 13.9 | 13.6 | |
| 27 | 100.7 | 95.0 | 123.6 | 32.5 | 16.3 | 30.8 | |
| 28 | 94.5 | 82.6 | 53.3 | 43.2 | 15.0 | 34.4 | |
| 29 | 14.8 | 8.5 | 3.7 | 10.1 | 2.3 | 6.4 | |
| 30 | 23.4 | 11.5 | 30.8 | 15.0 | 8.6 | 19.5 | |
| Farms | Frequency | Total Pools | ||
|---|---|---|---|---|
| True-Positive Pools (TP) | True-Negative Pools (TN) | False-Negative Pools (FN) | ||
| A | 12 | Not applicable | 24 | 36 |
| B | 6 | 0 | 4 | 10 |
| C | 9 | 1 | 0 | 10 |
| D | 2 | 7 | 1 | 10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamkasem, J.; Tattiyapong, P.; Gardner, I.A.; Surachetpong, W. Assessment and Performance of Pooled Serum Samples for Monitoring Farm-Level Immunity in Tilapia Infected with Tilapia Lake Virus. Viruses 2025, 17, 877. https://doi.org/10.3390/v17070877
Yamkasem J, Tattiyapong P, Gardner IA, Surachetpong W. Assessment and Performance of Pooled Serum Samples for Monitoring Farm-Level Immunity in Tilapia Infected with Tilapia Lake Virus. Viruses. 2025; 17(7):877. https://doi.org/10.3390/v17070877
Chicago/Turabian StyleYamkasem, Jidapa, Puntanat Tattiyapong, Ian A. Gardner, and Win Surachetpong. 2025. "Assessment and Performance of Pooled Serum Samples for Monitoring Farm-Level Immunity in Tilapia Infected with Tilapia Lake Virus" Viruses 17, no. 7: 877. https://doi.org/10.3390/v17070877
APA StyleYamkasem, J., Tattiyapong, P., Gardner, I. A., & Surachetpong, W. (2025). Assessment and Performance of Pooled Serum Samples for Monitoring Farm-Level Immunity in Tilapia Infected with Tilapia Lake Virus. Viruses, 17(7), 877. https://doi.org/10.3390/v17070877






