CRISPR-Cas12a/RPA Dual-Readout Assay for Rapid Field Detection of Porcine Rotavirus with Visualization
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid, Virus Strain, and Genome
2.2. RNA Extraction
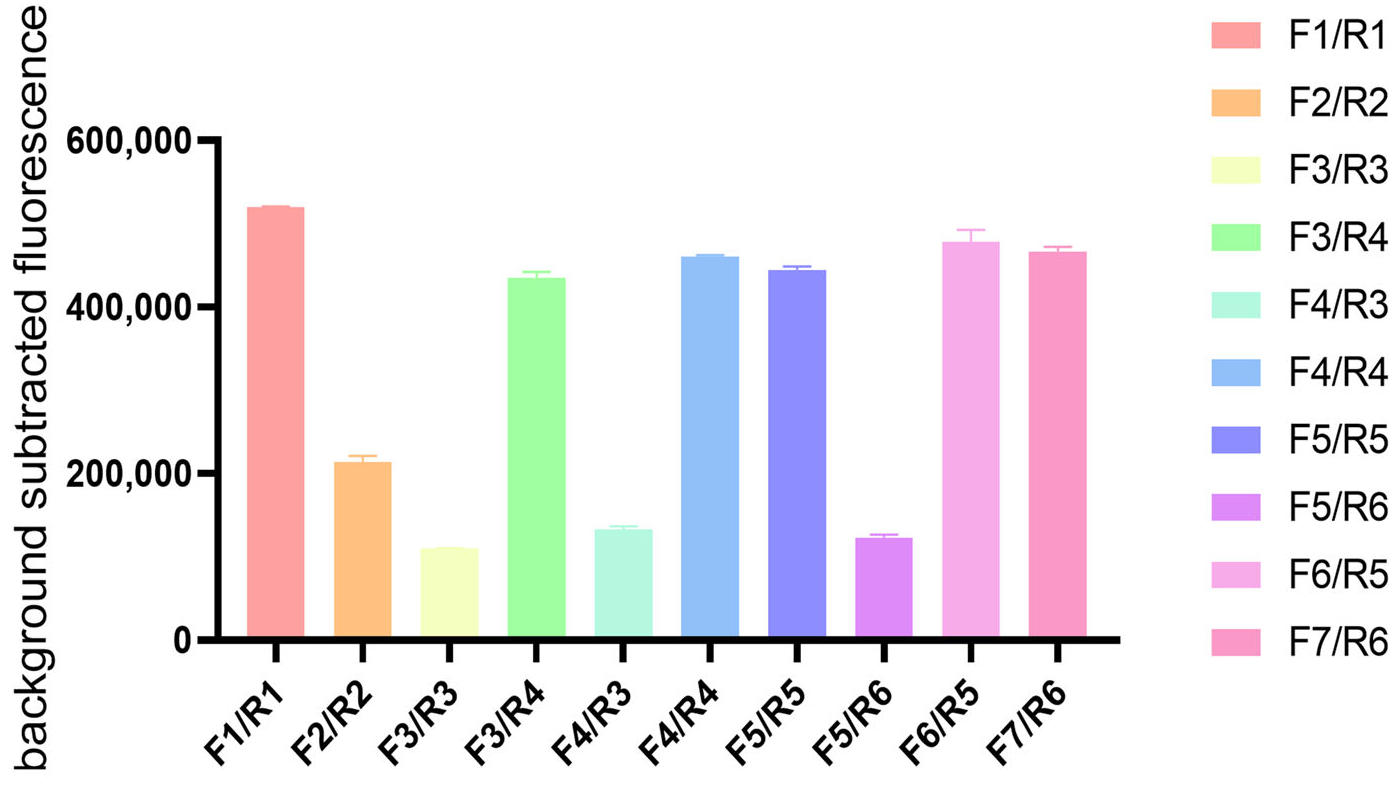
2.3. Recombinase Polymerase Amplification (RPA) Primer Screening
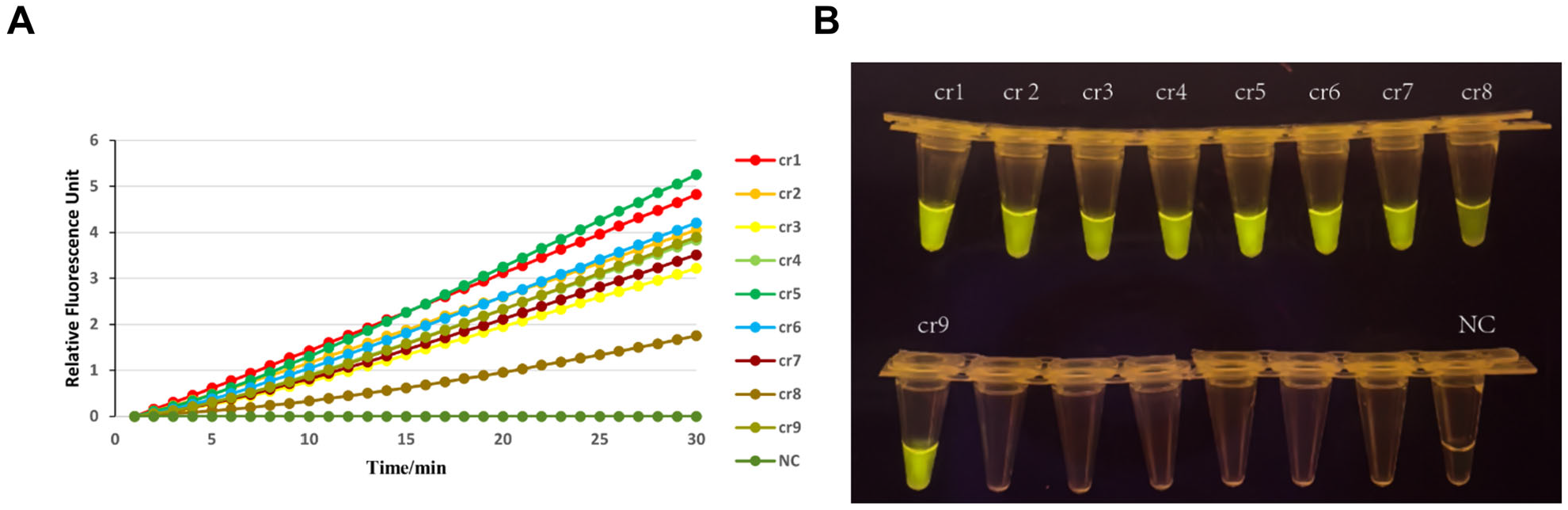
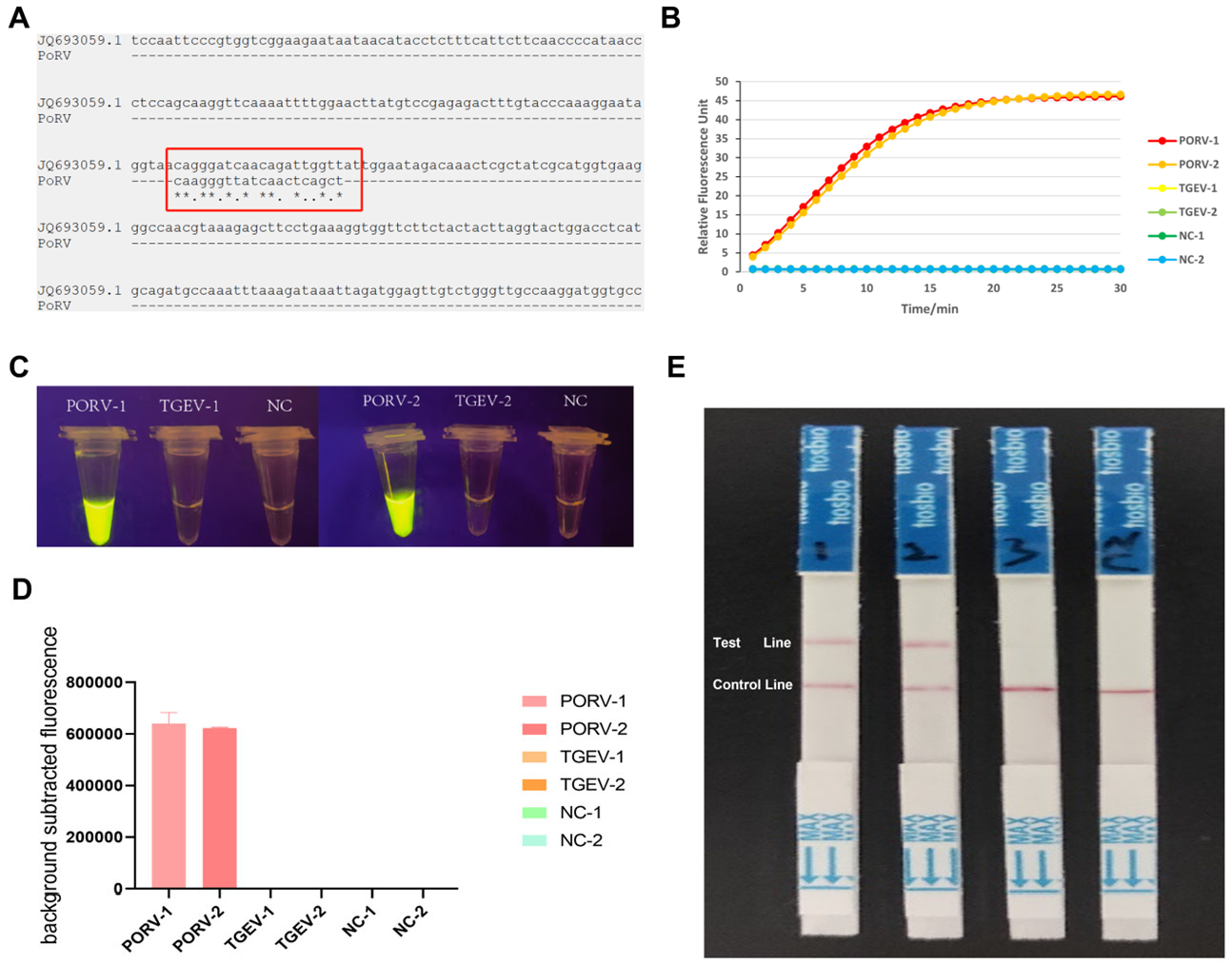
2.4. Design and Screening of crRNA Primers
2.5. Optimization of RT-RAA Product Volume
2.6. Specificity Evaluation
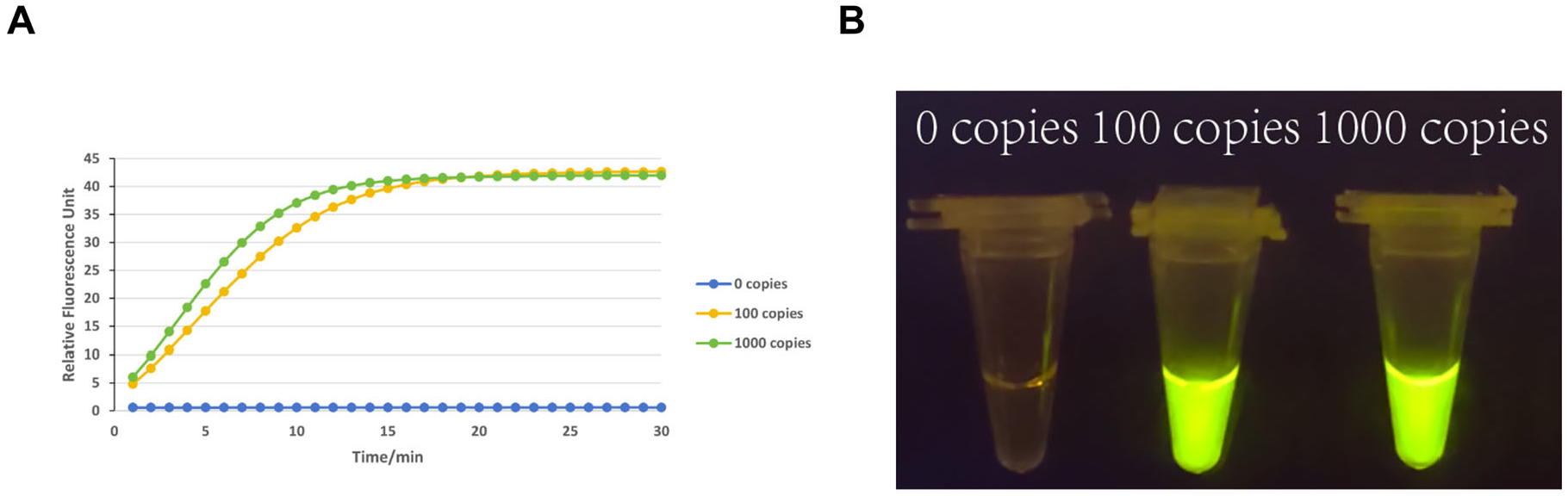
2.7. Sensitivity Assessment
2.8. Lateral Flow Dipstick (LFD) Validation
2.9. Clinical Sample Validation and Statistical Analysis
3. Results
3.1. Primer Screening and Optimization
3.2. Sensitivity Testing
3.3. Screening of Amplification Components: Primer Selection and crRNA Evaluation
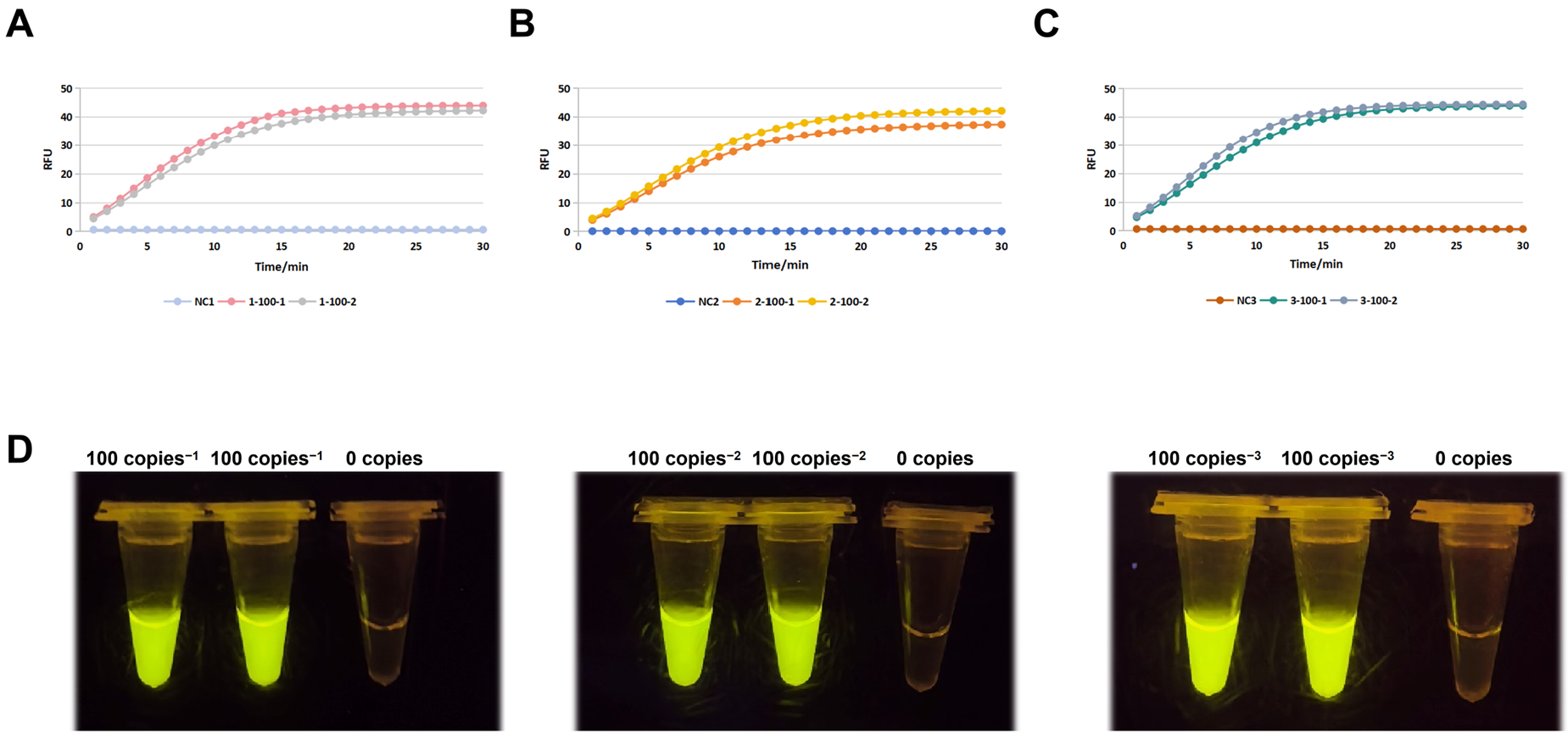
3.4. Sensitivity Confirmation and Optimization
3.5. Specificity Testing
3.6. Clinical Sample Test Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ghonaim, A.H.; Rouby, S.R.; Nageeb, W.M.; Elgendy, A.A.; Xu, R.; Jiang, C.; Ghonaim, N.H.; He, Q.; Li, W. Insights into recent advancements in human and animal rotavirus vaccines: Exploring new frontiers. Virol. Sin. 2025, 40, 1–14. [Google Scholar] [CrossRef]
- Huang, Y.; Zhu, Q.; Wang, Y.; Zhu, K. Bacterial-derived sialidases inhibit porcine rotavirus OSU replication by interfering with the early steps of infection. Microb. Pathog. 2024, 190, 106628. [Google Scholar] [CrossRef]
- Chen, Y.; He, Z.; Luo, Y.; Su, Q.; Wang, Q.; Wang, J.; He, J.; Yu, M.; You, H.; Chen, H. Tris stabilized AuNPs based lateral flow immunochromatography for the simultaneous detection of porcine epidemic diarrhea virus and rotavirus on-site. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2024, 320, 124670. [Google Scholar] [CrossRef]
- Yin, Y.; Zhu, L.; Liu, P.; Zhao, J.; Fan, Y.; Sun, X.; Xu, Z. Evaluation on the efficacy and immunogenicity of recombinant DNA plasmids expressing S gene from porcine epidemic diarrhea virus and VP7 gene from porcine rotavirus. Braz. J. Microbiol. 2019, 50, 279–286. [Google Scholar] [CrossRef]
- Jia, S.; Feng, B.; Wang, Z.; Ma, Y.; Gao, X.; Jiang, Y.; Cui, W.; Qiao, X.; Tang, L.; Li, Y.; et al. Dual priming oligonucleotide (DPO)-based real-time RT-PCR assay for accurate differentiation of four major viruses causing porcine viral diarrhea. Mol. Cell. Probes 2019, 47, 101435. [Google Scholar] [CrossRef]
- De La Cruz Hernandez, S.I.; Anaya Molina, Y.; Gomez Santiago, F.; Teran Vega, H.L.; Monroy Leyva, E.; Mendez Perez, H.; Garcia Lozano, H. Real-time RT-PCR, a necessary tool to support the diagnosis and surveillance of rotavirus in Mexico. Diagn. Microbiol. Infect. Dis. 2018, 90, 272–276. [Google Scholar] [CrossRef]
- Li, H.; Bello, A.; Smith, G.; Kielich, D.M.S.; Strong, J.E.; Pickering, B.S. Degenerate sequence-based CRISPR diagnostic for Crimean-Congo hemorrhagic fever virus. PLoS Negl. Trop. Dis. 2022, 16, e0010285. [Google Scholar] [CrossRef]
- Han, W.; Ma, Z.; Li, Z.; Chang, C.; Yuan, Y.; Li, Y.; Feng, R.; Zheng, C.; Shi, Z.; Tian, H.; et al. A novel double antibody sandwich quantitative ELISA for detecting porcine epidemic diarrhea virus infection. Appl. Microbiol. Biotechnol. 2024, 108, 482. [Google Scholar] [CrossRef]
- Huang, S.; Du, L.; Liu, S.; Yang, Q.; Lei, C.; Wang, H.; Yang, L.; Yang, X. Development and Validation of RAA-CRISPR/Cas12a-Based Assay for Detecting Porcine Rotavirus. Animals 2024, 14, 3387. [Google Scholar] [CrossRef]
- Shi, K.; Zhou, H.; Feng, S.; He, J.; Li, B.; Long, F.; Shi, Y.; Yin, Y.; Li, Z. Development of a Quadruplex RT-qPCR for the Detection of Porcine Rotaviruses and the Phylogenetic Analysis of Porcine RVH in China. Pathogens 2023, 12, 1091. [Google Scholar] [CrossRef]
- Pan, Y.; Li, Z.; Miao, Q.; Shi, H.; Guo, L.; Feng, L.; Tian, J. Phylogenitc analysis and immunogenicity comparison of porcine genotype G9 rotavirus in China from 2020–2023. Virol. Sin. 2025, 40, 176–185. [Google Scholar] [CrossRef]
- Almeida, P.R.; Lorenzetti, E.; Cruz, R.S.; Watanabe, T.T.; Zlotowski, P.; Alfieri, A.A.; Driemeier, D. Diarrhea caused by rotavirus A, B, and C in suckling piglets from southern Brazil: Molecular detection and histologic and immunohistochemical characterization. J. Vet. Diagn. Investig. 2018, 30, 370–376. [Google Scholar] [CrossRef]
- Shahni, S.N.; Albogami, S.; Azmi, I.; Pattnaik, B.; Chaudhuri, R.; Dev, K.; Iqbal, J.; Sharma, A.; Ahmad, T. Dual Detection of Hepatitis B and C Viruses Using CRISPR-Cas Systems and Lateral Flow Assay. J. Nucleic Acids 2024, 2024, 8819834. [Google Scholar] [CrossRef] [PubMed]
- Brogan, D.J.; Akbari, O.S. CRISPR Diagnostics: Advances toward the Point of Care. Biochemistry 2023, 62, 3488–3492. [Google Scholar] [CrossRef] [PubMed]
- Garrison, A.R.; Moresco, V.; Zeng, X.; Cline, C.R.; Ward, M.D.; Ricks, K.M.; Olschner, S.P.; Cazares, L.H.; Karaaslan, E.; Fitzpatrick, C.J.; et al. Nucleocapsid protein-specific monoclonal antibodies protect mice against Crimean-Congo hemorrhagic fever virus. Nat. Commun. 2024, 15, 1722. [Google Scholar] [CrossRef] [PubMed]
- Shariq, M.; Khan, M.F.; Raj, R.; Ahsan, N.; Singh, R.; Kumar, P. CRISPR-based diagnostic approaches: Implications for rapid management of future pandemics (Review). Mol. Med. Rep. 2023, 27, 118. [Google Scholar] [CrossRef]
- Zhou, L.; Simonian, A.L. CRISPR/Cas Technology: The Unique Synthetic Biology Genome-Editing Tool Shifting the Paradigm in Viral Diagnostics, Defense, and Therapeutics. Annu. Rev. Biomed. Eng. 2024, 26, 247–272. [Google Scholar] [CrossRef]
- Luo, T.; Li, K.; Li, C.; Xia, C.; Gao, C. Development of a triplex quantitative reverse transcription-polymerase chain reaction for the detection of porcine epidemic diarrhea virus, porcine transmissible gastroenteritis virus, and porcine rotavirus A. Front. Microbiol. 2024, 15, 1390328. [Google Scholar] [CrossRef]
- Kostyusheva, A.; Brezgin, S.; Babin, Y.; Vasilyeva, I.; Glebe, D.; Kostyushev, D.; Chulanov, V. CRISPR-Cas systems for diagnosing infectious diseases. Methods 2022, 203, 431–446. [Google Scholar] [CrossRef]
- Qu, H.; Zhang, W.; Li, J.; Fu, Q.; Li, X.; Wang, M.; Fu, G.; Cui, J. A rapid and sensitive CRISPR-Cas12a for the detection of Fusobacterium nucleatum. Microbiol. Spectr. 2024, 12, e0362923. [Google Scholar] [CrossRef]
- Liu, Q.; Zeng, H.; Wang, T.; Ni, H.; Li, Y.; Qian, W.; Fang, T.; Xu, G. Development of RPA-Cas12a assay for rapid and sensitive detection of Pneumocystis jirovecii. BMC Microbiol. 2024, 24, 314. [Google Scholar] [CrossRef]
- Kim, H.J.; Cho, I.S.; Choi, S.R.; Jeong, R.D. Identification of an Isolate of Citrus Tristeza Virus by Nanopore Sequencing in Korea and Development of a CRISPR/Cas12a-Based Assay for Rapid Visual Detection of the Virus. Phytopathology 2024, 114, 1421–1428. [Google Scholar] [CrossRef]
- Cao, X.; Chang, Y.; Tao, C.; Chen, S.; Lin, Q.; Ling, C.; Huang, S.; Zhang, H. Cas12a/Guide RNA-Based Platforms for Rapidly and Accurately Identifying Staphylococcus aureus and Methicillin-Resistant S. aureus. Microbiol. Spectr. 2023, 11, e0487022. [Google Scholar] [CrossRef]
- Jiao, J.; Kong, K.; Han, J.; Song, S.; Bai, T.; Song, C.; Wang, M.; Yan, Z.; Zhang, H.; Zhang, R.; et al. Field detection of multiple RNA viruses/viroids in apple using a CRISPR/Cas12a-based visual assay. Plant Biotechnol. J. 2021, 19, 394–405. [Google Scholar] [CrossRef] [PubMed]
- Ling, C.; Chang, Y.; Wang, X.; Cao, X.; Tu, Q.; Liu, B.; Huang, S. Two CRISPR/Cas12a-based methods for fast and accurate detection of single-base mutations. Anal. Chim. Acta 2023, 1247, 340881. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Liu, Y.; Zhang, X.; Gai, Z.; Lei, H.; Shen, X. A Rapid RPA-CRISPR/Cas12a Detection Method for Adulteration of Goat Milk Powder. Foods 2023, 12, 1569. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Xu, Z.; Zhou, H.; Li, T.; Du, X.; Hu, R.; Zhu, J.; Ou, G.; Li, Y.; Yang, Y. Integration of CRISPR/Cas12a and Multiplexed RPA for Fast Detection of Gene Doping. Anal. Chem. 2022, 94, 16481–16490. [Google Scholar] [CrossRef]
- Wang, P.; Guo, B.; Zhang, X.; Wang, Y.; Yang, G.; Shen, H.; Gao, S.; Zhang, L. One-Pot Molecular Diagnosis of Acute Hepatopancreatic Necrosis Disease by Recombinase Polymerase Amplification and CRISPR/Cas12a with Specially Designed crRNA. J. Agric. Food Chem. 2023, 71, 6490–6498. [Google Scholar] [CrossRef]
- Zeng, L.; Zheng, S.; Stejskal, V.; Opit, G.; Aulicky, R.; Li, Z. New and rapid visual detection assay for Trogoderma granarium everts based on recombinase polymerase amplification and CRISPR/Cas12a. Pest Manag. Sci. 2023, 79, 5304–5311. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, X.; Wang, J.; Zhang, Y.; Zeng, W.; Liu, Y.; Zhang, X. Photoactivatable CRISPR/Cas12a Strategy for One-Pot DETECTR Molecular Diagnosis. Anal. Chem. 2022, 94, 9724–9731. [Google Scholar] [CrossRef]
- Lin, K.; Guo, J.; Guo, X.; Li, Q.; Li, X.; Sun, Z.; Zhao, Z.; Weng, J.; Wu, J.; Zhang, R.; et al. Fast and visual detection of nucleic acids using a one-step RPA-CRISPR detection (ORCD) system unrestricted by the PAM. Anal. Chim. Acta 2023, 1248, 340938. [Google Scholar] [CrossRef]
- Li, X.; Liu, M.; Men, D.; Duan, Y.; Deng, L.; Zhou, S.; Hou, J.; Hou, C.; Huo, D. Rapid, portable, and sensitive detection of CaMV35S by RPA-CRISPR/Cas12a-G4 colorimetric assays with high accuracy deep learning object recognition and classification. Talanta 2024, 278, 126441. [Google Scholar] [CrossRef]
- He, Q.; Yu, D.; Bao, M.; Korensky, G.; Chen, J.; Shin, M.; Kim, J.; Park, M.; Qin, P.; Du, K. High-throughput and all-solution phase African Swine Fever Virus (ASFV) detection using CRISPR-Cas12a and fluorescence based point-of-care system. Biosens. Bioelectron. 2020, 154, 112068. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Z.; Daczkowski, C.M.; Gabel, C.; Mesecar, A.D.; Chang, L. Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins. Cell Host Microbe 2019, 25, 815–826.E4. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, X.; Xu, T.; Jin, X.; Jiang, J.; Guan, F. Rapid and Ultrasensitive Detection of H. aduncum via the RPA-CRISPR/Cas12a Platform. Molecules 2024, 29, 4789. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Qiu, Z.; Bi, Z.; Tian, T.; Jiang, Y.; Zhou, X. Photocontrolled crRNA activation enables robust CRISPR-Cas12a diagnostics. Proc. Natl. Acad. Sci. USA 2022, 119, e2202034119. [Google Scholar] [CrossRef] [PubMed]
- MacGregor, S.R.; McManus, D.P.; Sivakumaran, H.; Egwang, T.G.; Adriko, M.; Cai, P.; Gordon, C.A.; Duke, M.G.; French, J.D.; Collinson, N.; et al. Development of CRISPR/Cas13a-based assays for the diagnosis of Schistosomiasis. EBioMedicine 2023, 94, 104730. [Google Scholar] [CrossRef]

| Primer | Sequence (5′-3′) |
|---|---|
| F1 | GTAACTTACCTGTTAGGAATTGGACTTTTG |
| F2 | CCACAATCTGAAGCGTTAAGGAAGTTAGCAGG |
| F3 | ACGCACCAGCGAATATACAACAGTTTGAGCAT |
| F4 | GTTTGAGCATATTGTCCAACTTAGACGCGCAC |
| F5 | CAATTCGCTTATCATTTCAATTAATGCGTCC |
| F6 | TGCGTCCACCGAACATGACACCAGCTGTTAATGC |
| F7 | GCTGTGCGTCAAGAGTATGCTATACCAGTTGG |
| R1 | ACCTGCTAACTTCCTTAACGCTTCAGATTGTG |
| R2 | CTGATCCAGCATTGAGCCACATAGTTCCCATC |
| R3 | ATAACTGGGTTAAAGAACCATGTGGTTGCGCC |
| R4 | GGTCTTAAAATAACTGGGTTAAAGAACCATGTGG |
| R5 | ATACTCTTGACGCACAGCGGTCACATTCGCC |
| R6 | AACACGTTGCAAATTATCCTCTCTGGATGGTG |
| crRNA | Sequence (5′-3′) |
|---|---|
| crRNA1 | GGGUAAUUUCUACUAAGUGUAGAUGUCUAUUAGGCACAACACUC |
| crRNA2 | GGGUAAUUUCUACUAAGUGUAGAUUCUGAGAUUCUCUCGCCAUU |
| crRNA3 | GGGUAAUUUCUACUAAGUGUAGAUUGCAUCAGGCAACAAAGUUA |
| crRNA4 | GGGUAAUUUCUACUAAGUGUAGAUUUGCCUGAUGCAGAAAGAUU |
| crRNA5 | GGGUAAUUUCUACUAAGUGUAGAUCAAGGGUUAUCAACUCAGCU |
| crRNA6 | GGGUAAUUUCUACUAAGUGUAGAUCGCAAGCACAACCCUCUCAA |
| crRNA7 | GGGUAAUUUCUACUAAGUGUAGAUUGAAUCAGUGCUUGCGGAUG |
| crRNA8 | GGGUAAUUUCUACUAAGUGUAGAUCAUCCGCAAGCACUGAUUCA |
| crRNA9 | GGGUAAUUUCUACUAAGUGUAGAUCACCAGGCAUGAAUUGGACU |
| Sample Type | RT-RAA-CRISPR/Cas12a | Real-Time RT-PCR | ||
|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |
| Clinical sample | 2 | 48 | 2 | 48 |
| Artificially contaminated sample | 5 | 0 | 5 | 0 |
| Accuracy | 100% | 100% | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, X.; Huang, Y.; Jiang, Y.; Yang, G.; Zheng, X.; Gao, S. CRISPR-Cas12a/RPA Dual-Readout Assay for Rapid Field Detection of Porcine Rotavirus with Visualization. Viruses 2025, 17, 872. https://doi.org/10.3390/v17070872
Jiang X, Huang Y, Jiang Y, Yang G, Zheng X, Gao S. CRISPR-Cas12a/RPA Dual-Readout Assay for Rapid Field Detection of Porcine Rotavirus with Visualization. Viruses. 2025; 17(7):872. https://doi.org/10.3390/v17070872
Chicago/Turabian StyleJiang, Xinjie, Yun Huang, Yi Jiang, Guang Yang, Xiaocong Zheng, and Shuai Gao. 2025. "CRISPR-Cas12a/RPA Dual-Readout Assay for Rapid Field Detection of Porcine Rotavirus with Visualization" Viruses 17, no. 7: 872. https://doi.org/10.3390/v17070872
APA StyleJiang, X., Huang, Y., Jiang, Y., Yang, G., Zheng, X., & Gao, S. (2025). CRISPR-Cas12a/RPA Dual-Readout Assay for Rapid Field Detection of Porcine Rotavirus with Visualization. Viruses, 17(7), 872. https://doi.org/10.3390/v17070872







