Trafficking and Activation of Henipavirus, Parahenipavirus, and Henipa-like Virus Fusion Proteins
Abstract
:1. Introduction
2. Natural Reservoirs and Taxonomy of Henipaviruses, Parahenipaviruses, and Henipa-like Viruses
3. Glycoprotein Organization in the HNV Particle
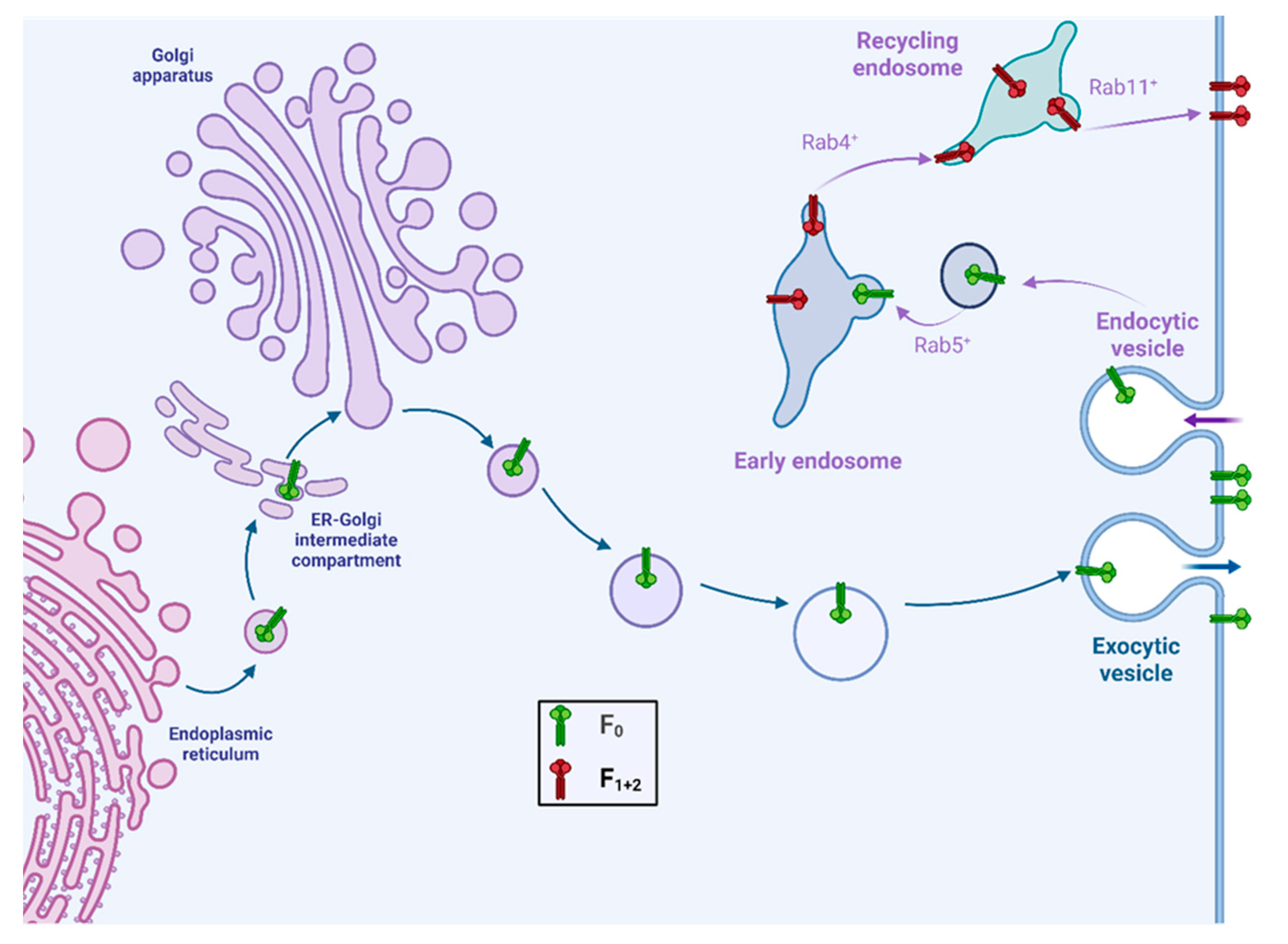
4. Fusion Protein Trafficking and Proteolytic Cleavage: Henipaviruses
5. Fusion Protein Trafficking and Proteolytic Cleavage: Parahenipaviruses and Henipa-like Viruses
6. Role of the Fusion Protein in Particle Assembly and Budding
7. Future Perspective
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AngV | Angavokely virus |
| CedV | Cedar virus |
| CFR | case fatality rate |
| CHV | Camp Hill virus |
| ChyV | Chodsigoa hypsibia henipavirus |
| CT | cytoplasmic tail |
| CtV | Crocidura tanakae henipavirus |
| DARV | Daeryong virus |
| DewV | Denwin virus |
| FP | fusion peptide |
| GAKV | Gamak virus |
| GhV | Ghana virus |
| HasV | Hasua virus |
| HeV | Hendra virus |
| HNV | henipavirus |
| HRA | heptad repeat A |
| HRB | heptad repeat B |
| ICTV | International Committee on Taxonomy of Viruses |
| JCS-1 | Jingmen Crocidura shantungensis henipavirus 1 |
| JCS-2 | Jingmen Crocidura shantungensis henipavirus 2 |
| LayV | Langya virus |
| LechV | Lechcodon virus |
| MEFs | mouse embryonic fibroblasts |
| MelV | Melian virus |
| MojV | Mojiang virus |
| NinExV | Ninorex virus |
| NiV | Nipah virus |
| ResV | Resua virus |
| RNP | ribonucleocapsid |
| SCtV | Shiyan Crocidura tanakae henipavirus |
| shRNA | small hairpin RNA |
| TM | transmembrane |
| VLP | virus-like paticle |
| WAa-1 | Wenzhou Apodemus agrarius henipavirus 1 |
| Wca-1 | Wufeng Crocidura attenuata henipavirus 1 |
| WCs-1 | Wufeng Chodsigoa smithii henipavirus 1 |
| WsV1 | Wenzhou shrew henipavirus 1 |
References
- Murray, K.; Rogers, R.; Selvey, L.; Selleck, P.; Hyatt, A.; Gould, A.; Gleeson, L.; Hooper, P.; Westbury, H. A novel morbillivirus pneumonia of horses and its transmission to humans. Emerg. Infect. Dis. 1995, 1, 31–33. [Google Scholar] [CrossRef] [PubMed]
- Chua, K.B.; Bellini, W.J.; Rota, P.A.; Harcourt, B.H.; Tamin, A.; Lam, S.K.; Ksiazek, T.G.; Rollin, P.E.; Zaki, S.R.; Shieh, W.; et al. Nipah virus: A recently emergent deadly paramyxovirus. Science 2000, 288, 1432–1435. [Google Scholar] [CrossRef]
- Wang, L.-F.; Mackenzie, J.; Broder, C. Henipaviruses. In Fields Virology; David, M.K., Peter, M.H., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 1, pp. 1070–1085. [Google Scholar]
- Luby, S.P.; Gurley, E.S. Epidemiology of henipavirus disease in humans. Curr. Top. Microbiol. Immunol. 2012, 359, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Geisbert, T.W.; Feldmann, H.; Broder, C.C. Animal challenge models of henipavirus infection and pathogenesis. Curr. Top. Microbiol. Immunol. 2012, 359, 153–177. [Google Scholar] [CrossRef] [PubMed]
- Eaton, B.T.; Broder, C.C.; Middleton, D.; Wang, L.F. Hendra and Nipah viruses: Different and dangerous. Nat. Rev. Microbiol. 2006, 4, 23–35. [Google Scholar] [CrossRef]
- Li, H.; Kim, J.V.; Pickering, B.S. Henipavirus zoonosis: Outbreaks, animal hosts and potential new emergence. Front. Microbiol. 2023, 14, 1167085. [Google Scholar] [CrossRef]
- Halpin, K.; Hyatt, A.D.; Fogarty, R.; Middleton, D.; Bingham, J.; Epstein, J.H.; Rahman, S.A.; Hughes, T.; Smith, C.; Field, H.E.; et al. Pteropid bats are confirmed as the reservoir hosts of henipaviruses: A comprehensive experimental study of virus transmission. Am. J. Trop. Med. Hyg. 2011, 85, 946–951. [Google Scholar] [CrossRef]
- Wacharapluesadee, S.; Lumlertdacha, B.; Boongird, K.; Wanghongsa, S.; Chanhome, L.; Rollin, P.; Stockton, P.; Rupprecht, C.E.; Ksiazek, T.G.; Hemachudha, T. Bat Nipah virus, Thailand. Emerg. Infect. Dis. 2005, 11, 1949–1951. [Google Scholar] [CrossRef]
- Iehle, C.; Razafitrimo, G.; Razainirina, J.; Andriaholinirina, N.; Goodman, S.M.; Faure, C.; Georges-Courbot, M.C.; Rousset, D.; Reynes, J.M. Henipavirus and Tioman virus antibodies in pteropodid bats, Madagascar. Emerg. Infect. Dis. 2007, 13, 159–161. [Google Scholar] [CrossRef]
- Epstein, J.H.; Prakash, V.; Smith, C.S.; Daszak, P.; McLaughlin, A.B.; Meehan, G.; Field, H.E.; Cunningham, A.A. Henipavirus infection in fruit bats (Pteropus giganteus), India. Emerg. Infect. Dis. 2008, 14, 1309–1311. [Google Scholar] [CrossRef]
- Hayman, D.T.; Suu-Ire, R.; Breed, A.C.; McEachern, J.A.; Wang, L.; Wood, J.L.; Cunningham, A.A. Evidence of henipavirus infection in West African fruit bats. PLoS ONE 2008, 3, e2739. [Google Scholar] [CrossRef] [PubMed]
- Yob, J.M.; Field, H.; Rashdi, A.M.; Morrissy, C.; van der Heide, B.; Rota, P.; bin Adzhar, A.; White, J.; Daniels, P.; Jamaluddin, A.; et al. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg. Infect. Dis. 2001, 7, 439–441. [Google Scholar] [CrossRef] [PubMed]
- Drexler, J.F.; Corman, V.M.; Muller, M.A.; Maganga, G.D.; Vallo, P.; Binger, T.; Gloza-Rausch, F.; Cottontail, V.M.; Rasche, A.; Yordanov, S.; et al. Bats host major mammalian paramyxoviruses. Nat. Commun. 2012, 3, 796. [Google Scholar] [CrossRef]
- Breed, A.C.; Yu, M.; Barr, J.A.; Crameri, G.; Thalmann, C.M.; Wang, L.F. Prevalence of henipavirus and rubulavirus antibodies in pteropid bats, Papua New Guinea. Emerg. Infect. Dis. 2010, 16, 1997–1999. [Google Scholar] [CrossRef] [PubMed]
- Gomez Roman, R.; Wang, L.F.; Lee, B.; Halpin, K.; de Wit, E.; Broder, C.C.; Rahman, M.; Kristiansen, P.; Saville, M. Nipah@20: Lessons Learned from Another Virus with Pandemic Potential. mSphere 2020, 5, e00602-20. [Google Scholar] [CrossRef]
- Soman Pillai, V.; Krishna, G.; Valiya Veettil, M. Nipah Virus: Past Outbreaks and Future Containment. Viruses 2020, 12, 465. [Google Scholar] [CrossRef]
- Khan, S.; Akbar, S.M.F.; Mahtab, M.A.; Uddin, M.N.; Rashid, M.M.; Yahiro, T.; Hashimoto, T.; Kimitsuki, K.; Nishizono, A. Twenty-five years of Nipah outbreaks in Southeast Asia: A persistent threat to global health. IJID Reg. 2024, 13, 100434. [Google Scholar] [CrossRef]
- Navaratnarajah, C.K.; Generous, A.R.; Yousaf, I.; Cattaneo, R. Receptor-mediated cell entry of paramyxoviruses: Mechanisms, and consequences for tropism and pathogenesis. J. Biol. Chem. 2020, 295, 2771–2786. [Google Scholar] [CrossRef]
- Bonaparte, M.I.; Dimitrov, A.S.; Bossart, K.N.; Crameri, G.; Mungall, B.A.; Bishop, K.A.; Choudhry, V.; Dimitrov, D.S.; Wang, L.F.; Eaton, B.T.; et al. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc. Natl. Acad. Sci. USA 2005, 102, 10652–10657. [Google Scholar] [CrossRef]
- Negrete, O.A.; Levroney, E.L.; Aguilar, H.C.; Bertolotti-Ciarlet, A.; Nazarian, R.; Tajyar, S.; Lee, B. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature 2005, 436, 401–405. [Google Scholar] [CrossRef]
- Laing, E.D.; Navaratnarajah, C.K.; Cheliout Da Silva, S.; Petzing, S.R.; Xu, Y.; Sterling, S.L.; Marsh, G.A.; Wang, L.-F.; Amaya, M.; Nikolov, D.B.; et al. Structural and functional analyses reveal promiscuous and species specific use of ephrin receptors by Cedar virus. Proc. Natl. Acad. Sci. USA 2019, 116, 20707–20715. [Google Scholar] [CrossRef] [PubMed]
- Pager, C.T.; Dutch, R.E. Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J. Virol. 2005, 79, 12714–12720. [Google Scholar] [CrossRef] [PubMed]
- Meulendyke, K.A.; Wurth, M.A.; McCann, R.O.; Dutch, R.E. Endocytosis plays a critical role in proteolytic processing of the Hendra virus fusion protein. J. Virol. 2005, 79, 12643–12649. [Google Scholar] [CrossRef] [PubMed]
- Diederich, S.; Moll, M.; Klenk, H.D.; Maisner, A. The Nipah virus fusion protein is cleaved within the endosomal compartment. J. Biol. Chem. 2005, 280, 29899–29903. [Google Scholar] [CrossRef]
- Pager, C.T.; Craft, W.W., Jr.; Patch, J.; Dutch, R.E. A mature and fusogenic form of the Nipah virus fusion protein requires proteolytic processing by cathepsin L. Virology 2006, 346, 251–257. [Google Scholar] [CrossRef]
- Diederich, S.; Thiel, L.; Maisner, A. Role of endocytosis and cathepsin-mediated activation in Nipah virus entry. Virology 2008, 375, 391–400. [Google Scholar] [CrossRef]
- Vogt, C.; Eickmann, M.; Diederich, S.; Moll, M.; Maisner, A. Endocytosis of the Nipah virus glycoproteins. J. Virol. 2005, 79, 3865–3872. [Google Scholar] [CrossRef]
- Halpin, K.; Young, P.L.; Field, H.E.; Mackenzie, J.S. Isolation of Hendra virus from pteropid bats: A natural reservoir of Hendra virus. J. Gen. Virol. 2000, 81, 1927–1932. [Google Scholar] [CrossRef]
- Kane, Y.; Nalikka, B.; Tendu, A.; Omondi, V.; Bienes, K.M.; Padane, A.; Duong, V.; Berthet, N.; Wong, G. Genetic Diversity and Geographic Spread of Henipaviruses. Emerg. Infect. Dis. 2025, 31, 427–437. [Google Scholar] [CrossRef]
- Vanmechelen, B.B.-B.A.; Drexler, J.F.; Duprex, P.W.; Lee, B.; Plemper, R.K.; Maes, P. Taxonomic Reorganization of the Family Paramyxoviridae. 2023. Available online: https://ictv.global/ictv/proposals/2023.018M.Paramyxoviridae_reorg.zip (accessed on 1 April 2025).
- Drexler, J.F.; Corman, V.M.; Gloza-Rausch, F.; Seebens, A.; Annan, A.; Ipsen, A.; Kruppa, T.; Muller, M.A.; Kalko, E.K.; Adu-Sarkodie, Y.; et al. Henipavirus RNA in African bats. PLoS ONE 2009, 4, e6367. [Google Scholar] [CrossRef]
- Marsh, G.A.; de Jong, C.; Barr, J.A.; Tachedjian, M.; Smith, C.; Middleton, D.; Yu, M.; Todd, S.; Foord, A.J.; Haring, V.; et al. Cedar virus: A novel Henipavirus isolated from Australian bats. PLoS Pathog. 2012, 8, e1002836. [Google Scholar] [CrossRef]
- Rodriguez, J.J.; Parisien, J.P.; Horvath, C.M. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 2002, 76, 11476–11483. [Google Scholar] [CrossRef]
- Rodriguez, J.J.; Wang, L.F.; Horvath, C.M. Hendra virus V protein inhibits interferon signaling by preventing STAT1 and STAT2 nuclear accumulation. J. Virol. 2003, 77, 11842–11845. [Google Scholar] [CrossRef]
- Shaw, M.L.; Cardenas, W.B.; Zamarin, D.; Palese, P.; Basler, C.F. Nuclear localization of the Nipah virus W protein allows for inhibition of both virus- and toll-like receptor 3-triggered signaling pathways. J. Virol. 2005, 79, 6078–6088. [Google Scholar] [CrossRef]
- Lieu, K.G.; Marsh, G.A.; Wang, L.F.; Netter, H.J. The non-pathogenic Henipavirus Cedar paramyxovirus phosphoprotein has a compromised ability to target STAT1 and STAT2. Antivir. Res. 2015, 124, 69–76. [Google Scholar] [CrossRef]
- Madera, S.; Kistler, A.; Ranaivoson, H.C.; Ahyong, V.; Andrianiaina, A.; Andry, S.; Raharinosy, V.; Randriambolamanantsoa, T.H.; Ravelomanantsoa, N.A.F.; Tato, C.M.; et al. Discovery and Genomic Characterization of a Novel Henipavirus, Angavokely Virus, from Fruit Bats in Madagascar. J. Virol. 2022, 96, e0092122. [Google Scholar] [CrossRef]
- Irving, A.T.; Ahn, M.; Goh, G.; Anderson, D.E.; Wang, L.F. Lessons from the host defences of bats, a unique viral reservoir. Nature 2021, 589, 363–370. [Google Scholar] [CrossRef]
- Wu, Z.; Yang, L.; Yang, F.; Ren, X.; Jiang, J.; Dong, J.; Sun, L.; Zhu, Y.; Zhou, H.; Jin, Q. Novel Henipa-like virus, Mojiang Paramyxovirus, in rats, China, 2012. Emerg. Infect. Dis. 2014, 20, 1064–1066. [Google Scholar] [CrossRef]
- Lee, S.H.; Kim, K.; Kim, J.; No, J.S.; Park, K.; Budhathoki, S.; Lee, S.H.; Lee, J.; Cho, S.H.; Cho, S.; et al. Discovery and Genetic Characterization of Novel Paramyxoviruses Related to the Genus Henipavirus in Crocidura Species in the Republic of Korea. Viruses 2021, 13, 2020. [Google Scholar] [CrossRef]
- Zhang, X.A.; Li, H.; Jiang, F.C.; Zhu, F.; Zhang, Y.F.; Chen, J.J.; Tan, C.W.; Anderson, D.E.; Fan, H.; Dong, L.Y.; et al. A Zoonotic Henipavirus in Febrile Patients in China. N. Engl. J. Med. 2022, 387, 470–472. [Google Scholar] [CrossRef]
- Parry, R.H.; Yamada, K.Y.H.; Hood, W.R.; Zhao, Y.; Lu, J.Y.; Seluanov, A.; Gorbunova, V.; Modhiran, N.; Watterson, D.; Isaacs, A. Henipavirus in Northern Short-Tailed Shrew, Alabama, USA. Emerg. Infect. Dis. 2025, 31, 392–394. [Google Scholar] [CrossRef]
- Caruso, S.; Edwards, S.J. Recently Emerged Novel Henipa-like Viruses: Shining a Spotlight on the Shrew. Viruses 2023, 15, 2407. [Google Scholar] [CrossRef]
- Sabir, A.J.; Rong, L.; Broder, C.C.; Amaya, M. Cedar virus biology and its applications as a surrogate for highly pathogenic henipaviruses. Cell Insight 2024, 3, 100181. [Google Scholar] [CrossRef]
- Madeira, F.; Madhusoodanan, N.; Lee, J.; Eusebi, A.; Niewielska, A.; Tivey, A.R.N.; Lopez, R.; Butcher, S. The EMBL-EBI Job Dispatcher sequence analysis tools framework in 2024. Nucleic Acids Res. 2024, 52, W521–W525. [Google Scholar] [CrossRef]
- Vanmechelen, B.; Meurs, S.; Horemans, M.; Loosen, A.; Joly Maes, T.; Laenen, L.; Vergote, V.; Koundouno, F.R.; Magassouba, N.; Konde, M.K.; et al. The characterization of multiple novel paramyxoviruses highlights the diverse nature of the subfamily Orthoparamyxovirinae. Virus Evol. 2022, 8, veac061. [Google Scholar] [CrossRef]
- Horemans, M.; Van Bets, J.; Joly Maes, T.; Maes, P.; Vanmechelen, B. Discovery and genome characterization of six new orthoparamyxoviruses in small Belgian mammals. Virus Evol. 2023, 9, vead065. [Google Scholar] [CrossRef]
- Kaza, B.; Aguilar, H.C. Pathogenicity and virulence of henipaviruses. Virulence 2023, 14, 2273684. [Google Scholar] [CrossRef]
- Meier, K.; Olejnik, J.; Hume, A.J.; Muhlberger, E. A Comparative Assessment of the Pathogenic Potential of Newly Discovered Henipaviruses. Pathogens 2024, 13, 587. [Google Scholar] [CrossRef]
- Hyatt, A.D.; Zaki, S.R.; Goldsmith, C.S.; Wise, T.G.; Hengstberger, S.G. Ultrastructure of Hendra virus and Nipah virus within cultured cells and host animals. Microbes Infect. 2001, 3, 297–306. [Google Scholar] [CrossRef]
- Goldsmith, C.S.; Whistler, T.; Rollin, P.E.; Ksiazek, T.G.; Rota, P.A.; Bellini, W.J.; Daszak, P.; Wong, K.T.; Shieh, W.J.; Zaki, S.R. Elucidation of Nipah virus morphogenesis and replication using ultrastructural and molecular approaches. Virus Res. 2003, 92, 89–98. [Google Scholar] [CrossRef]
- Gui, L.; Jurgens, E.M.; Ebner, J.L.; Porotto, M.; Moscona, A.; Lee, K.K. Electron tomography imaging of surface glycoproteins on human parainfluenza virus 3: Association of receptor binding and fusion proteins before receptor engagement. mBio 2015, 6, e02393-14. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, J.; Luo, Y.; Kliemke, V.; Matta, G.L.; Wang, J.; Liu, Q. The nanoscale organization of the Nipah virus fusion protein informs new membrane fusion mechanisms. eLife 2025, 13, RP97017. [Google Scholar] [CrossRef]
- Michalski, W.P.; Crameri, G.; Wang, L.; Shiell, B.J.; Eaton, B. The cleavage activation and sites of glycosylation in the fusion protein of Hendra virus. Virus Res. 2000, 69, 83–93. [Google Scholar] [CrossRef]
- Dietzel, E.; Kolesnikova, L.; Sawatsky, B.; Heiner, A.; Weis, M.; Kobinger, G.P.; Becker, S.; von Messling, V.; Maisner, A. Nipah Virus Matrix Protein Influences Fusogenicity and Is Essential for Particle Infectivity and Stability. J. Virol. 2015, 90, 2514–2522. [Google Scholar] [CrossRef]
- Fischer, K.; Groschup, M.H.; Diederich, S. Importance of Endocytosis for the Biological Activity of Cedar Virus Fusion Protein. Cells 2020, 9, 2054. [Google Scholar] [CrossRef]
- Aguilar, H.C.; Iorio, R.M. Henipavirus membrane fusion and viral entry. Curr. Top. Microbiol. Immunol. 2012, 359, 79–94. [Google Scholar] [CrossRef]
- Steffen, D.L.; Xu, K.; Nikolov, D.B.; Broder, C.C. Henipavirus mediated membrane fusion, virus entry and targeted therapeutics. Viruses 2012, 4, 280–308. [Google Scholar] [CrossRef]
- Pager, C.T.; Wurth, M.A.; Dutch, R.E. Subcellular localization and calcium and pH requirements for proteolytic processing of the Hendra virus fusion protein. J. Virol. 2004, 78, 9154–9163. [Google Scholar] [CrossRef]
- McGrath, M.E. The lysosomal cysteine proteases. Annu. Rev. Biophys. Biomol. Struct. 1999, 28, 181–204. [Google Scholar] [CrossRef]
- Diederich, S.; Sauerhering, L.; Weis, M.; Altmeppen, H.; Schaschke, N.; Reinheckel, T.; Erbar, S.; Maisner, A. Activation of the Nipah virus fusion protein in MDCK cells is mediated by cathepsin B within the endosome-recycling compartment. J. Virol. 2012, 86, 3736–3745. [Google Scholar] [CrossRef]
- Erbar, S.; Maisner, A. Nipah virus infection and glycoprotein targeting in endothelial cells. Virol. J. 2010, 7, 305. [Google Scholar] [CrossRef]
- Weise, C.; Erbar, S.; Lamp, B.; Vogt, C.; Diederich, S.; Maisner, A. Tyrosine residues in the cytoplasmic domains affect sorting and fusion activity of the Nipah virus glycoproteins in polarized epithelial cells. J. Virol. 2010, 84, 7634–7641. [Google Scholar] [CrossRef]
- Mattera, R.; Farias, G.G.; Mardones, G.A.; Bonifacino, J.S. Co-assembly of viral envelope glycoproteins regulates their polarized sorting in neurons. PLoS Pathog. 2014, 10, e1004107. [Google Scholar] [CrossRef]
- Lawrence, P.; Escudero Perez, B.; Drexler, J.F.; Corman, V.M.; Muller, M.A.; Drosten, C.; Volchkov, V. Surface glycoproteins of the recently identified African Henipavirus promote viral entry and cell fusion in a range of human, simian and bat cell lines. Virus Res. 2014, 181, 77–80. [Google Scholar] [CrossRef]
- Weis, M.; Behner, L.; Hoffmann, M.; Kruger, N.; Herrler, G.; Drosten, C.; Drexler, J.F.; Dietzel, E.; Maisner, A. Characterization of African bat henipavirus GH-M74a glycoproteins. J. Gen. Virol. 2014, 95, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Biniossek, M.L.; Nagler, D.K.; Becker-Pauly, C.; Schilling, O. Proteomic identification of protease cleavage sites characterizes prime and non-prime specificity of cysteine cathepsins B, L, and S. J. Proteome Res. 2011, 10, 5363–5373. [Google Scholar] [CrossRef]
- Rissanen, I.; Ahmed, A.A.; Azarm, K.; Beaty, S.; Hong, P.; Nambulli, S.; Duprex, W.P.; Lee, B.; Bowden, T.A. Idiosyncratic Mojiang virus attachment glycoprotein directs a host-cell entry pathway distinct from genetically related henipaviruses. Nat. Commun. 2017, 8, 16060. [Google Scholar] [CrossRef]
- Wang, Z.; McCallum, M.; Yan, L.; Gibson, C.A.; Sharkey, W.; Park, Y.J.; Dang, H.V.; Amaya, M.; Person, A.; Broder, C.C.; et al. Structure and design of Langya virus glycoprotein antigens. Proc. Natl. Acad. Sci. USA 2024, 121, e2314990121. [Google Scholar] [CrossRef]
- Cheliout Da Silva, S.; Yan, L.; Dang, H.V.; Xu, K.; Epstein, J.H.; Veesler, D.; Broder, C.C. Functional Analysis of the Fusion and Attachment Glycoproteins of Mojiang Henipavirus. Viruses 2021, 13, 517. [Google Scholar] [CrossRef]
- Wang, C.; Li, M.; Wang, Y.; Ding, Q.; Fan, S.; Lan, J. Structural insights into the Langya virus attachment glycoprotein. Structure 2024, 32, 1090–1098 e1093. [Google Scholar] [CrossRef]
- Takimoto, T.; Portner, A. Molecular mechanism of paramyxovirus budding. Virus Res. 2004, 106, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Patch, J.R.; Crameri, G.; Wang, L.F.; Eaton, B.T.; Broder, C.C. Quantitative analysis of Nipah virus proteins released as virus-like particles reveals central role for the matrix protein. Virol. J. 2007, 4, 1. [Google Scholar] [CrossRef]
- Johnston, G.P.; Contreras, E.M.; Dabundo, J.; Henderson, B.A.; Matz, K.M.; Ortega, V.; Ramirez, A.; Park, A.; Aguilar, H.C. Cytoplasmic Motifs in the Nipah Virus Fusion Protein Modulate Virus Particle Assembly and Egress. J. Virol. 2017, 91, e02150-16. [Google Scholar] [CrossRef] [PubMed]
- Cifuentes-Munoz, N.; Sun, W.; Ray, G.; Schmitt, P.T.; Webb, S.; Gibson, K.; Dutch, R.E.; Schmitt, A.P. Mutations in the transmembrane domain and cytoplasmic tail of Hendra virus fusion protein disrupt virus-like particle assembly. J. Virol. 2017, 91, e00152-17. [Google Scholar] [CrossRef] [PubMed]
- Johnston, G.P.; Bradel-Tretheway, B.; Piehowski, P.D.; Brewer, H.M.; Lee, B.N.R.; Usher, N.T.; Zamora, J.L.R.; Ortega, V.; Contreras, E.M.; Teuton, J.R.; et al. Nipah Virus-Like Particle Egress Is Modulated by Cytoskeletal and Vesicular Trafficking Pathways: A Validated Particle Proteomics Analysis. mSystems 2019, 4, e00194-19. [Google Scholar] [CrossRef]

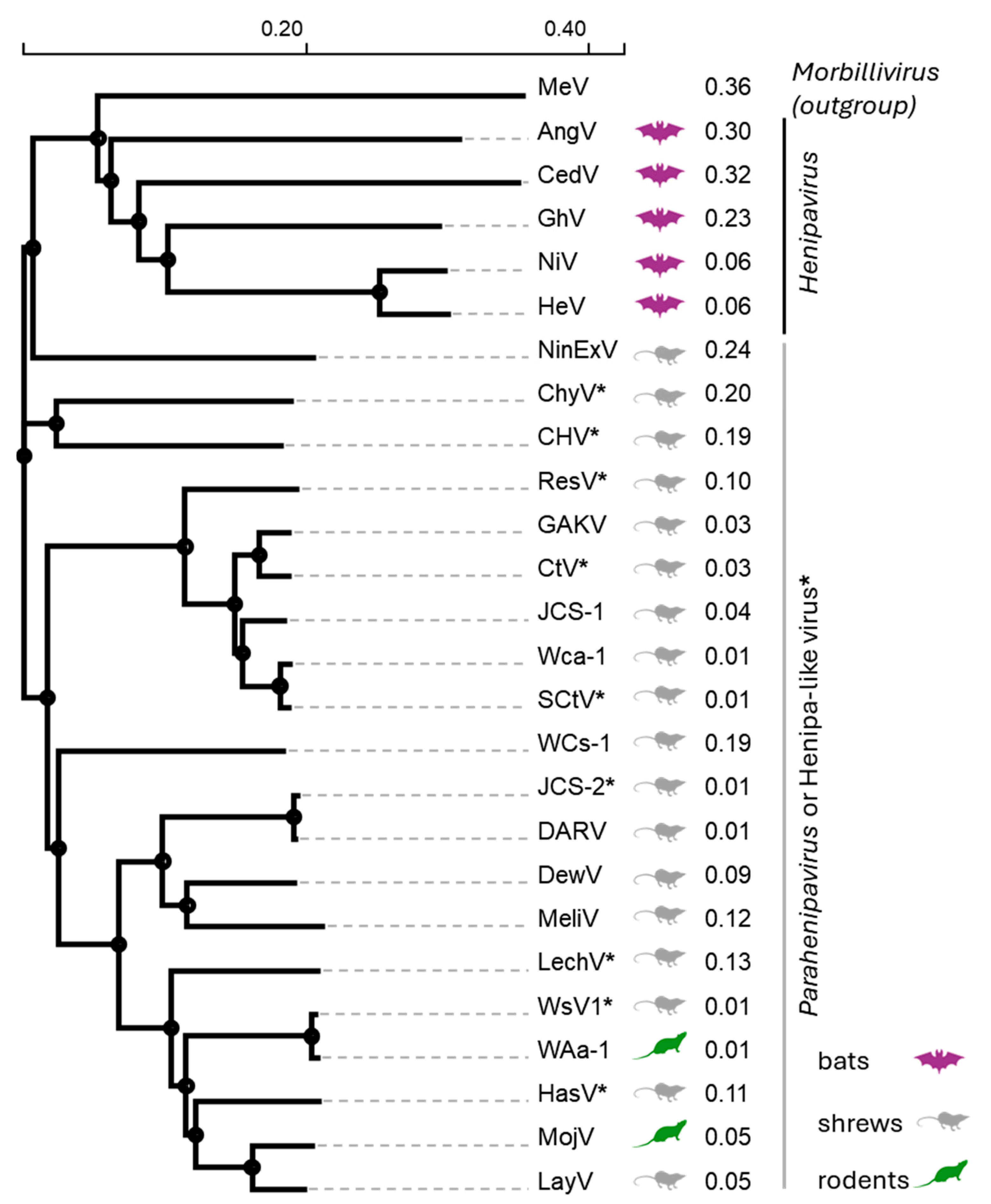

| Virus Name 1 | Genus/Status 2 | Natural Reservoir | Geographic Location | Year | F Protein Accession # |
|---|---|---|---|---|---|
| Angavokely virus (AngV) | Henipavirus | Madagascar fruit bats | Madagascar (Angavokely Cave) | 2019 | UVG43988.1 |
| Camp Hill virus | Henipa-like virus | Northern short-tailed shrews | USA (Alabama) | 2025 | XJU75810.1 |
| Cedar virus (CedV) | Henipavirus | Bats (Pteropus spp.) | Australia (Cedar Grove, Queensland) | 2009 | AFP87278.1 |
| Daeryong virus (DARV) | Parahenipavirus | Shrews (Crocidura lasiura and Crocidura shantungensis) | Republic of Korea | 2018 | QYO90531.1 |
| Chodsigoa hypsibia henipavirus | Henipa-like virus | Shrew (Chodsigoa hypsibia) | China | 2022 | WEU70826.1 |
| Crocidura tanakae henipavirus | Henipa-like virus | Shrew (Crocidura lasiura) | China | 2023 | WZI33221.1 |
| Denwin virus (DewV) | Parahenipavirus | Greater white-toothed shrews (C. russula) | Belgium | 2022 | WPS63725.1 |
| Gamak virus (GAKV) | Parahenipavirus | Shrews (Crocidura lasiura and Crocidura shantungensis) | Republic of Korea | 2018 | QYO90517.1 |
| Ghana virus (GhV) | Henipavirus | African straw-colored fruit bats (Eidolon helvum) | Ghana (Kumasi) | 2008 | AFH96010.1 |
| Hasua virus (HasV) | Henipa-like virus 3 | Crocidura suaveolens shrews | Germany (north-eastern) | 2023 | WPS63719.1 |
| Hendra virus (HeV) | Henipavirus | Bats (Pteropus spp.) | Australia (Hendra, Brisbane). | 1994 | NP_047111.2 |
| Jingmen Crocidura shantungensis henipavirus 1 | Parahenipavirus | Shrews (Crocidura shantungensis) | China (Jingmen) | 2021 | UOX72983.1 |
| Jingmen Crocidura shantungensis henipavirus 2 | Henipa-like virus | Shrews (Crocidura shantungensis) | China (Jingmen) | 2021 | UOX72990.1 |
| Langya virus (LayV) | Parahenipavirus | Shrews (Crocidura spp.) | China (Eastern China) | 2022 | UUV47205.1 |
| Lechcodon virus (LechV) | Henipa-like virus 3 | Shrews (Crocidura leucodon) | Germany (southern) | 2023 | WPS63705.1 |
| Melian virus (MeliV) | Parahenipavirus | Large-headed forest shrews (C. grandiceps) | Guinea | 2022 | UQM99518.1 |
| Mojiang virus (MojV) | Parahenipavirus | Cave rats (Rattus flavipectus) | China (Mòjiāng Hani Autonomous County, Yunnan Province) | 2012 | YP_009094094.1 |
| Ninorex virus (NinExV) | Parahenipavirus | Eurasian pygmy shrews (Sorex minutus) | Belgium | 2022 | WJL29504.1 |
| Nipah virus (NiV) | Henipavirus | Bats (Pteropus spp.) | Malaysia (Peninsular Malaysia) | 1998 | NP_112026.1 |
| Peixe-Boi virus (PBV) | Henipa-like virus | Brazilian opossum (Marmosa demerarae) | Brazil | 2015 | Not available |
| Resua virus (ResV) | Henipa-like virus 3 | Shrews (Crocidura suaveolens) | Germany (north-eastern) | 2023 | WPS63698.1 |
| Shiyan Crocidura tanakae henipavirus (SCtV) | Henipa-like virus | Taiwanese gray shrews (C. tanakae) | China (Taiwan, Shiyan) | 2024 | WIU81496.1 |
| Wenzhou Apodemus agrarius henipavirus 1 | Parahenipavirus | Striped field mouse (Apodemus agrarius) | China (Wenzhou) | 2021 | UBB42272.1 |
| Wenzhou shrew henipavirus 1 | Henipa-like virus | Shrew | China (Wenzhou, Zhejiang) | 2023 | WPV62326.1 |
| Wufeng Crocidura attenuata henipavirus 1 | Parahenipavirus | Shrews (Crocidura attenuate) | China (Wufeng) | 2021 | UOX73004.1 |
| Wufeng Chodsigoa smithii henipavirus 1 | Parahenipavirus | Shrews (Chodsigoa smithii) | China (Wufeng) | 2022 | UOX72997.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Navaratnarajah, C.K.; Cattaneo, R. Trafficking and Activation of Henipavirus, Parahenipavirus, and Henipa-like Virus Fusion Proteins. Viruses 2025, 17, 866. https://doi.org/10.3390/v17060866
Navaratnarajah CK, Cattaneo R. Trafficking and Activation of Henipavirus, Parahenipavirus, and Henipa-like Virus Fusion Proteins. Viruses. 2025; 17(6):866. https://doi.org/10.3390/v17060866
Chicago/Turabian StyleNavaratnarajah, Chanakha K., and Roberto Cattaneo. 2025. "Trafficking and Activation of Henipavirus, Parahenipavirus, and Henipa-like Virus Fusion Proteins" Viruses 17, no. 6: 866. https://doi.org/10.3390/v17060866
APA StyleNavaratnarajah, C. K., & Cattaneo, R. (2025). Trafficking and Activation of Henipavirus, Parahenipavirus, and Henipa-like Virus Fusion Proteins. Viruses, 17(6), 866. https://doi.org/10.3390/v17060866






