Abstract
Background: Infectious uveitis is a potentially sight-threatening condition that requires timely and accurate pathogen identification to guide effective therapy. However, conventional microbiological tests (CMTs) often lack sensitivity and the inclusiveness of pathogen detection. Metagenomic next-generation sequencing (mNGS) offers an unbiased approach to detecting a broad range of pathogens. This review evaluates its diagnostic performance in detecting infectious uveitis. Methods: A systematic search across multiple databases identified studies assessing the use of mNGS for diagnosing infectious uveitis. The included studies compared mNGS to CMTs, including polymerase chain reaction (PCR), culture, serology, and the IGRA (Interferon-Gamma Release Assay). The study characteristics; the detection rates; and the sensitivity, specificity, and predictive values were extracted. The sensitivity and specificity of mNGS were calculated using CMTs as a reference. Results: Twelve studies comprising 859 patients were included. The sensitivity of mNGS compared to that of CMTs ranged from 38.4% to 100%, while specificity varied between 15.8% and 100%. The commonly detected pathogens included varicella-zoster virus, cytomegalovirus, Toxoplasma gondii, and herpes simplex virus. In some cases, mNGS outperformed PCR in viral detection, aiding diagnosis when the standard methods failed. However, contamination risks and inconsistent diagnostic thresholds were noted. Conclusions: mNGS enables the diagnosis of infectious uveitis, particularly for viral causes, but its variable performance and standardization challenges warrant further investigation.
1. Introduction
Infectious uveitis is a serious intraocular inflammatory condition that can cause severe vision impairment and blindness if not promptly identified and treated. This condition is often difficult to diagnose because it can be caused by various pathogens, including viruses, bacteria, fungi, and parasites [1]. These infections usually exhibit overlapping clinical features, which can mimic non-infectious uveitis [1]. In this scenario, one of the primary challenges in diagnosing infectious uveitis is accurately identifying the causative pathogen to guide appropriate treatment. However, the small sample size of the intraocular specimens often limits the sensitivity and specificity of the traditional diagnostic methods, such as culture and intraocular antibody detection, which are also time-consuming [1,2,3]. Moreover, the eye’s immune privilege and localized immunological mechanisms can further hinder pathogen detection, contributing to diagnostic challenges [4].
Metagenomic next-generation sequencing (mNGS) is a novel technique that has the potential to improve pathogen detection in infectious uveitis [5]. It allows for the comprehensive identification of a broad spectrum of pathogens, including those difficult to culture or identify using the traditional methods [5,6]. Its ability to generate vast sequencing data for unbiased pathogen detection has led to its increasing global adoption as a diagnostic tool [7,8]. However, mNGS also presents challenges, such as a poor specificity, which can lead to false positives due to contamination and the detection of clinically nonsignificant organisms. Additionally, interpreting mNGS data requires advanced bioinformatics tools [3,5,9,10]. Here, we performed a comprehensive systemic review to assess the clinical performance of mNGS compared to that of the conventional microbiological tests (CMTs) in diagnosing infectious uveitis and to identify the barriers that currently hinder the broad applicability of mNGS as a diagnostic tool for infectious uveitis.
2. Materials and Methods
2.1. Systematic Literature Review and Inclusion and Exclusion Criteria
This review followed the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) statement [11]. Institutional Review Board approval was not required. This study was registered on Prospero (https://www.crd.york.ac.uk/PROSPERO/) on 7 December 2024 (registration number CRD42024618873). In this study, we defined infectious uveitis as inflammation involving all three anatomical regions of the uveal tract—anterior, intermediate, and posterior segments—resulting from infections by various pathogens, including bacteria, viruses, fungi, and parasites. The complete search strategy is demonstrated in Supplementary S1 [12]. We defined metagenomic next-generation sequencing (mNGS) as a laboratory method that uses shotgun next-generation sequencing to detect any genome present in a clinical sample [5]. The typical mNGS workflow involves three key phases. First, during sample preparation, ocular fluids (aqueous or vitreous humor) undergo DNA/RNA extraction using commercial kits, followed by the generation of cDNA libraries (for RNA-seq) and sequencing library preparation using fragmentation for short-read sequencing (not performed for long-read sequencing) and adapter ligation. Because of the small size of intraocular specimens, most methods employed for the sequencing of intraocular fluids rely on the use of PCR to amplify and concentrate the sequencing libraries. Second, libraries are quantified and pooled, and sequencing is performed on platforms such as Illumina (short-read and high-accuracy) or Oxford Nanopore (long-read and real-time), generating from hundreds of thousands to millions (1–20 million) of reads of per samples depending on the method/platform used and the number of samples pooled in one run. Third, bioinformatic analysis filters human sequences and aligns the remaining reads to pathogen databases [9,13,14]. On the other hand, the conventional microbiological tests (CMTs) included antigenic tests, culture, the enzyme-linked immunosorbent assay (ELISA) for the detection of local antibody production in aqueous humor, PCR (polymerase chain reaction), and the IGRA (Interferon-Gamma Release Assay). We defined a positive result in the CMT group if at least one of the diagnostic methods mentioned was positive in a single sample.
This review included manuscripts published up to 17 February 2025. There were no language restrictions. The inclusion criteria for studies were original research manuscripts; published in peer-reviewed, scientific journals; case series studies; observational study designs; and involved individuals that had infectious uveitis and were submitted to mNGS-based pathogen detection.
Commentaries, case reports, studies on pediatric populations, studies with overlapping samples of patients, and non-peer-reviewed studies were excluded. Studies in which there was no comparison between mNGS and a CMT were also excluded.
2.2. Search Strategy
We performed literature searches in PubMed, Embase (Elsevier Platform), the Cochrane Central Register of Controlled Trials, Web of Science, the Cumulative Index to Nursing and Allied Health (CINAHL), and Scopus. The complete search strategy is demonstrated in Supplementary S1. Also, we assessed the reference lists of included articles to find papers not identified from the initial literature search.
This study employed the PICO framework [15]. We focused on patients with infectious uveitis (P) to compare metagenomic next-generation sequencing (I) against conventional microbiological tests (C). Our primary outcomes (O) were diagnostic performance, clinical impact, and possible barriers to the broad implementation of mNGS for routine clinical use. To filter the 4467 studies acquired from the databases, the titles and/or abstracts were evaluated by two investigators (I.P. and A.R.M.). Studies with overlapping patients, incomplete data, or that did not meet the PICO inclusion criteria were excluded using Rayyan app through consensus [16]. After the first filtration, 15 studies were fully read, and 12 were included in this systematic literature review (Figure 1).
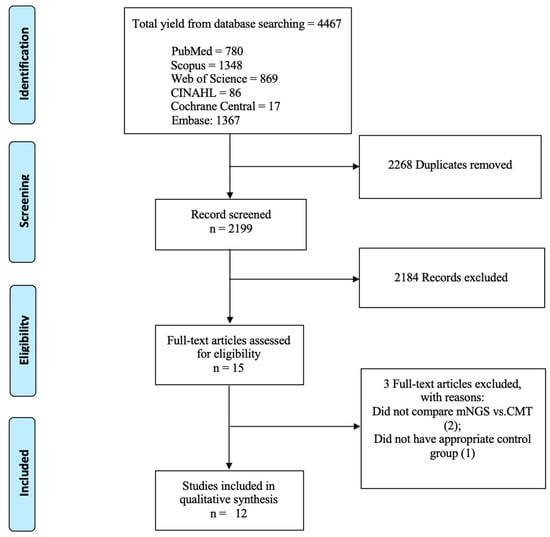
Figure 1.
Flow diagram of literature search adapted from PRISMA flow chart.
2.3. Data Abstraction and Quality Assessment
Of six independent reviewers (I.P., P.S.M., M.C.G., J.M.G.F., M.K.H., and Y.L.), two independently extracted the data for each article using a standardized abstraction form (Supplementary S2). The reviewers resolved disagreements by unanimity. The standardized abstraction form included a record of the data on study design, publication year and calendar time, demographic characteristics of the selected population, mNGS methodology, analyzed CMTs, sample types, detection rates and accuracy comparison between methods, commonly identified pathogens, mNGS advantages, limitations, clinical impact, and conclusions.
We assessed the risk of bias and methodological quality of the included studies using a modified Downs and Black scale [17]. All the original questions from this scale were applied, except for question #27, which was adapted to a yes/no response, resulting in a maximum achievable score of 28 points. Two reviewers independently scored each study, resolving disagreements through consensus. The scores were categorized as follows: good quality (>18 points), fair quality (15–18 points), and poor quality (<15 points).
3. Results
3.1. Characteristics of Included Studies in This Systematic Literature Review
Twelve studies [18,19,20,21,22,23,24,25,26,27,28,29] were included in this systematic review (Figure 1 and Table 1). Five studies were retrospective cohort studies [21,22,25,27,29], three were prospective cohort studies [24,26,28], three were case series [18,19,20], and one was a transversal study [23]. Most studies were performed in China (six studies) [18,25,26,27,28,29], followed by the USA (three studies) [20,21,22], the Netherlands (one study) [19], Japan (one study) [23], and South Korea (one study) [24]. These studies were conducted between 2010 and 2023, varying from 5 to 70 months in duration [18,19,20,21,22,23,24,25,26,27,28,29].

Table 1.
Summary of characteristics of studies included in this systematic literature review.
In qualitative analysis, twelve studies including 859 patients, of whom 525 (61.1%) had infectious uveitis, evaluated the diagnostic value of mNGS. Of 459 infectious uveitis cases with a reported etiology, 206 (44.9%) were viral [20,21,23,24,25,26,27,28,29]. The majority of the studies used the Illumina platform to perform mNGS (eight studies) [19,20,21,22,25,26,27,29], followed by Oxford Nanopore Technology sequencers (two studies) [23,24], the Beijing Genomics Institute (one study) [28], and MGI Sequencers (one study) [18]. Of the twelve studies evaluated, nine focused on DNA testing [18,19,21,23,24,25,26,27,29], while three examined RNA testing [20,22,28]. The vast majority of the studies sampled both aqueous and vitreous humors (seven studies) [18,20,22,24,25,26,27], although four studies solely focused on sampling aqueous humor [19,23,28,29], and one study on sampling vitreous humor [21]. There was a vast range of sample volume used for mNGS testing (20–100 µL) [18,20,21,22,23,24,26,27,28,29], with a mode of 50 µL [21,24,28,29], which was similar to the amounts used for each CMT sample.
Eleven studies compared the positivity rates of mNGS and a CMT, defined as the proportion of samples yielding positive results indicative of infection [18,19,20,21,22,24,25,27,28,29]. In three studies, mNGS yielded a higher positivity rate than the CMT [18,21,22]; in three studies, the positivity rates were equivalent [19,20,24]; and in five studies, the positivity rates were higher with the CMT [23,25,27,28,29]. Overall, the positivity rates ranged from 9.7% to 100% for mNGS and from 19.2% to 100% for the CMT [18,19,20,21,22,23,24,25,27,28,29]. One study did not separately report rates for each method, providing only comparative sensitivity and specificity values [25].
3.2. Sensitivity and Specificity of mNGS Using Conventional Microbiological Tests (CMTs) as Reference
Using CMTs as the reference, the sensitivity of mNGS ranged from as low as 38.4% [29] to as high as 100%, depending on the comparator and the study design [18,19,20]. Specificity also varied widely, ranging from 15.8% [26] to 100% [19]. When using culture-based methods as the reference, mNGS generally demonstrated more sensitivity, with studies such as Doan 2016 and Lee 2023 reporting 100% sensitivity [20,24]. In contrast, when targeted PCR was used as the comparator, mNGS sensitivity was more variable, ranging from 40% to 100% across multiple studies [18,19,21,22,23,24,25,26,27,28,29]. Additionally, one study specifically evaluated the diagnostic performance of mNGS for viral infections in intraocular fluid samples, reporting good sensitivity (90.7%), specificity (100%), a positive predictive value (100%), and a negative predictive value (81.0%) [27]. Positive predictive values (PPVs) and negative predictive values (NPVs) were less consistently reported, but also varied considerably, reflecting differences in the patient populations, pathogen load, and the laboratory procedures.
3.3. Sensitivity and Specificity of mNGS and CMTs Using Clinical Diagnosis as Reference
Two studies quantitatively compared mNGS and the clinical diagnosis [18,26]. Sensitivity ranged from 87.8% to 96.9%, while specificity varied from 58.4% to 69.2% [18,26]. Two studies compared CMTs and the clinical diagnosis. Sensitivity ranged from 59.4% to 75.0%, while specificity varied from 96.2% to 100% [18,20].
3.4. Pathogens Identified
The most commonly detected pathogens using mNGS included varicella-zoster virus (VZV), cytomegalovirus (CMV), Toxoplasma gondii, herpes simplex virus (HSV-1 and HSV-2), Epstein–Barr virus (EBV), and Klebsiella pneumoniae. Some studies, such as Doan 2016 and Doan 2021, reported the unique identification of Cryptococcus neoformans and Pithomyces chartarum, respectively, which were not detected using the conventional methods [20,22]. Notably, Qian 2023 highlighted the difficulty of mNGS in detecting Mycobacterium tuberculosis, indicating that protocol optimization is necessary for intracellular bacterial and fungal pathogens [25]. Doan 2016 highlighted how mNGS aided in the diagnosis of a bilateral chronic rubella uveitis in a patient without a previous known etiology [20].
One study evaluated the difference in the detection rates of vitreous humor and aqueous humor using mNGS. The vitreous humor samples showed a slightly increased pathogen detection rate (85.7–100.0%) when compared to that of aqueous humor using mNGS (80.0–88.9%) in most types of infection, except for viral infection, with 80.0% and 86.4% rates, respectively. However, statistically, both the samples are suitable for mNGS testing [26].
Two studies evaluated the diagnostic value of mNGS in identifying coinfections in uveitis [27,29]. Sun et al. reported two cases of coinfection, one involving VZV and CMV, and another involving HSV-2 and Treponema pallidum. When comparing mNGS and PCR, mNGS successfully identified both the pathogens in each coinfection. Although the PCR panel test correctly detected both VZV and CMV in the first case, it failed to identify Treponema pallidum in the second coinfection, detecting only HSV-2 [27]. Yu et al. reported one case with both HSV-1 and Epstein–Barr virus, where mNGS was positive for both pathogens, while the ELISA assay of aqueous humor only detected HSV-1. On the other hand, one study reported a false positive call from mNGS (Klebsiella pneumoniae) for a potential VZV-Klebsiella coinfection in a patient with only chronic VZV, correctly confirmed by the ELISA [29].
Regarding methodological quality assessed by the modified Downs and Black scale, two studies were classified as good [24,26], six as fair [21,23,25,27,28,29], and four as poor [18,19,20,22].
4. Discussion
mNGS has emerged as a valuable diagnostic tool for identifying pathogens in cases of infectious uveitis. In this systematic literature review, mNGS demonstrated wide ranges of sensitivity (38–100%) and specificity (15.8–100%) when compared to those of the CMTs as the reference standard. Notably, when using clinical diagnosis as the reference, mNGS showed more sensitivity (87.8–96.9%), but less specificity (58.4–69.2%), whereas CMTs had less sensitivity (59.4–75.0%), but more specificity (96.2–100%).
mNGS showed equal or superior positivity rates compared to those of the CMT in six out of eleven studies that reported this value (54.5%), supporting its potential role as a complementary or alternative diagnostic approach for infectious uveitis. The particularly high detection rates for viruses such as CMV, VZV, HSV-1, and HSV-2 reinforce the diagnostic utility of mNGS, especially for viral etiologies of infectious uveitis. However, the detection rate of mNGS among the infected patients ranged from 9.7% to 100%, while the CMTs showed a detection rate range from 19.2% to 100%. The majority of included studies (eight out of twelve; 66.7%) were classified as having good or fair methodological quality, strengthening our confidence in these results.
4.1. Sensitivity and Specificity of mNGS in Viral Detection Using CMTs as Reference
mNGS demonstrated more sensitivity in detecting viral pathogens using CMTs as reference. Studies by Doan et al. (2016, 2017, 2021), Sun 2024, and Qian 2023 reported sensitivity rates above 80% for viral detection [20,21,22,25,27]. For instance, Sun 2024 observed a sensitivity of 90.7% for mNGS in viral infectious uveitis, supporting the reliability of mNGS in identifying intraocular viral pathogens [27].
4.2. Advantages over Conventional Methods
A significant advantage of mNGS is its unbiased, hypothesis-free approach, which enables the detection of a wide array of pathogens without prior assumptions. Unlike PCR, which requires specific primers, mNGS can identify both known and novel viruses (Table 2). This was evident in Doan 2017, where mNGS uncovered 6 additional pathogens either not detected or not tested with pathogen-directed PCRs in 8 samples (22%) [21]. Also, Doan 2021 showed that mNGS identified pathogens not considered in the differential diagnosis in 9.7% of cases [21,22]. Beyond common viruses, mNGS identified rare or unexpected agents, such as rubella virus and human T-cell leukemia virus (HTLV-1). In Doan et al., 2016 study, mNGS was able to detect rubella virus in a case of chronic bilateral uveitis, suggesting the presence of long-term viral replication in the eye [20]. This highlights the potential of metagenomic to uncover latent viral infections that may not be detected by routine diagnostic tests. However, it is important to note that the ability to detect some of these viruses depends on whether RNA sequencing is included in the mNGS workflow, as DNA sequencing alone would not capture the RNA viruses detected in the studies.

Table 2.
Comparative diagnostic performance of mNGS vs. CMTs.
In terms of diagnostic performance, mNGS has also demonstrated superiority in sensitivity when compared to CMTs using clinical diagnosis as the reference standard. Notably, mNGS showed more sensitivity (87.8–96.9%), but less specificity (58.4–69.2%) [15,23] whereas CMTs exhibited less sensitivity (56.4–75.0%), but more specificity (96.2–100%) [15,17]. This trade-off underscores mNGS strength in identifying a broader spectrum of pathogens, including those potentially missed by the CMTs (Table 2), though it may also yield more false positives due to its heightened sensitivity.
Moreover, mNGS can simultaneously detect atypical infections, a capability that is especially valuable in immunocompromised patients who are at higher risk for viral coinfections. Consistent with the findings from our systematic review, Nguyen et al. reported a case in which mNGS was highly valuable for detecting intraocular Monkeypox virus in an immunocompromised patient with chronic lymphocytic leukemia who presented without classic prodromal symptoms or skin lesions [30]. Also, Xu et al. and Hu et al. demonstrated that the metagenomic sequencing of vitreous fluid diagnosed acute retinal necrosis associated with pseudorabies virus infection. As this disease has rapid progression and can cause severe visual impairment, mNGS’s fast and unbiased diagnostic capability can be crucial [31,32]. The capacity of mNGS to simultaneously detect atypical pathogens is therefore particularly relevant in immunocompromised populations. In contrast, within this review, mNGS showed limited and variable sensitivity in detecting M. tuberculosis, emphasizing the need for methodological improvements in ocular tuberculosis diagnosis. This challenge is further supported by the minimal sensitivity observed with multiplex PCR for detecting M. tuberculosis DNA in intraocular samples from clinically suspected cases, likely due to the paucibacillary nature of this disease [33].
Furthermore, mNGS provides genomic insights beyond pathogen identification. In one study included in our review, Koyanagi et al. demonstrated that mNGS can infer drug resistance genes, supporting targeted therapy decisions [23]. External studies not included in this review also highlight its potential. Gu et al. showed how metagenomics helped detect gene variations, such as loss-of-function variations in the RNR gene in VZV, that might lead to impaired virulence with an increased duration of the plateau phase during treatment [34]. Doan et al. (2017) also reported that mNGS could infer the phenotypic behavior of identified pathogens [21]. Three out of seven CMV samples analyzed in this study had mutations in the gene UL97 (phosphotransferase), which confers ganciclovir and valganciclovir resistance [21]. Such capability enhances patient management by enabling precision medicine approaches by predicting drug resistance profiles based on the genomic data.
4.3. Challenges and Limitations
Despite its promising applications, mNGS faces several limitations. One primary concern is the risk of false positive results due to contamination from the laboratory environment, kit reagents, or the periocular and ocular surface microbiota microbes that can be introduced during sample collection. Multiple studies, including those by Cai 2024, Koyanagi 2023, Qian 2024, and Sun 2024, highlighted difficulties in differentiating true pathogens from contaminants [18,23,26,27]. Qian 2024 found that while mNGS exhibited 82.4% sensitivity, its specificity was only 15.8% [26], suggesting a tendency to over-detect pathogens. The identification of non-pathogenic or commensal microbes can complicate clinical interpretation, necessitating robust bioinformatics filtering and threshold settings. Variations in sequencing platforms, such as Illumina and Oxford Nanopore, which have substantial difference in the total sequencing outputs, could also contribute to discrepancies in sensitivity and specificity.
Additionally, the high cost, labor-intensive workflow, and complex bioinformatics analyses required for mNGS implementation were identified as barriers to its widespread clinical adoption [22,23]. The turnaround times, although improving, remain longer than those of PCR, making mNGS less feasible for urgent clinical decision making.
Also, while the testing of vitreous humor demonstrated slightly higher pathogen detection rates, particularly for non-viral infections [26], testing aqueous humor samples may represent a more feasible and less invasive alternative for mNGS-based diagnostics in routine clinical practice. The vast range of sample volumes used in mNGS testing (20–100 µL) [18,20,21,22,23,24,26,27,28,29], a quantity similar for each CMT sample, shows an area of possible improvement for mNGS, as this method could potentially streamline the diagnosis of infectious uveitis by supporting the use of a single and comprehensive test to allow for broad pathogen detection. As ocular samples are limited, the reduction in the amount of fluid required for mNGS detection can become an essential advantage over CMT.
4.4. Implications for Future Research and Clinical Practice
This study has several limitations. All the studies included in this analysis were non-randomized, which may introduce potential bias in our findings. A lack of randomization increases the risk of confounding variables, preventing us from establishing causal relationships. Additionally, the limited number of studies that reported data comparing mNGS to clinical diagnosis prevented us from conducting meta-analysis with the pooled sensitivity and specificity values [18,26]. Furthermore, most studies did not provide sufficient independent information regarding the mNGS performance differences according to the anatomical classification of uveitis. Lastly, it is important to acknowledge that the variability in sample types (aqueous vs. vitreous humor), sequencing platforms (e.g., Illumina and Oxford Nanopore), and sequencing depth across the included studies may have influenced the reported sensitivity and specificity. This underscores the need for methodological standardization to ensure a more consistent diagnostic performance. Consequently, this study highlights the need for further research to expand the scope of mNGS assessment of the diagnosis of infectious uveitis.
To maximize the clinical utility of mNGS in diagnosing infectious uveitis, several advancements are needed. First, the standardization of diagnostic thresholds and bioinformatics pipelines is essential to improve specificity and reduce false positives. Second, future research should focus on optimizing the sequencing depth and pathogen detection sensitivity, particularly for low-viral-load infections. Lastly, cost reduction strategies, such as targeted mNGS panels, may enhance feasibility for routine clinical practice. Future studies should address these issues and clarify specific clinical scenarios where mNGS could be preferred over the traditional methods, such as in immunocompromised patients, atypical cases, or when conventional diagnostics are inconclusive. Additionally, there is a lack of studies directly comparing the mNGS diagnostic performance against clinical diagnosis as the reference standard. This gap limits our ability to fully assess mNGS clinical diagnostic accuracy, especially regarding its sensitivity and specificity in clinically confirmed infectious uveitis cases. Across the studies, the nucleic acid-based tests (mNGS and PCR) demonstrated more agreement and a better performance in the early or acute phases of infection. In contrast, the antibody-based tests showed prolonged detection windows, especially in chronic cases. This disparity highlights the importance of choosing diagnostic tools based on the disease stage. Addressing this limitation should be a priority in future research to better define the diagnostic utility of mNGS in routine ophthalmologic practice.
5. Conclusions
In conclusion, this systematic review highlights the clinical value of mNGS in diagnosing infectious uveitis, particularly for viral etiologies and in cases where the traditional diagnostic methods have failed. mNGS has demonstrated the ability to identify unexpected pathogens and viral coinfections, offering a hypothesis-free diagnostic approach. While some studies reported sensitivity levels approaching 100%, this suggests that mNGS may achieve a diagnostic performance comparable to that of PCR in certain contexts. However, given that CMTs served as the reference standard in most included studies, the true comparative accuracy of mNGS versus PCR remains difficult to establish. Nevertheless, several barriers, including specificity concerns, contamination control, costs, and the need for methodological standardization, particularly in bioinformatics analyses, still need to be addressed to enhance its broader clinical applicability.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v17060757/s1, File: Supplemental Material mNGS, Supplementary S1: Search terms and strategy; Supplementary S2: Standardized Data Abstract Form.
Author Contributions
Conceptualization, I.P. and A.R.M.; Rayyan software, I.P. and A.R.M.; investigation, I.P., P.S.M., M.C.G., J.M.G.F., M.K.H. and Y.L.; writing—original draft preparation I.P., A.R.M. and L.P.S.F.; writing—review and editing J.R.R.P., L.V.R., P.J.M.B., T.K., D.J.D. and M.B.E.; supervision, A.R.M. and L.P.S.F.; project administration, I.P. and A.R.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data are available in the References and the Supplemental Material.
Acknowledgments
We thank Kioko Kusuki de Oliveira, a librarian from Instituto Israelita de Ensino e Pesquisa of Hospital Israelita Albert Einstein, for her crucial assistance in defining our research strategy.
Conflicts of Interest
Dr. Paulo Bispo was supported by the National Eye Institute of the National Institutes of Health (grants EY032231, EY036137, and EY036444). The remaining authors do not have any financial support to disclose.
References
- de-la-Torre, A.; Valdés-Camacho, J.; De Mesa, C.L.; Uauy-Nazal, A.; Zuluaga, J.D.; Ramírez-Páez, L.M.; Durán, F.; Torres-Morales, E.; Triviño, J.; Murillo, M.; et al. Coinfections and Differential Diagnosis in Immunocompetent Patients with Uveitis of Infectious Origin. BMC Infect. Dis. 2019, 19, 91. [Google Scholar] [CrossRef]
- Miller, J.M.; Binnicker, M.J.; Campbell, S.; Carroll, K.C.; Chapin, K.C.; Gilligan, P.H.; Gonzalez, M.D.; Jerris, R.C.; Kehl, S.C.; Patel, R.; et al. A Guide to Utilization of the Microbiology Laboratory for Diagnosis of Infectious Diseases: 2018 Update by the Infectious Diseases Society of America and the American Society for Microbiologya. Clin. Infect. Dis. 2018, 67, e1–e94. [Google Scholar] [CrossRef]
- Ma, L.; Jakobiec, F.A.; Dryja, T.P. A Review of Next-Generation Sequencing (NGS): Applications to the Diagnosis of Ocular Infectious Diseases. Semin. Ophthalmol. 2019, 34, 223–231. [Google Scholar] [CrossRef]
- Forrester, J.V.; Xu, H. Good News—Bad News: The Yin and Yang of Immune Privilege in the Eye. Front. Immunol. 2012, 3, 338. [Google Scholar] [CrossRef]
- Chiu, C.Y.; Miller, S.A. Clinical Metagenomics. Nat. Rev. Genet. 2019, 20, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Simner, P.J.; Miller, S.; Carroll, K.C. Understanding the Promises and Hurdles of Metagenomic Next-Generation Sequencing as a Diagnostic Tool for Infectious Diseases. Clin. Infect. Dis. 2018, 66, 778–788. [Google Scholar] [CrossRef]
- Batool, M.; Galloway-Peña, J. Clinical Metagenomics—Challenges and Future Prospects. Front. Microbiol. 2023, 14, 1186424. [Google Scholar] [CrossRef]
- Duan, H.; Li, X.; Mei, A.; Li, P.; Liu, Y.; Li, X.; Li, W.; Wang, C.; Xie, S. The Diagnostic Value of Metagenomic Next-generation Sequencing in Infectious Diseases. BMC Infect. Dis. 2021, 21, 62. [Google Scholar] [CrossRef]
- Greninger, A.L. The Challenge of Diagnostic Metagenomics. Expert Rev. Mol. Diagn. 2018, 18, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.; Chiu, C. The Role of Metagenomics and Next-Generation Sequencing in Infectious Disease Diagnosis. Clin. Chem. 2021, 68, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. The PRISMA Group Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [PubMed]
- Lin, P. Infectious Uveitis. Curr. Ophthalmol. Rep. 2015, 3, 170–183. [Google Scholar] [CrossRef]
- Wilson, M.R.; Sample, H.A.; Zorn, K.C.; Arevalo, S.; Yu, G.; Neuhaus, J.; Federman, S.; Stryke, D.; Briggs, B.; Langelier, C.; et al. Clinical Metagenomic Sequencing for Diagnosis of Meningitis and Encephalitis. N. Engl. J. Med. 2019, 380, 2327–2340. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.; Naccache, S.N.; Samayoa, E.; Messacar, K.; Arevalo, S.; Federman, S.; Stryke, D.; Pham, E.; Fung, B.; Bolosky, W.J.; et al. Laboratory Validation of a Clinical Metagenomic Sequencing Assay for Pathogen Detection in Cerebrospinal Fluid. Genome Res. 2019, 29, 831–842. [Google Scholar] [CrossRef]
- Schardt, C.; Adams, M.B.; Owens, T.; Keitz, S.; Fontelo, P. Utilization of the PICO Framework to Improve Searching PubMed for Clinical Questions. BMC Med. Inform. Decis. Mak. 2007, 7, 16. [Google Scholar] [CrossRef]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—A Web and Mobile App for Systematic Reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef]
- Downs, S.H.; Black, N. The Feasibility of Creating a Checklist for the Assessment of the Methodological Quality Both of Randomised and Non-Randomised Studies of Health Care Interventions. J. Epidemiol. Community Health 1998, 52, 377–384. [Google Scholar] [CrossRef]
- Cai, Z.; Zhang, X.; Song, Y.; Jiang, Y.; Jiang, L.; Li, T.; Sun, X. Performance of Metagenomic Next-Generation Sequencing for Microbiological Diagnosis of Infectious Uveitis. J. Med. Microbiol. 2024, 73, 001879. [Google Scholar] [CrossRef] [PubMed]
- de Groot-Mijnes, J.D.F. Next Generation Sequencing for the Diagnosis of Infectious Uveitis. In Proceedings of the Netherlands Ophthalmological Society (NOG) Annual Congress, Maastricht, The Netherlands, 29–31 March 2017. [Google Scholar]
- Doan, T.; Wilson, M.R.; Crawford, E.D.; Chow, E.D.; Khan, L.M.; Knopp, K.A.; O’Donovan, B.D.; Xia, D.; Hacker, J.K.; Stewart, J.M.; et al. Illuminating Uveitis: Metagenomic Deep Sequencing Identifies Common and Rare Pathogens. Genome Med. 2016, 8, 90. [Google Scholar] [CrossRef]
- Doan, T.; Acharya, N.R.; Pinsky, B.A.; Sahoo, M.K.; Chow, E.D.; Banaei, N.; Budvytiene, I.; Cevallos, V.; Zhong, L.; Zhou, Z.; et al. Metagenomic DNA Sequencing for the Diagnosis of Intraocular Infections. Ophthalmology 2017, 124, 1247–1248. [Google Scholar] [CrossRef]
- Doan, T.; Sahoo, M.K.; Ruder, K.; Huang, C.; Zhong, L.; Chen, C.; Hinterwirth, A.; Lin, C.; Gonzales, J.A.; Pinsky, B.A.; et al. Comprehensive Pathogen Detection for Ocular Infections. J. Clin. Virol. 2021, 136, 104759. [Google Scholar] [CrossRef]
- Koyanagi, Y.; Sajiki, A.F.; Ushida, H.; Kawano, K.; Fujita, K.; Okuda, D.; Kawabe, M.; Yamada, K.; Suzumura, A.; Kachi, S.; et al. Metagenomic Profiling of Long-Read Sequencing for Clinical Diagnosis of Ocular Inflammation. Nvestigative Ophthalmol. Vis. Sci. 2025, 66, 50. [Google Scholar] [CrossRef]
- Lee, J.; Jeong, H.; Kang, H.G.; Park, J.; Choi, E.Y.; Lee, C.S.; Byeon, S.H.; Kim, M. Rapid Pathogen Detection in Infectious Uveitis Using Nanopore Metagenomic Next-Generation Sequencing: A Preliminary Study. Ocul. Immunol. Inflamm. 2024, 32, 463–469. [Google Scholar] [CrossRef]
- Qian, Z.; Zhang, Y.; Wang, L.; Li, Z.; Wang, H.; Kang, H.; Feng, J.; Hu, X.; Tao, Y. Application of Metagenomic Next-Generation Sequencing in Suspected Intraocular Infections. Eur. J. Ophthalmol. 2023, 33, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Xia, H.; Zhou, J.; Wang, R.; Zhu, D.; Chen, L.; Kang, H.; Feng, J.; Hu, X.; Wang, L.; et al. Performance of Metagenomic Next-Generation Sequencing of Cell-Free DNA from Vitreous and Aqueous Humor for Diagnoses of Intraocular Infections. J. Infect. Dis. 2024, 229, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.-B.; Chen, Y.; Li, J.; Xiao, Q.; Liu, G.; Liu, Z. Metagenomic Next-Generation Sequencing for the Diagnosis of Viral Infectious Uveitis and Its Mimics. Ocul. Immunol. Inflamm. 2024, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Z.; Ma, J.; Li, Q.; Chen, X.; Chen, Y.; Sun, X. Comparison of Intraocular Antibody Measurement, Quantitative Pathogen PCR, and Metagenomic Deep Sequencing of Aqueous Humor in Secondary Glaucoma Associated with Anterior Segment Uveitis. Ocul. Immunol. Inflamm. 2022, 30, 153–159. [Google Scholar] [CrossRef]
- Yu, J.; Gao, Y.; Bi, H.; Zhang, Y.; Tang, K.; Guo, D.; Xie, X. Preliminary Exploration of Metagenomic Sequencing for Pathogenic Identification in Infectious Uveitis. J. Ophthalmic Inflamm. Infect. 2024, 14, 70. [Google Scholar] [CrossRef]
- Nguyen, M.T.; Mentreddy, A.; Schallhorn, J.; Chan, M.; Aung, S.; Doernberg, S.B.; Babik, J.; Miles, K.; Yang, K.; Lydon, E.; et al. Isolated Ocular Mpox without Skin Lesions, United States. Emerg. Infect. Dis. 2023, 29, 1285–1288. [Google Scholar] [CrossRef]
- Hu, F.; Wang, J.; Peng, X.-Y. Bilateral Necrotizing Retinitis Following Encephalitis Caused by the Pseudorabies Virus Confirmed by Next-Generation Sequencing. Ocul. Immunol. Inflamm. 2021, 29, 922–925. [Google Scholar] [CrossRef]
- Xu, G.; Hou, B.; Xue, C.; Xu, Q.; Qu, L.; Hao, X.; Liu, Y.; Wang, D.; Li, Z.; Jin, X. Acute Retinal Necrosis Associated with Pseudorabies Virus Infection: A Case Report and Literature Review. Ocul. Immunol. Inflamm. 2024, 32, 594–601. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Xu, M.; Chen, F.; Chen, H.; Du, W.; Yu, J. Detection of Mycobacterium Tuberculosis DNA in Intraocular Fluid of 11 Suspected Tuberculous Uveitis Patients by Multiplex PCR. BMC Ophthalmol. 2025, 25, 7. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Lei, B.; Wang, Z.; Zhang, T.; Jiang, T.; Zhang, P.; Chen, W.; Zhang, Y.; Jiang, R.; Xu, G.; et al. Dynamic Viral Load Monitoring and Metagenomic Sequencing in Acute Retinal Necrosis Caused by Varicella-Zoster Virus. Retina 2024, 44, 1966–1975. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).