HTLV-I Basic Leucine Zipper Factor (sHBZ) Actively Associates with Nucleophosmin (B23) in the Nucleolus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture of Cell Lines
2.2. Plasmids
2.3. HTLV-1 Infection of HeLa Cells by Coculture with MT2 Cells
2.4. Cell Transfection
2.5. Nucleolus Isolation and Cell Fractionation
2.6. Western Blot Analysis
2.7. Co-Immunoprecipitation Experiments
2.8. Immunofluorescence
3. Results
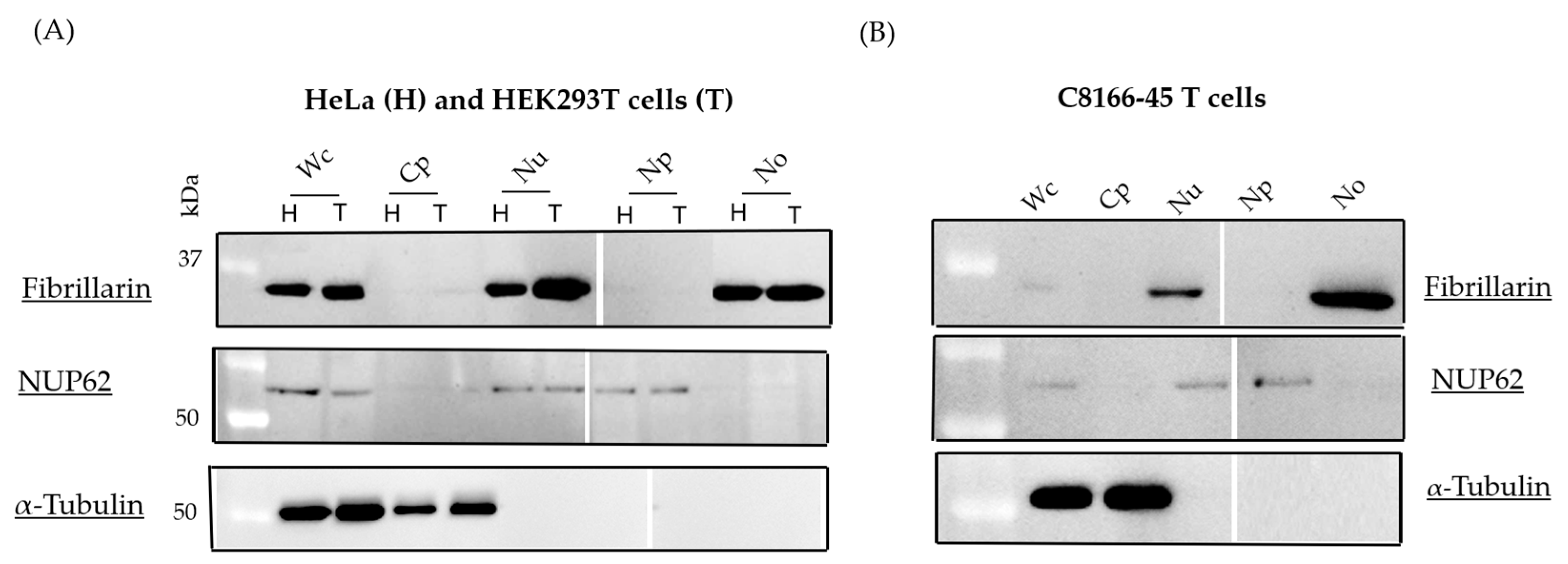
3.1. Isolation of Nucleolar Fraction
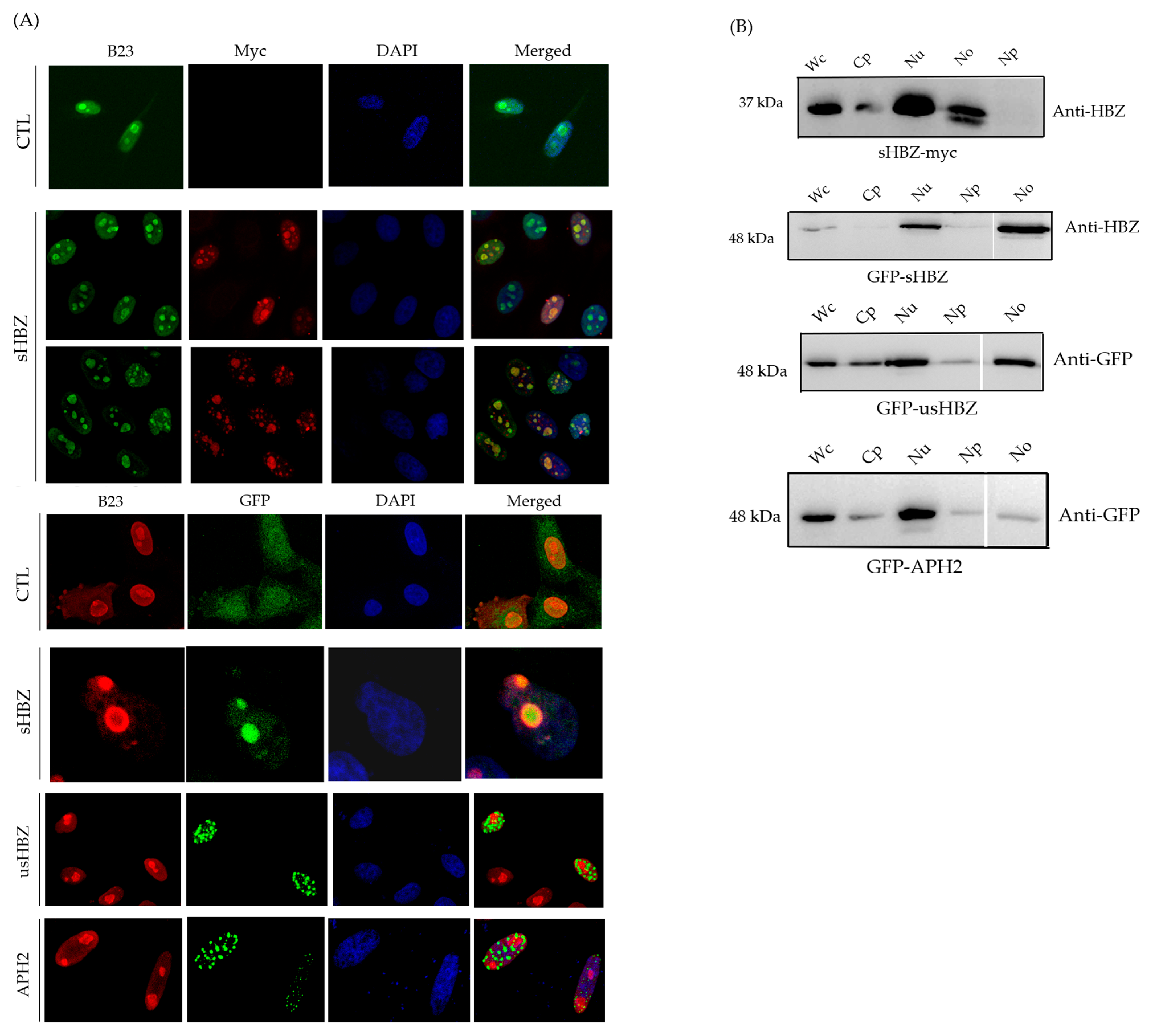
3.2. Subcellular Localization of sHBZ Was Maintained During Nucleolus Isolation
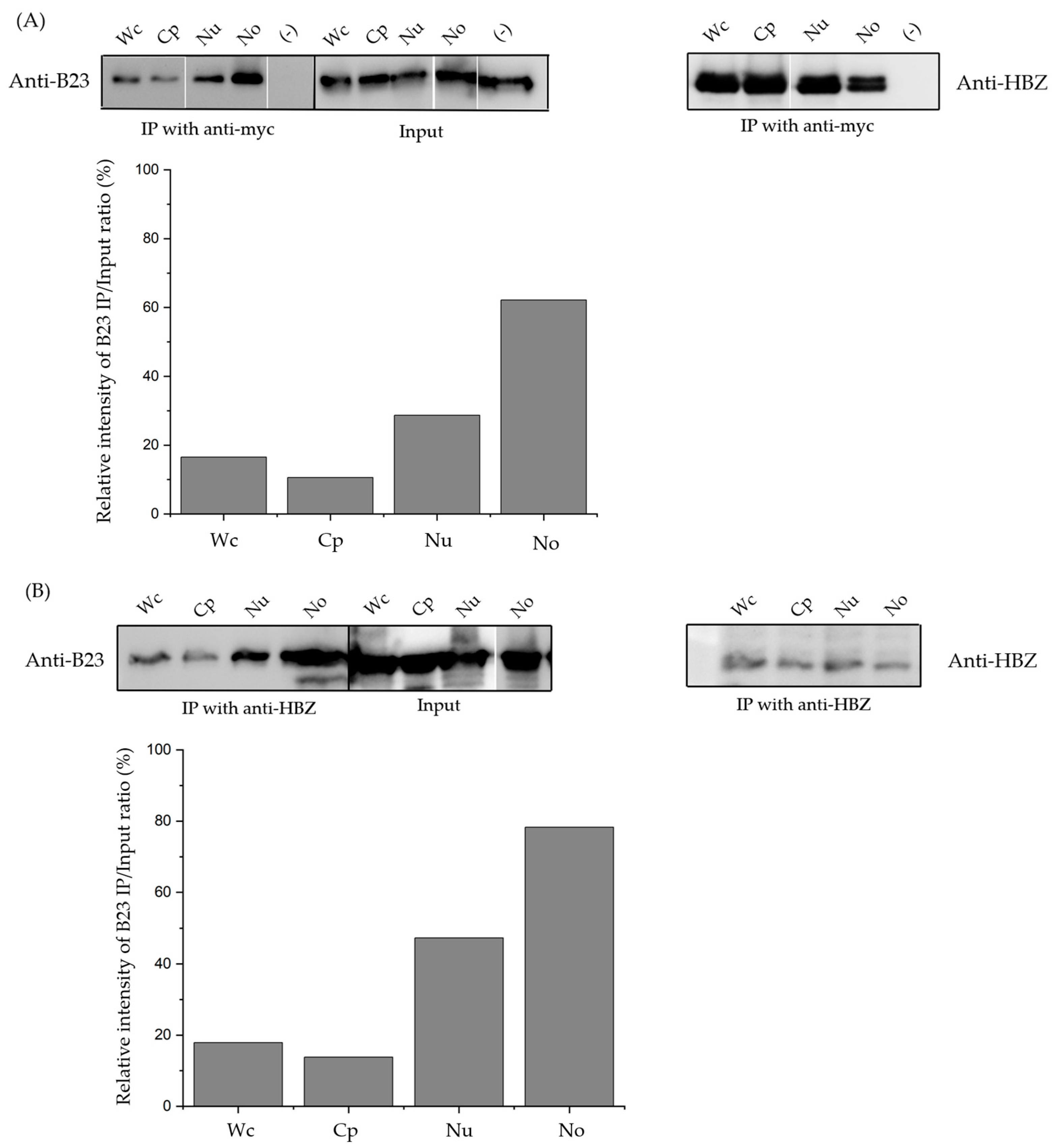
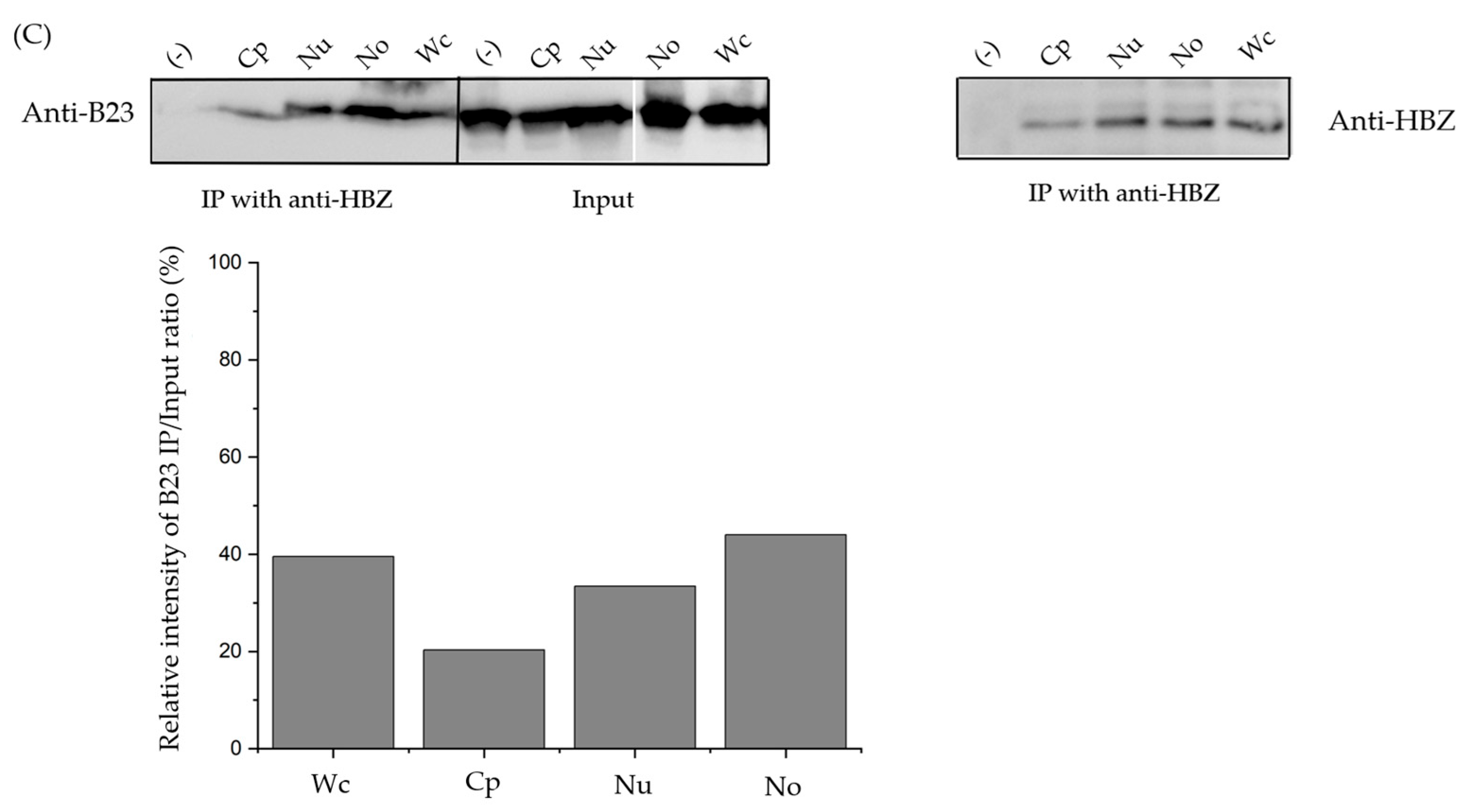
3.3. The sHBZ-B23 Association Occurs Predominantly in the Nucleolus
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Poiesz, B.J.; Ruscetti, F.W.; Gazdart, A.F.; Bunnt, P.A.; Minnat, J.D.; Gallo, R.C. Detection and Isolation of Type C Retrovirus Particles from Fresh and Cultured Lymphocytes of a Patient with Cutaneous T-Cell Lymphoma. Proc. Natl. Acad. Sci. USA 1980, 77, 7415–7419. [Google Scholar] [CrossRef] [PubMed]
- Gessain, A.; Cassar, O. Epidemiological Aspects and World Distribution of HTLV-1 Infection. Front. Microbiol. 2012, 3, 388. [Google Scholar] [CrossRef]
- Mortreux, F.; Gabet, A.S.; Wattel, E. Molecular and Cellular Aspects of HTLV-1 Associated Leukemogenesis in Vivo. Leukemia 2003, 17, 26–38. [Google Scholar] [CrossRef]
- Yasunaga, J.I. Strategies of Human T-Cell Leukemia Virus Type 1 for Persistent Infection: Implications for Leukemogenesis of Adult T-Cell Leukemia-Lymphoma. Front. Microbiol. 2020, 11, 979. [Google Scholar] [CrossRef]
- Yamagochi, K.; Kiyokawa, T.; Nakada, K.; Yol, L.S.; Asou, N.; Ishii, T.; Sanada, I.; Seiki, M.; Yoshida, M.; Matutes, E.; et al. Polyclonal Integration of HTLV-I Proviral DNA in Lymphocytes from HTLV-I Seropositive Individuals: An Intermediate State between the Healthy Carrier State and Smouldering ATL. Br. J. Haematol. 1988, 68, 169–174. [Google Scholar] [CrossRef]
- Fujikawa, D.; Nakagawa, S.; Hori, M.; Kurokawa, N.; Soejima, A.; Nakano, K.; Yamochi, T.; Nakashima, M.; Kobayashi, S.; Tanaka, Y.; et al. Polycomb-Dependent Epigenetic Landscape in Adult T-Cell Leukemia. Blood J. Am. Soc. Hematol. 2016, 127, 1790–1802. [Google Scholar] [CrossRef]
- Matsuoka, M.; Jeang, K.T. Human T-Cell Leukaemia Virus Type 1 (HTLV-1) Infectivity and Cellular Transformation. Nat. Rev. Cancer 2007, 7, 270–280. [Google Scholar] [CrossRef]
- Grassmann, R.; Aboud, M.; Jeang, K.T. Molecular Mechanisms of Cellular Transformation by HTLV-1 Tax. Oncogene 2005, 24, 5976–5985. [Google Scholar] [CrossRef]
- Larocca, D.; Chao, L.A.; Seto, M.H.; Brunck, T.K. Human T-cell leukemia virus minus strand transcription in infected T-cells. Biochem. Biophys. Res. Commun. 1989, 163, 1006–1013. [Google Scholar] [CrossRef]
- Gaudray, G.; Gachon, F.; Basbous, J.; Biard-Piechaczyk, M.; Devaux, C.; Mesnard, J.-M. The Complementary Strand of the Human T-Cell Leukemia Virus Type 1 RNA Genome Encodes a BZIP Transcription Factor That Down-Regulates Viral Transcription. J. Virol. 2002, 76, 12813–12822. [Google Scholar] [CrossRef]
- Arnold, J.; Yamamoto, B.; Li, M.; Phipps, A.J.; Younis, I.; Lairmore, M.D.; Green, P.L. Enhancement of Infectivity and Persistence in Vivo by HBZ, a Natural Antisense Coded Protein of HTLV-1. Blood 2006, 107, 3976–3982. [Google Scholar] [CrossRef] [PubMed]
- Satou, Y.; Yasunaga, J.-I.; Yoshida, M.; Matsuoka, M. HTLV-I Basic Leucine Zipper Factor Gene mRNA Supports Proliferation of Adult T Cell Leukemia. Proc. Natl. Acad. Sci. USA 2006, 103, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Hagiya, K.; Yasunaga, J.I.; Satou, Y.; Ohshima, K.; Matsuoka, M. ATF3, an HTLV-1 BZip Factor Binding Protein, Promotes Proliferation of Adult T-Cell Leukemia Cells. Retrovirology 2011, 8, 19. [Google Scholar] [CrossRef]
- Basbous, J.; Arpin, C.; Gaudray, G.; Piechaczyk, M.; Devaux, C.; Mesnard, J.M. The HBZ Factor of Human T-Cell Leukemia Virus Type I Dimerizes with Transcription Factors JunB and c-Jun and Modulates Their Transcriptional Activity. J. Biol. Chem. 2003, 278, 43620–43627. [Google Scholar] [CrossRef]
- Thébault, S.; Basbous, J.; Hivin, P.; Devaux, C.; Mesnard, J.M. HBZ Interacts with JunD and Stimulates Its Transcriptional Activity. FEBS Lett. 2004, 562, 165–170. [Google Scholar] [CrossRef]
- Matsuoka, M.; Yasunaga, J.I. Human T-Cell Leukemia Virus Type 1: Replication, Proliferation and Propagation by Tax and HTLV-1 BZIP Factor. Curr. Opin. Virol. 2013, 3, 684–691. [Google Scholar] [CrossRef]
- Zhao, T.; Yasunaga, J.-I.; Satou, Y.; Nakao, M.; Takahashi, M.; Fujii, M.; Matsuoka, M. Human T-Cell Leukemia Virus Type 1 BZIP Factor Selectively Suppresses the Classical Pathway of NF-κB. Blood 2009, 113, 2755–2764. [Google Scholar] [CrossRef]
- Wright, D.G.; Marchal, C.; Hoang, K.; Ankney, J.A.; Nguyen, S.T.; Rushing, A.W.; Polakowski, N.; Miotto, B.; Lemasson, I. Human T-Cell Leukemia Virus Type-1-Encoded Protein HBZ Represses P53 Function by Inhibiting the Acetyltransferase Activity of P300/CBP and HBO1. Oncotarget 2015, 7, 1687–1706. [Google Scholar] [CrossRef]
- Kawatsuki, A.; Yasunaga, J.I.; Mitobe, Y.; Green, P.L.; Matsuoka, M. HTLV-1 BZIP Factor Protein Targets the Rb/E2F-1 Pathway to Promote Proliferation and Apoptosis of Primary CD4+ T Cells. Oncogene 2016, 35, 4509–4517. [Google Scholar] [CrossRef]
- Murata, K.; Hayashibara, T.; Sugahara, K.; Uemura, A.; Yamaguchi, T.; Harasawa, H.; Hasegawa, H.; Tsuruda, K.; Okazaki, T.; Koji, T.; et al. A Novel Alternative Splicing Isoform of Human T-Cell Leukemia Virus Type 1 BZIP Factor (HBZ-SI) Targets Distinct Subnuclear Localization. J. Virol. 2006, 80, 2495–2505. [Google Scholar] [CrossRef]
- Cavanagh, M.H.; Landry, S.; Audet, B.; Arpin-André, C.; Hivin, P.; Paré, M.È.; Thête, J.; Wattel, É.; Marriott, S.J.; Mesnard, J.M.; et al. HTLV-I Antisense Transcripts Initiating in the 3’LTR Are Alternatively Spliced and Polyadenylated. Retrovirology 2006, 3, 15. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, M.; Satou, Y.; Yasunaga, J.; Fujisawa, J.; Matsuoka, M. Transcriptional Control of Spliced and Unspliced Human T-Cell Leukemia Virus Type 1 BZIP Factor (HBZ) Gene. J. Virol. 2008, 82, 9359–9368. [Google Scholar] [CrossRef]
- Matsuoka, M.; Green, P.L. The HBZ Gene, a Key Player in HTLV-1 Pathogenesis. Retrovirology 2009, 6, 71. [Google Scholar] [CrossRef]
- Olson, M.O.; Dundr, M.; Szebeni, A. The Nucleolus: An Old Factory with Unexpected Capabilities. Trend. Cell Biol. 2000, 10, 189–196. [Google Scholar] [CrossRef]
- Pederson, T. The Plurifunctional Nucleolus. Nucleic Acids Res. 1998, 26, 3871–3876. [Google Scholar] [CrossRef]
- Maggi, L.B.; Weber, J.D. Nucleolar Adaptation in Human Cancer. Cancer Investig. 2005, 23, 599–608. [Google Scholar] [CrossRef]
- Liu, Z.; Larocque, É.; Xie, Y.; Xiao, Y.; Lemay, G.; Peloponese, J.M.; Mesnard, J.M.; Rassart, É.; Lin, R.; Zhou, S.; et al. A Newly Identified Interaction between Nucleolar NPM1/B23 and the HTLV-I Basic Leucine Zipper Factor in HTLV-1 Infected Cells. Front. Microbiol. 2022, 13, 988944. [Google Scholar] [CrossRef]
- Box, J.K.; Paquet, N.; Adams, M.N.; Boucher, D.; Bolderson, E.; O’Byrne, K.J.; Richard, D.J. Nucleophosmin: From Structure and Function to Disease Development. BMC Mol. Biol. 2016, 17, 19. [Google Scholar] [CrossRef]
- Hivin, P.; Frédéric, M.; Arpin-André, C.; Basbous, J.; Gay, B.; Thébault, S.; Mesnard, J.M. Nuclear Localization of HTLV-I BZIP Factor (HBZ) Is Mediated by Three Distinct Motifs. J. Cell Sci. 2005, 118, 1355–1362. [Google Scholar] [CrossRef]
- Liu, M.; Yang, L.; Zhang, L.; Liu, B.; Merling, R.; Xia, Z.; Giam, C.-Z. Human T-Cell Leukemia Virus Type 1 Infection Leads to Arrest in the G 1 Phase of the Cell Cycle. J. Virol. 2008, 82, 8442–8455. [Google Scholar] [CrossRef]
- Chamousset, D.; Mamane, S.; Boisvert, F.M.; Trinkle-Mulcahy, L. Efficient Extraction of Nucleolar Proteins for Interactome Analyses. Proteomics 2010, 10, 3045–3050. [Google Scholar] [CrossRef] [PubMed]
- Douceron, E.; Kaidarova, Z.; Miyazato, P.; Matsuoka, M.; Murphy, E.L.; Mahieux, R. HTLV-2 APH-2 Expression Is Correlated with Proviral Load but APH-2 Does Not Promote Lymphocytosis. J. Infect. Dis. 2012, 205, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Forlani, G.; Shallak, M.; Tedeschi, A.; Cavallari, I.; Marçais, A.; Hermine, O.; Accolla, R.S. Dual Cytoplasmic and Nuclear Localization of HTLV-1-Encoded HBZ Protein Is a Unique Feature of Adult T-Cell Leukemia. Haematologica 2021, 106, 2076–2085. [Google Scholar] [CrossRef] [PubMed]
- Baratella, M.; Forlani, G.; Raval, G.U.; Tedeschi, A.; Gout, O.; Gessain, A.; Tosi, G.; Accolla, R.S. Cytoplasmic Localization of HTLV-1 HBZ Protein: A Biomarker of HTLV-1-Associated Myelopathy/Tropical Spastic Paraparesis (HAM/TSP). PLoS Negl. Trop. Dis. 2017, 11, e0005285. [Google Scholar] [CrossRef]
- Greco, A. Involvement of the Nucleolus in Replication of Human Viruses. Rev. Med. Virol. 2009, 19, 201–214. [Google Scholar] [CrossRef]
- Lobaina, Y.; Perera, Y. Implication of B23/Nucleophosmin in Viral Infections, Potential Uses of B23/NPM1 Inhibitors as Antiviral Therapy. Infect. Disord. Drug Targets 2018, 19, 2–16. [Google Scholar] [CrossRef]
- Zakaryan, H.; Stamminger, T. Nuclear Remodelling during Viral Infections. Cell Microbiol. 2011, 13, 806–813. [Google Scholar] [CrossRef]
- Takemura, M.; Ohoka, F.; Perpelescu, M.; Ogawa, M.; Matsushita, H.; Takaba, T.; Akiyama, T.; Umekawa, H.; Furuichi, Y.; Cook, P.R.; et al. Phosphorylation-Dependent Migration of Retinoblastoma Protein into the Nucleolus Triggered by Binding to Nucleophosmin/B23. Exp. Cell Res. 2002, 276, 233–241. [Google Scholar] [CrossRef]
- Lindström, M.S.; Zhang, Y. B23 and ARF: Friends or Foes? Cell Biochem. Biophys. 2006, 46, 79–90. [Google Scholar] [CrossRef]
- Liu, X.; Liu, Z.; Jang, S.-W.; Ma, Z.; Shinmura, K.; Kang, S.; Dong, S.; Chen, J.; Fukasawa, K.; Ye, K. Sumoylation of Nucleophosmin/B23 Regulates Its Subcellular Localization, Mediating Cell Proliferation and Survival. Proc. Natl. Acad. Sci. USA 2007, 104, 9679–9684. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moghadam, N.; Xiao, Y.; Dragon, F.; Barbeau, B. HTLV-I Basic Leucine Zipper Factor (sHBZ) Actively Associates with Nucleophosmin (B23) in the Nucleolus. Viruses 2025, 17, 727. https://doi.org/10.3390/v17050727
Moghadam N, Xiao Y, Dragon F, Barbeau B. HTLV-I Basic Leucine Zipper Factor (sHBZ) Actively Associates with Nucleophosmin (B23) in the Nucleolus. Viruses. 2025; 17(5):727. https://doi.org/10.3390/v17050727
Chicago/Turabian StyleMoghadam, Nahid, Yong Xiao, Francois Dragon, and Benoit Barbeau. 2025. "HTLV-I Basic Leucine Zipper Factor (sHBZ) Actively Associates with Nucleophosmin (B23) in the Nucleolus" Viruses 17, no. 5: 727. https://doi.org/10.3390/v17050727
APA StyleMoghadam, N., Xiao, Y., Dragon, F., & Barbeau, B. (2025). HTLV-I Basic Leucine Zipper Factor (sHBZ) Actively Associates with Nucleophosmin (B23) in the Nucleolus. Viruses, 17(5), 727. https://doi.org/10.3390/v17050727





