Multidimensional Regulatory Mechanisms and Targeting Strategies of the eEF1 Family in RNA Virus Infection
Abstract
1. Introduction
2. Structural and Functional Architecture of the eEF1 Family
2.1. Domain Organization and Isoform Diversity of eEF1A
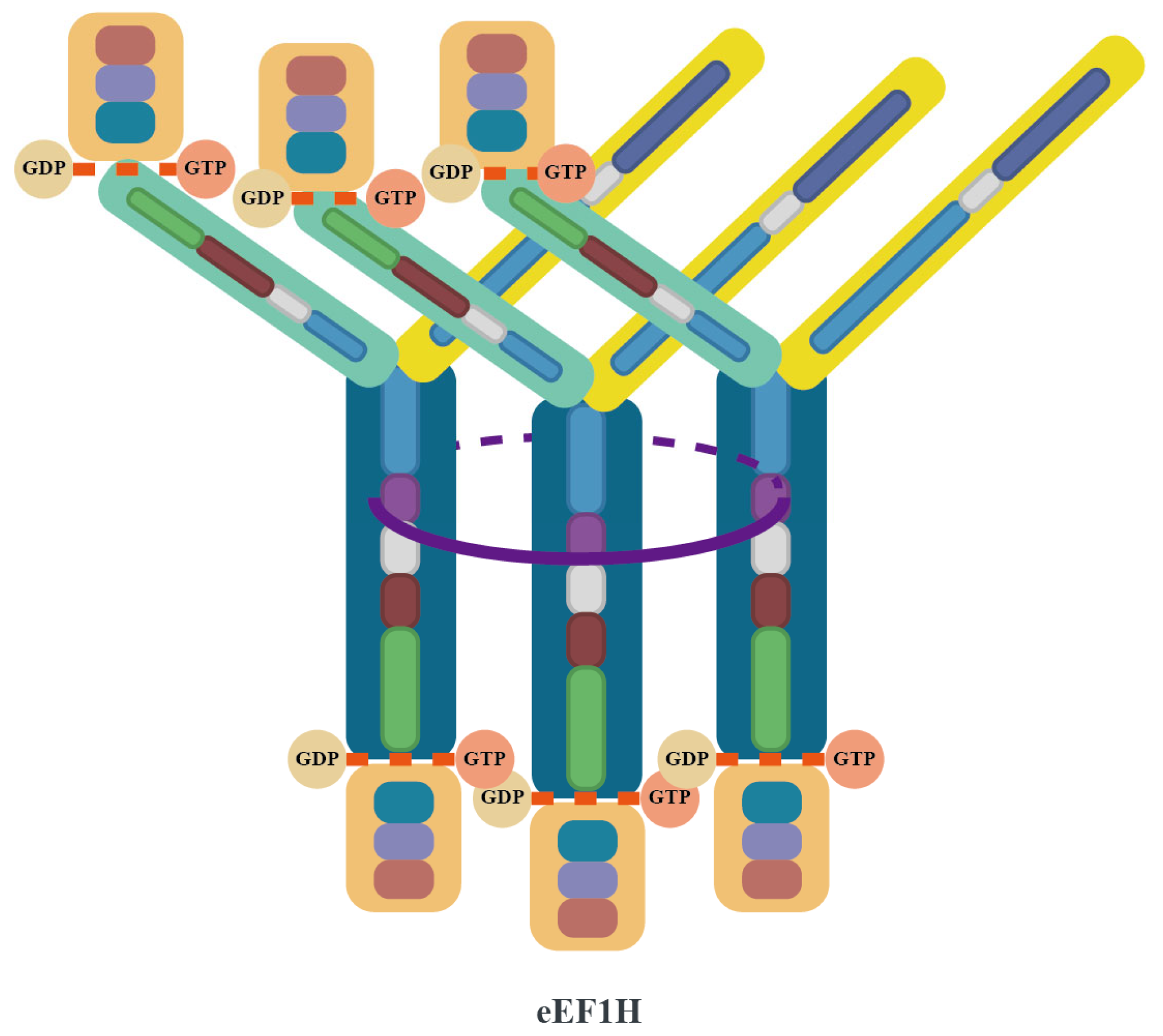
2.2. Subunit Composition and Assembly Mechanisms of the eEF1B Complex
2.3. Canonical Roles of the eEF1 Family in Protein Synthesis
2.4. Non-Canonical Functions of the eEF1 Family
2.4.1. Cytoskeletal Dynamics
2.4.2. Modulation of Apoptosis and Cell Survival Pathways
3. Mechanistic Roles of the eEF1 Family in Viral Pathogenesis
3.1. eEF1A in Viral Replication and Immune Evasion
3.1.1. eEF1A-Driven Viral Genome Replication Mechanisms
3.1.2. Viral Assembly and Release Mediated by eEF1A
3.1.3. Suppression of Host Immunity and Apoptotic Pathways
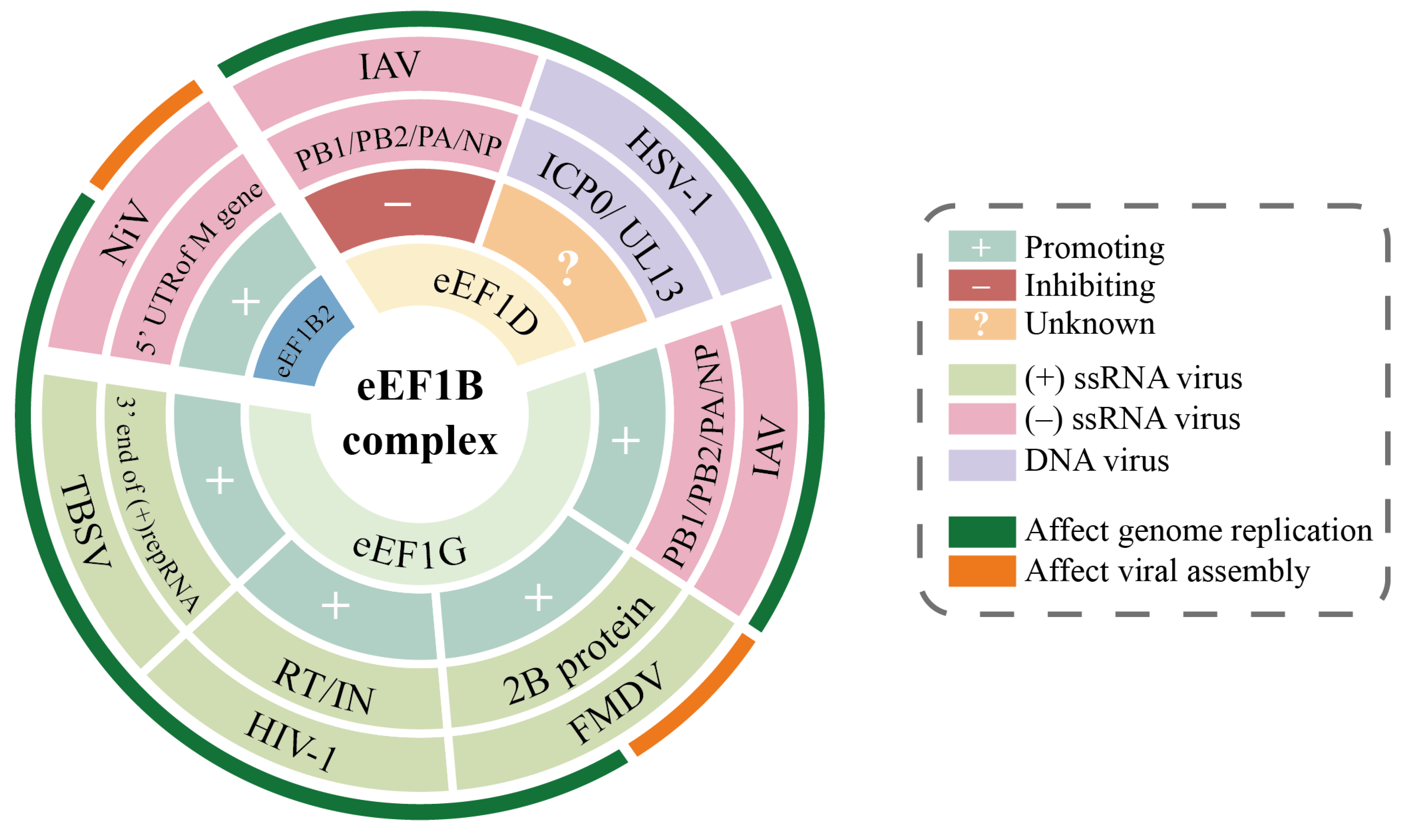
3.2. eEF1B Subunits in Viral Lifecycle Regulation
3.2.1. eEF1B2 Enhances Viral mRNA Translation Efficiency
3.2.2. eEF1D Coordinates Nuclear Transport and Post-Translational Modifications
3.2.3. eEF1G Facilitates Viral Replication Complex Assembly
4. Strategies and Challenges in Developing eEF1-Targeted Antiviral Drugs
5. Innovative Directions for eEF1-Targeted Antiviral Therapies
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dever, T.E.; Dinman, J.D.; Green, R. Translation Elongation and Recoding in Eukaryotes. Cold Spring Harb. Perspect. Biol. 2018, 10, a032649. [Google Scholar] [CrossRef]
- Negrutskii, B.S.; El’skaya, A.V. Eukaryotic translation elongation factor 1 alpha: Structure, expression, functions, and possible role in aminoacyl-tRNA channeling. Prog. Nucleic Acid Res. Mol. Biol. 1998, 60, 47–78. [Google Scholar] [CrossRef] [PubMed]
- Negrutskii, B.S.; Shalak, V.F.; Novosylna, O.V.; Porubleva, L.V.; Lozhko, D.M.; El’skaya, A.V. The eEF1 family of mammalian translation elongation factors. BBA Adv. 2023, 3, 100067. [Google Scholar] [CrossRef]
- Bondarchuk, T.V.; Shalak, V.F.; Lozhko, D.M.; Fatalska, A.; Szczepanowski, R.H.; Liudkovska, V.; Tsuvariev, O.Y.; Dadlez, M.; El’skaya, A.V.; Negrutskii, B.S. Quaternary organization of the human eEF1B complex reveals unique multi-GEF domain assembly. Nucleic Acids Res. 2022, 50, 9490–9504. [Google Scholar] [CrossRef] [PubMed]
- Morita, K.; Bunai, F.; Numata, O. Roles of three domains of Tetrahymena eEF1A in bundling F-actin. Zoolog. Sci. 2008, 25, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Sasikumar, A.N.; Perez, W.B.; Kinzy, T.G. The many roles of the eukaryotic elongation factor 1 complex. Wiley Interdiscip. Rev. RNA 2012, 3, 543–555. [Google Scholar] [CrossRef]
- Mingot, J.M.; Vega, S.; Cano, A.; Portillo, F.; Nieto, M.A. eEF1A mediates the nuclear export of SNAG-containing proteins via the Exportin5-aminoacyl-tRNA complex. Cell Rep. 2013, 5, 727–737. [Google Scholar] [CrossRef]
- Chang, R.; Wang, E. Mouse translation elongation factor eEF1A-2 interacts with Prdx-I to protect cells against apoptotic death induced by oxidative stress. J. Cell. Biochem. 2007, 100, 267–278. [Google Scholar] [CrossRef]
- Abbas, W.; Khan, K.A.; Kumar, A.; Tripathy, M.K.; Dichamp, I.; Keita, M.; Mahlknecht, U.; Rohr, O.; Herbein, G. Blockade of BFA-mediated apoptosis in macrophages by the HIV-1 Nef protein. Cell Death Dis. 2014, 5, e1080. [Google Scholar] [CrossRef]
- Talapatra, S.; Wagner, J.D.; Thompson, C.B. Elongation factor-1 alpha is a selective regulator of growth factor withdrawal and ER stress-induced apoptosis. Cell Death Differ. 2002, 9, 856–861. [Google Scholar] [CrossRef]
- Cui, H.; Li, H.; Wu, H.; Du, F.; Xie, X.; Zeng, S.; Zhang, Z.; Dong, K.; Shang, L.; Jing, C.; et al. A novel 3′tRNA-derived fragment tRF-Val promotes proliferation and inhibits apoptosis by targeting EEF1A1 in gastric cancer. Cell Death Dis. 2022, 13, 471. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jiang, S.; Yang, P.Y.; Zhang, Y.F.; Li, T.J.; Rui, Y.C. EF1A1/HSC70 Cooperatively Suppress Brain Endothelial Cell Apoptosis via Regulating JNK Activity. CNS Neurosci. Ther. 2016, 22, 836–844. [Google Scholar] [CrossRef] [PubMed]
- Abbas, W.; Kumar, A.; Herbein, G. The eEF1A Proteins: At the Crossroads of Oncogenesis, Apoptosis, and Viral Infections. Front. Oncol. 2015, 5, 75. [Google Scholar] [CrossRef] [PubMed]
- Prommahom, A.; Dharmasaroja, P. Effects of eEF1A2 knockdown on autophagy in an MPP(+)-induced cellular model of Parkinson’s disease. Neurosci. Res. 2021, 164, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Liu, L.; Song, G. Functions and Regulation of Translation Elongation Factors. Front. Mol. Biosci. 2021, 8, 816398. [Google Scholar] [CrossRef]
- Andersen, G.R.; Pedersen, L.; Valente, L.; Chatterjee, I.; Kinzy, T.G.; Kjeldgaard, M.; Nyborg, J. Structural basis for nucleotide exchange and competition with tRNA in the yeast elongation factor complex eEF1A:eEF1Balpha. Mol. Cell 2000, 6, 1261–1266. [Google Scholar] [CrossRef]
- Mateyak, M.K.; Kinzy, T.G. eEF1A: Thinking outside the ribosome. J. Biol. Chem. 2010, 285, 21209–21213. [Google Scholar] [CrossRef]
- Mills, A.; Gago, F. On the Need to Tell Apart Fraternal Twins eEF1A1 and eEF1A2, and Their Respective Outfits. Int. J. Mol. Sci. 2021, 22, 6973. [Google Scholar] [CrossRef]
- Shao, S.; Murray, J.; Brown, A.; Taunton, J.; Ramakrishnan, V.; Hegde, R.S. Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes. Cell 2016, 167, 1229–1240.e1215. [Google Scholar] [CrossRef]
- Soares, D.C.; Barlow, P.N.; Newbery, H.J.; Porteous, D.J.; Abbott, C.M. Structural models of human eEF1A1 and eEF1A2 reveal two distinct surface clusters of sequence variation and potential differences in phosphorylation. PLoS ONE 2009, 4, e6315. [Google Scholar] [CrossRef]
- Lee, S.; Francoeur, A.M.; Liu, S.; Wang, E. Tissue-specific expression in mammalian brain, heart, and muscle of S1, a member of the elongation factor-1 alpha gene family. J. Biol. Chem. 1992, 267, 24064–24068. [Google Scholar] [CrossRef]
- Le Sourd, F.; Boulben, S.; Le Bouffant, R.; Cormier, P.; Morales, J.; Belle, R.; Mulner-Lorillon, O. eEF1B: At the dawn of the 21st century. Biochim. Biophys. Acta 2006, 1759, 13–31. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, M.; Uritani, M.; Fujimura, K.; Yamakatsu, H.; Kageyama, T.; Takahashi, K. Peptide elongation factor 1 from yeasts: Purification and biochemical characterization of peptide elongation factors 1 alpha and 1 beta (gamma) from Saccharomyces carlsbergensis and Schizosaccharomyces pombe. J. Biochem. 1988, 103, 508–521. [Google Scholar] [CrossRef] [PubMed]
- Ejiri, S.; Ebata, N.; Kawamura, R.; Katsumata, T. Occurrence of four subunits in high molecular weight forms of polypeptide chain elongation factor 1 from wheat embryo. J. Biochem. 1983, 94, 319–322. [Google Scholar] [CrossRef] [PubMed]
- Minella, O.; Mulner-Lorillon, O.; Bec, G.; Cormier, P.; Bellé, R. Multiple phosphorylation sites and quaternary organization of guanine-nucleotide exchange complex of elongation factor-1 (EF-1betagammadelta/ValRS) control the various functions of EF-1alpha. Biosci. Rep. 1998, 18, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Maessen, G.D.; Amons, R.; Zeelen, J.P.; Möller, W. Primary structure of elongation factor 1 gamma from Artemia. FEBS Lett. 1987, 223, 181–186. [Google Scholar] [CrossRef]
- Cormier, P.; Osborne, H.B.; Morales, J.; Bassez, T.; Poulhe, R.; Mazabraud, A.; Mulner-Lorillon, O.; Bellé, R. Molecular cloning of Xenopus elongation factor 1 gamma, major M-phase promoting factor substrate. Nucleic Acids Res. 1991, 19, 6644. [Google Scholar] [CrossRef]
- Sanders, J.; Maassen, J.A.; Möller, W. Elongation factor-1 messenger-RNA levels in cultured cells are high compared to tissue and are not drastically affected further by oncogenic transformation. Nucleic Acids Res. 1992, 20, 5907–5910. [Google Scholar] [CrossRef]
- Kumabe, T.; Sohma, Y.; Yamamoto, T. Human cDNAs encoding elongation factor 1 gamma and the ribosomal protein L19. Nucleic Acids Res. 1992, 20, 2598. [Google Scholar] [CrossRef]
- Koonin, E.V.; Mushegian, A.R.; Tatusov, R.L.; Altschul, S.F.; Bryant, S.H.; Bork, P.; Valencia, A. Eukaryotic translation elongation factor 1 gamma contains a glutathione transferase domain--study of a diverse, ancient protein superfamily using motif search and structural modeling. Protein Sci. 1994, 3, 2045–2054. [Google Scholar] [CrossRef]
- Pérez, J.M.; Kriek, J.; Dijk, J.; Canters, G.W.; Möller, W. Expression, purification, and spectroscopic studies of the guanine nucleotide exchange domain of human elongation factor, EF-1beta. Protein Expr. Purif. 1998, 13, 259–267. [Google Scholar] [CrossRef] [PubMed]
- van Damme, H.T.; Amons, R.; Karssies, R.; Timmers, C.J.; Janssen, G.M.; Möller, W. Elongation factor 1 beta of artemia: Localization of functional sites and homology to elongation factor 1 delta. Biochim. Biophys. Acta 1990, 1050, 241–247. [Google Scholar] [CrossRef]
- Guerrucci, M.A.; Monnier, A.; Delalande, C.; Bellé, R. The elongation factor-1delta (EF-1delta) originates from gene duplication of an EF-1beta ancestor and fusion with a protein-binding domain. Gene 1999, 233, 83–87. [Google Scholar] [CrossRef]
- Alber, T. Structure of the leucine zipper. Curr. Opin. Genet. Dev. 1992, 2, 205–210. [Google Scholar] [CrossRef]
- Bec, G.; Kerjan, P.; Waller, J.P. Reconstitution in vitro of the valyl-tRNA synthetase-elongation factor (EF) 1 beta gamma delta complex. Essential roles of the NH2-terminal extension of valyl-tRNA synthetase and of the EF-1 delta subunit in complex formation. J. Biol. Chem. 1994, 269, 2086–2092. [Google Scholar] [CrossRef] [PubMed]
- Sang Lee, J.; Gyu Park, S.; Park, H.; Seol, W.; Lee, S.; Kim, S. Interaction network of human aminoacyl-tRNA synthetases and subunits of elongation factor 1 complex. Biochem. Biophys. Res. Commun. 2002, 291, 158–164. [Google Scholar] [CrossRef]
- Merrick, W.C.; Nyborg, J. The protein biosynthesis elongation cycle. Cold Spring Harb. Monogr. Arch. 2000, 39, 89–125. [Google Scholar] [CrossRef]
- Clark, B.F.C.; Grunberg-Manago, M.; Gupta, N.K.; Hershey, J.W.B.; Voorma, H.O. Prokaryotic and eukaryotic translation actors: International Union of Biochemistry and Molecular Biology (IUBMB). Biochimie 1996, 78, 1119–1122. [Google Scholar] [CrossRef]
- Li, D.; Wei, T.; Abbott, C.M.; Harrich, D. The unexpected roles of eukaryotic translation elongation factors in RNA virus replication and pathogenesis. Microbiol. Mol. Biol. Rev. 2013, 77, 253–266. [Google Scholar] [CrossRef]
- Carvalho, M.D.; Carvalho, J.F.; Merrick, W.C. Biological characterization of various forms of elongation factor 1 from rabbit reticulocytes. Arch. Biochem. Biophys. 1984, 234, 603–611. [Google Scholar] [CrossRef]
- Taylor, D.J.; Frank, J.; Kinzy, T.G. Structure and function of the eukaryotic ribosome and elongation factors. Cold Spring Harb. Monogr. Arch. 2007, 48, 59–85. [Google Scholar] [CrossRef]
- Pittman, Y.R.; Valente, L.; Jeppesen, M.G.; Andersen, G.R.; Patel, S.; Kinzy, T.G. Mg2+ and a key lysine modulate exchange activity of eukaryotic translation elongation factor 1B alpha. J. Biol. Chem. 2006, 281, 19457–19468. [Google Scholar] [CrossRef] [PubMed]
- Mansilla, F.; Friis, I.; Jadidi, M.; Nielsen, K.M.; Clark, B.F.; Knudsen, C.R. Mapping the human translation elongation factor eEF1H complex using the yeast two-hybrid system. Biochem. J. 2002, 365, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Motorin Yu, A.; Wolfson, A.D.; Orlovsky, A.F.; Gladilin, K.L. Mammalian valyl-tRNA synthetase forms a complex with the first elongation factor. FEBS Lett. 1988, 238, 262–264. [Google Scholar] [CrossRef]
- Yang, F.; Demma, M.; Warren, V.; Dharmawardhane, S.; Condeelis, J. Identification of an actin-binding protein from Dictyostelium as elongation factor 1a. Nature 1990, 347, 494–496. [Google Scholar] [CrossRef] [PubMed]
- Demma, M.; Warren, V.; Hock, R.; Dharmawardhane, S.; Condeelis, J. Isolation of an abundant 50,000-dalton actin filament bundling protein from Dictyostelium amoebae. J. Biol. Chem. 1990, 265, 2286–2291. [Google Scholar] [CrossRef]
- Edmonds, B.T.; Bell, A.; Wyckoff, J.; Condeelis, J.; Leyh, T.S. The effect of F-actin on the binding and hydrolysis of guanine nucleotide by Dictyostelium elongation factor 1A. J. Biol. Chem. 1998, 273, 10288–10295. [Google Scholar] [CrossRef]
- Liu, G.; Tang, J.; Edmonds, B.T.; Murray, J.; Levin, S.; Condeelis, J. F-actin sequesters elongation factor 1alpha from interaction with aminoacyl-tRNA in a pH-dependent reaction. J. Cell Biol. 1996, 135, 953–963. [Google Scholar] [CrossRef]
- Munshi, R.; Kandl, K.A.; Carr-Schmid, A.; Whitacre, J.L.; Adams, A.E.; Kinzy, T.G. Overexpression of translation elongation factor 1A affects the organization and function of the actin cytoskeleton in yeast. Genetics 2001, 157, 1425–1436. [Google Scholar] [CrossRef]
- Pittman, Y.R.; Kandl, K.; Lewis, M.; Valente, L.; Kinzy, T.G. Coordination of eukaryotic translation elongation factor 1A (eEF1A) function in actin organization and translation elongation by the guanine nucleotide exchange factor eEF1Balpha. J. Biol. Chem. 2009, 284, 4739–4747. [Google Scholar] [CrossRef]
- Furukawa, R.; Jinks, T.M.; Tishgarten, T.; Mazzawi, M.; Morris, D.R.; Fechheimer, M. Elongation factor 1beta is an actin-binding protein. Biochim. Biophys. Acta 2001, 1527, 130–140. [Google Scholar] [CrossRef]
- Kim, S.; Wong, P.; Coulombe, P.A. A keratin cytoskeletal protein regulates protein synthesis and epithelial cell growth. Nature 2006, 441, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kellner, J.; Lee, C.H.; Coulombe, P.A. Interaction between the keratin cytoskeleton and eEF1Bgamma affects protein synthesis in epithelial cells. Nat. Struct. Mol. Biol. 2007, 14, 982–983. [Google Scholar] [CrossRef] [PubMed]
- Boulben, S.; Monnier, A.; Le Breton, M.; Morales, J.; Cormier, P.; Bellé, R.; Mulner-Lorillon, O. Sea urchin elongation factor 1delta (EF1delta) and evidence for cell cycle-directed localization changes of a sub-fraction of the protein at M phase. Cell Mol. Life Sci. 2003, 60, 2178–2188. [Google Scholar] [CrossRef]
- Sivan, G.; Aviner, R.; Elroy-Stein, O. Mitotic modulation of translation elongation factor 1 leads to hindered tRNA delivery to ribosomes. J. Biol. Chem. 2011, 286, 27927–27935. [Google Scholar] [CrossRef]
- Stapulionis, R.; Kolli, S.; Deutscher, M.P. Efficient mammalian protein synthesis requires an intact F-actin system. J. Biol. Chem. 1997, 272, 24980–24986. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Coulombe, P.A. Emerging role for the cytoskeleton as an organizer and regulator of translation. Nat. Rev. Mol. Cell Biol. 2010, 11, 75–81. [Google Scholar] [CrossRef]
- Ruest, L.B.; Marcotte, R.; Wang, E. Peptide elongation factor eEF1A-2/S1 expression in cultured differentiated myotubes and its protective effect against caspase-3-mediated apoptosis. J. Biol. Chem. 2002, 277, 5418–5425. [Google Scholar] [CrossRef]
- Abbott, C.M.; Newbery, H.J.; Squires, C.E.; Brownstein, D.; Griffiths, L.A.; Soares, D.C. eEF1A2 and neuronal degeneration. Biochem. Soc. Trans. 2009, 37, 1293–1297. [Google Scholar] [CrossRef]
- Schlaeger, C.; Longerich, T.; Schiller, C.; Bewerunge, P.; Mehrabi, A.; Toedt, G.; Kleeff, J.; Ehemann, V.; Eils, R.; Lichter, P.; et al. Etiology-dependent molecular mechanisms in human hepatocarcinogenesis. Hepatology 2008, 47, 511–520. [Google Scholar] [CrossRef]
- Pellegrino, R.; Calvisi, D.F.; Neumann, O.; Kolluru, V.; Wesely, J.; Chen, X.; Wang, C.; Wuestefeld, T.; Ladu, S.; Elgohary, N.; et al. EEF1A2 inactivates p53 by way of PI3K/AKT/mTOR-dependent stabilization of MDM4 in hepatocellular carcinoma. Hepatology 2014, 59, 1886–1899. [Google Scholar] [CrossRef] [PubMed]
- Kaitsuka, T.; Tomizawa, K.; Matsushita, M. Transformation of eEF1Bδ into heat-shock response transcription factor by alternative splicing. EMBO Rep. 2011, 12, 673–681. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, Q.; Yao, D.; Wang, T.; Ni, M.; Xu, Y.; Tang, Z.; Liu, Z. Prenatal LPS Exposure Promotes Allergic Airway Inflammation via Long Coding RNA NONMMUT033452.2, and Protein Binding Partner, Eef1D. Am. J. Respir. Cell Mol. Biol. 2023, 68, 610–624. [Google Scholar] [CrossRef] [PubMed]
- Rawle, D.J.; Li, D.; Wu, Z.; Wang, L.; Choong, M.; Lor, M.; Reid, R.C.; Fairlie, D.P.; Harris, J.; Tachedjian, G.; et al. Oxazole-Benzenesulfonamide Derivatives Inhibit HIV-1 Reverse Transcriptase Interaction with Cellular eEF1A and Reduce Viral Replication. J. Virol. 2019, 93, e00239-19. [Google Scholar] [CrossRef]
- Yamaji, Y.; Kobayashi, T.; Hamada, K.; Sakurai, K.; Yoshii, A.; Suzuki, M.; Namba, S.; Hibi, T. In vivo interaction between Tobacco mosaic virus RNA-dependent RNA polymerase and host translation elongation factor 1A. Virology 2006, 347, 100–108. [Google Scholar] [CrossRef]
- Wei, T.; Li, D.; Marcial, D.; Khan, M.; Lin, M.H.; Snape, N.; Ghildyal, R.; Harrich, D.; Spann, K. The eukaryotic elongation factor 1A is critical for genome replication of the paramyxovirus respiratory syncytial virus. PLoS ONE 2014, 9, e114447. [Google Scholar] [CrossRef]
- Snape, N.; Li, D.; Wei, T.; Jin, H.; Lor, M.; Rawle, D.J.; Spann, K.M.; Harrich, D. The eukaryotic translation elongation factor 1A regulation of actin stress fibers is important for infectious RSV production. Virol. J. 2018, 15, 182. [Google Scholar] [CrossRef]
- Li, Z.; Pogany, J.; Panavas, T.; Xu, K.; Esposito, A.M.; Kinzy, T.G.; Nagy, P.D. Translation elongation factor 1A is a component of the tombusvirus replicase complex and affects the stability of the p33 replication co-factor. Virology 2009, 385, 245–260. [Google Scholar] [CrossRef]
- Li, Z.; Pogany, J.; Tupman, S.; Esposito, A.M.; Kinzy, T.G.; Nagy, P.D. Translation elongation factor 1A facilitates the assembly of the tombusvirus replicase and stimulates minus-strand synthesis. PLoS Pathog. 2010, 6, e1001175. [Google Scholar] [CrossRef]
- Blumenthal, T.; Landers, T.A.; Weber, K. Bacteriophage Q replicase contains the protein biosynthesis elongation factors EF Tu and EF Ts. Proc. Natl. Acad. Sci. USA 1972, 69, 1313–1317. [Google Scholar] [CrossRef]
- Li, D.; Wei, T.; Jin, H.; Rose, A.; Wang, R.; Lin, M.H.; Spann, K.; Harrich, D. Binding of the eukaryotic translation elongation factor 1A with the 5′UTR of HIV-1 genomic RNA is important for reverse transcription. Virol. J. 2015, 12, 118. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, J.L.; Brinton, M.A. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J. Virol. 1997, 71, 6433–6444. [Google Scholar] [CrossRef] [PubMed]
- Davis, W.G.; Blackwell, J.L.; Shi, P.Y.; Brinton, M.A. Interaction between the cellular protein eEF1A and the 3′-terminal stem-loop of West Nile virus genomic RNA facilitates viral minus-strand RNA synthesis. J. Virol. 2007, 81, 10172–10187. [Google Scholar] [CrossRef] [PubMed]
- Dreher, T.W.; Goodwin, J.B. Transfer RNA mimicry among tymoviral genomic RNAs ranges from highly efficient to vestigial. Nucleic Acids Res. 1998, 26, 4356–4364. [Google Scholar] [CrossRef]
- Joshi, R.L.; Ravel, J.M.; Haenni, A.L. Interaction of turnip yellow mosaic virus Val-RNA with eukaryotic elongation factor EF-1 [alpha]. Search for a function. Embo J. 1986, 5, 1143–1148. [Google Scholar] [CrossRef]
- Dreher, T.W.; Uhlenbeck, O.C.; Browning, K.S. Quantitative assessment of EF-1alpha.GTP binding to aminoacyl-tRNAs, aminoacyl-viral RNA, and tRNA shows close correspondence to the RNA binding properties of EF-Tu. J. Biol. Chem. 1999, 274, 666–672. [Google Scholar] [CrossRef]
- Matsuda, D.; Yoshinari, S.; Dreher, T.W. eEF1A binding to aminoacylated viral RNA represses minus strand synthesis by TYMV RNA-dependent RNA polymerase. Virology 2004, 321, 47–56. [Google Scholar] [CrossRef]
- Matsuda, D.; Dreher, T.W. The tRNA-like structure of Turnip yellow mosaic virus RNA is a 3′-translational enhancer. Virology 2004, 321, 36–46. [Google Scholar] [CrossRef]
- Li, S.; Feng, S.; Wang, J.H.; He, W.R.; Qin, H.Y.; Dong, H.; Li, L.F.; Yu, S.X.; Li, Y.; Qiu, H.J. eEF1A Interacts with the NS5A Protein and Inhibits the Growth of Classical Swine Fever Virus. Viruses 2015, 7, 4563–4581. [Google Scholar] [CrossRef]
- Fan, M. The Effect and Mechanism of Host Factor eEF1A1 on Influenza Virus Propagation. Master’s Thesis, Huazhong Agricultural University, Wuhan, China, 2017. [Google Scholar]
- Moerdyk-Schauwecker, M.; Hwang, S.I.; Grdzelishvili, V.Z. Analysis of virion associated host proteins in vesicular stomatitis virus using a proteomics approach. Virol. J. 2009, 6, 166. [Google Scholar] [CrossRef]
- Neuman, B.W.; Joseph, J.S.; Saikatendu, K.S.; Serrano, P.; Chatterjee, A.; Johnson, M.A.; Liao, L.; Klaus, J.P.; Yates, J.R., 3rd; Wüthrich, K.; et al. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 2008, 82, 5279–5294. [Google Scholar] [CrossRef] [PubMed]
- Cimarelli, A.; Luban, J. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 1999, 73, 5388–5401. [Google Scholar] [CrossRef]
- Chertova, E.; Chertov, O.; Coren, L.V.; Roser, J.D.; Trubey, C.M.; Bess, J.W., Jr.; Sowder, R.C., 2nd; Barsov, E.; Hood, B.L.; Fisher, R.J.; et al. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 2006, 80, 9039–9052. [Google Scholar] [CrossRef] [PubMed]
- Ott, D.E.; Coren, L.V.; Johnson, D.G.; Kane, B.P.; Sowder, R.C., 2nd; Kim, Y.D.; Fisher, R.J.; Zhou, X.Z.; Lu, K.P.; Henderson, L.E. Actin-binding cellular proteins inside human immunodeficiency virus type 1. Virology 2000, 266, 42–51. [Google Scholar] [CrossRef]
- Chung, C.S.; Chen, C.H.; Ho, M.Y.; Huang, C.Y.; Liao, C.L.; Chang, W. Vaccinia virus proteome: Identification of proteins in vaccinia virus intracellular mature virion particles. J. Virol. 2006, 80, 2127–2140. [Google Scholar] [CrossRef] [PubMed]
- Resch, W.; Hixson, K.K.; Moore, R.J.; Lipton, M.S.; Moss, B. Protein composition of the vaccinia virus mature virion. Virology 2007, 358, 233–247. [Google Scholar] [CrossRef]
- Kattenhorn, L.M.; Mills, R.; Wagner, M.; Lomsadze, A.; Makeev, V.; Borodovsky, M.; Ploegh, H.L.; Kessler, B.M. Identification of proteins associated with murine cytomegalovirus virions. J. Virol. 2004, 78, 11187–11197. [Google Scholar] [CrossRef]
- Varnum, S.M.; Streblow, D.N.; Monroe, M.E.; Smith, P.; Auberry, K.J.; Pasa-Tolic, L.; Wang, D.; Camp, D.G., 2nd; Rodland, K.; Wiley, S.; et al. Identification of proteins in human cytomegalovirus (HCMV) particles: The HCMV proteome. J. Virol. 2004, 78, 10960–10966. [Google Scholar] [CrossRef]
- Gan, H.; Zhou, X.; Lei, Q.; Wu, L.; Niu, J.; Zheng, Q. RNA-dependent RNA polymerase of SARS-CoV-2 regulate host mRNA translation efficiency by hijacking eEF1A factors. Biochim. Biophys. Acta Mol. Basis Dis. 2024, 1870, 166871. [Google Scholar] [CrossRef]
- Warren, K.; Wei, T.; Li, D.; Qin, F.; Warrilow, D.; Lin, M.H.; Sivakumaran, H.; Apolloni, A.; Abbott, C.M.; Jones, A.; et al. Eukaryotic elongation factor 1 complex subunits are critical HIV-1 reverse transcription cofactors. Proc. Natl. Acad. Sci. USA 2012, 109, 9587–9592. [Google Scholar] [CrossRef]
- Wang, Y.E.; Park, A.; Lake, M.; Pentecost, M.; Torres, B.; Yun, T.E.; Wolf, M.C.; Holbrook, M.R.; Freiberg, A.N.; Lee, B. Ubiquitin-regulated nuclear-cytoplasmic trafficking of the Nipah virus matrix protein is important for viral budding. PLoS Pathog. 2010, 6, e1001186. [Google Scholar] [CrossRef] [PubMed]
- Uchida, S.; Sato, H.; Yoneda, M.; Kai, C. Eukaryotic elongation factor 1-beta interacts with the 5′ untranslated region of the M gene of Nipah virus to promote mRNA translation. Arch. Virol. 2016, 161, 2361–2368. [Google Scholar] [CrossRef]
- Gao, Q.; Yang, C.; Ren, C.; Zhang, S.; Gao, X.; Jin, M.; Chen, H.; Ma, W.; Zhou, H. Eukaryotic Translation Elongation Factor 1 Delta Inhibits the Nuclear Import of the Nucleoprotein and PA-PB1 Heterodimer of Influenza A Virus. J. Virol. 2020, 95, e01391-20. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, Y.; Kato, K.; Tanaka, M.; Kanamori, M.; Nishiyama, Y.; Yamanashi, Y. Conserved protein kinases encoded by herpesviruses and cellular protein kinase cdc2 target the same phosphorylation site in eukaryotic elongation factor 1delta. J. Virol. 2003, 77, 2359–2368. [Google Scholar] [CrossRef]
- Kawaguchi, Y.; Matsumura, T.; Roizman, B.; Hirai, K. Cellular elongation factor 1delta is modified in cells infected with representative alpha-, beta-, or gammaherpesviruses. J. Virol. 1999, 73, 4456–4460. [Google Scholar] [CrossRef]
- Kawaguchi, Y.; Bruni, R.; Roizman, B. Interaction of herpes simplex virus 1 alpha regulatory protein ICP0 with elongation factor 1delta: ICP0 affects translational machinery. J. Virol. 1997, 71, 1019–1024. [Google Scholar] [CrossRef] [PubMed]
- Sammaibashi, S.; Yamayoshi, S.; Kawaoka, Y. Strain-Specific Contribution of Eukaryotic Elongation Factor 1 Gamma to the Translation of Influenza A Virus Proteins. Front. Microbiol. 2018, 9, 1446. [Google Scholar] [CrossRef]
- Watanabe, T.; Kawakami, E.; Shoemaker, J.E.; Lopes, T.J.; Matsuoka, Y.; Tomita, Y.; Kozuka-Hata, H.; Gorai, T.; Kuwahara, T.; Takeda, E.; et al. Influenza virus-host interactome screen as a platform for antiviral drug development. Cell Host Microbe 2014, 16, 795–805. [Google Scholar] [CrossRef]
- Zhang, Z.; Pan, L.; Ding, Y.; Lv, J.; Zhou, P.; Fang, Y.; Liu, X.; Zhang, Y.; Wang, Y. eEF1G interaction with foot-and-mouth disease virus nonstructural protein 2B: Identification by yeast two-hybrid system. Microb. Pathog. 2017, 112, 111–116. [Google Scholar] [CrossRef]
- Sasvari, Z.; Izotova, L.; Kinzy, T.G.; Nagy, P.D. Synergistic roles of eukaryotic translation elongation factors 1Bγ and 1A in stimulation of tombusvirus minus-strand synthesis. PLoS Pathog. 2011, 7, e1002438. [Google Scholar] [CrossRef]
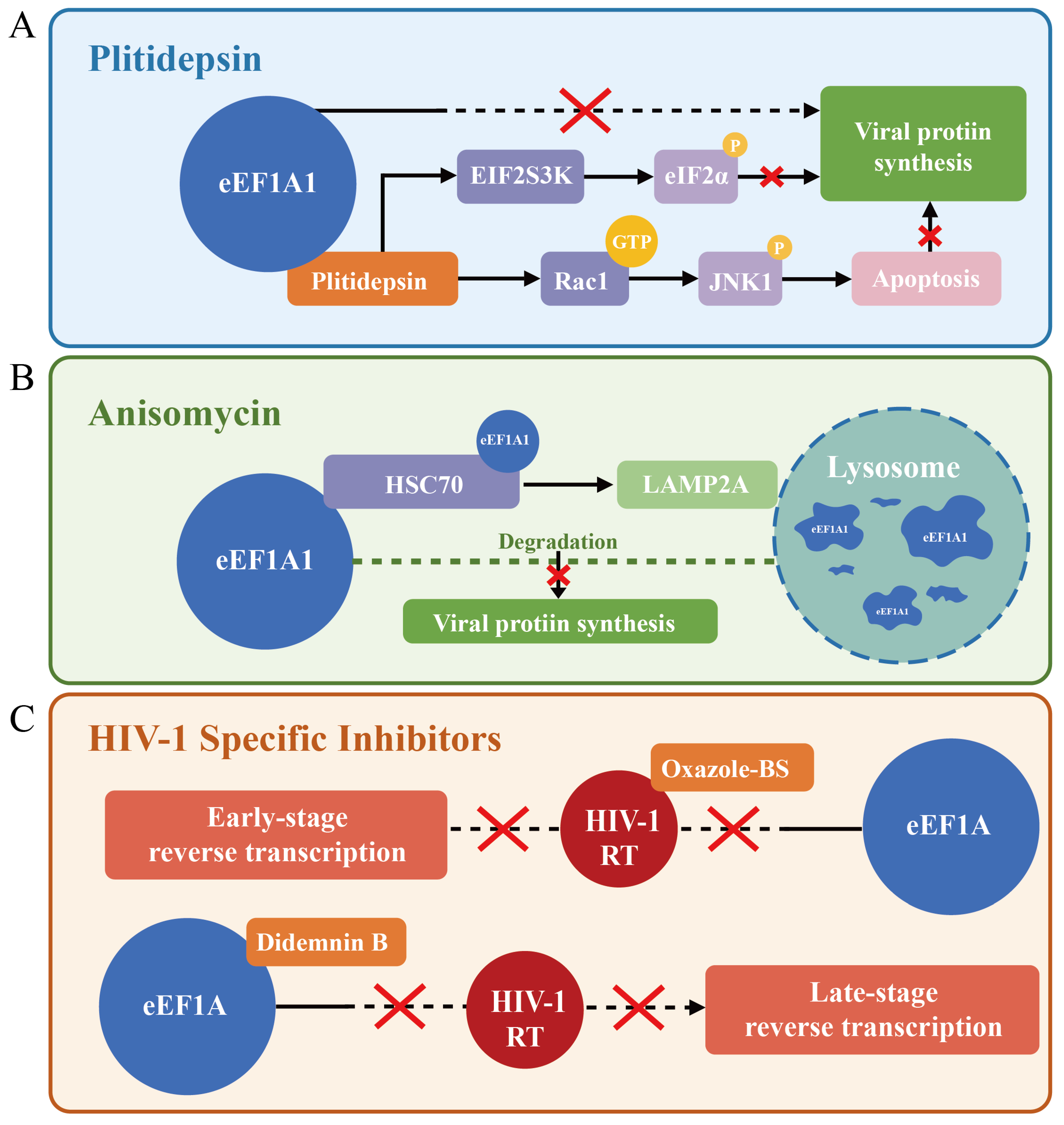
- Papapanou, M.; Papoutsi, E.; Giannakas, T.; Katsaounou, P. Plitidepsin: Mechanisms and Clinical Profile of a Promising Antiviral Agent against COVID-19. J. Pers. Med. 2021, 11, 668. [Google Scholar] [CrossRef]
- Losada, A.; Berlanga, J.J.; Molina-Guijarro, J.M.; Jiménez-Ruiz, A.; Gago, F.; Avilés, P.; de Haro, C.; Martínez-Leal, J.F. Generation of endoplasmic reticulum stress and inhibition of autophagy by plitidepsin induces proteotoxic apoptosis in cancer cells. Biochem. Pharmacol. 2020, 172, 113744. [Google Scholar] [CrossRef]
- Gordiyenko, Y.; Llácer, J.L.; Ramakrishnan, V. Structural basis for the inhibition of translation through eIF2α phosphorylation. Nat. Commun. 2019, 10, 2640. [Google Scholar] [CrossRef]
- Molina Molina, E.; Bech-Serra, J.J.; Franco-Trepat, E.; Jarne, I.; Perez-Zsolt, D.; Badia, R.; Riveira-Muñoz, E.; Garcia-Vidal, E.; Revilla, L.; Franco, S.; et al. Targeting eEF1A reprograms translation and uncovers broad-spectrum antivirals against cap or m(6)A protein synthesis routes. Nat. Commun. 2025, 16, 1087. [Google Scholar] [CrossRef]
- Brönstrup, M.; Sasse, F. Natural products targeting the elongation phase of eukaryotic protein biosynthesis. Nat. Prod. Rep. 2020, 37, 752–762. [Google Scholar] [CrossRef]
- Reina, J. Plitidepsin, an inhibitor of the cell elongation factor eEF1a, and molnupiravir an analogue of the ribonucleoside cytidine, two new chemical compounds with intense activity against SARS-CoV-2. Rev. Esp. Quimioter. 2021, 34, 402–407. [Google Scholar] [CrossRef]
- White, K.M.; Rosales, R.; Yildiz, S.; Kehrer, T.; Miorin, L.; Moreno, E.; Jangra, S.; Uccellini, M.B.; Rathnasinghe, R.; Coughlan, L.; et al. Plitidepsin has potent preclinical efficacy against SARS-CoV-2 by targeting the host protein eEF1A. Science 2021, 371, 926–931. [Google Scholar] [CrossRef]
- Shao, E.; Zhao, S.; Dong, Y.; Wang, Y.; Fei, Y.; Li, S.; Wang, L.; Bashir, T.; Luan, T.; Lin, L.; et al. Anisomycin inhibits Coxsackievirus B replication by promoting the lysosomal degradation of eEF1A1. Antivir. Res. 2023, 215, 105621. [Google Scholar] [CrossRef]
- Li, D.; Wei, T.; Rawle, D.J.; Qin, F.; Wang, R.; Soares, D.C.; Jin, H.; Sivakumaran, H.; Lin, M.H.; Spann, K.; et al. Specific Interaction between eEF1A and HIV RT Is Critical for HIV-1 Reverse Transcription and a Potential Anti-HIV Target. PLoS Pathog. 2015, 11, e1005289. [Google Scholar] [CrossRef]
- Mateos, M.V.; Prosper, F.; Martin Sánchez, J.; Ocio, E.M.; Oriol, A.; Motlló, C.; Michot, J.M.; Jarque, I.; Iglesias, R.; Solé, M.; et al. Phase I study of plitidepsin in combination with bortezomib and dexamethasone in patients with relapsed/refractory multiple myeloma. Cancer Med. 2023, 12, 3999–4009. [Google Scholar] [CrossRef]
- Mateos, M.V.; Cibeira, M.T.; Richardson, P.G.; Prosper, F.; Oriol, A.; de la Rubia, J.; Lahuerta, J.J.; García-Sanz, R.; Extremera, S.; Szyldergemajn, S.; et al. Phase II clinical and pharmacokinetic study of plitidepsin 3-hour infusion every two weeks alone or with dexamethasone in relapsed and refractory multiple myeloma. Clin. Cancer Res. 2010, 16, 3260–3269. [Google Scholar] [CrossRef] [PubMed]
- Spicka, I.; Ocio, E.M.; Oakervee, H.E.; Greil, R.; Banh, R.H.; Huang, S.Y.; D’Rozario, J.M.; Dimopoulos, M.A.; Martínez, S.; Extremera, S.; et al. Randomized phase III study (ADMYRE) of plitidepsin in combination with dexamethasone vs. dexamethasone alone in patients with relapsed/refractory multiple myeloma. Ann. Hematol. 2019, 98, 2139–2150. [Google Scholar] [CrossRef] [PubMed]
- Landete, P.; Caliman-Sturdza, O.A.; Lopez-Martin, J.A.; Preotescu, L.; Luca, M.C.; Kotanidou, A.; Villares, P.; Iglesias, S.P.; Guisado-Vasco, P.; Saiz-Lou, E.M.; et al. A Phase III Randomized Controlled Trial of Plitidepsin, a Marine-Derived Compound, in Hospitalized Adults with Moderate COVID-19. Clin. Infect. Dis. 2024, 79, 910–919. [Google Scholar] [CrossRef]

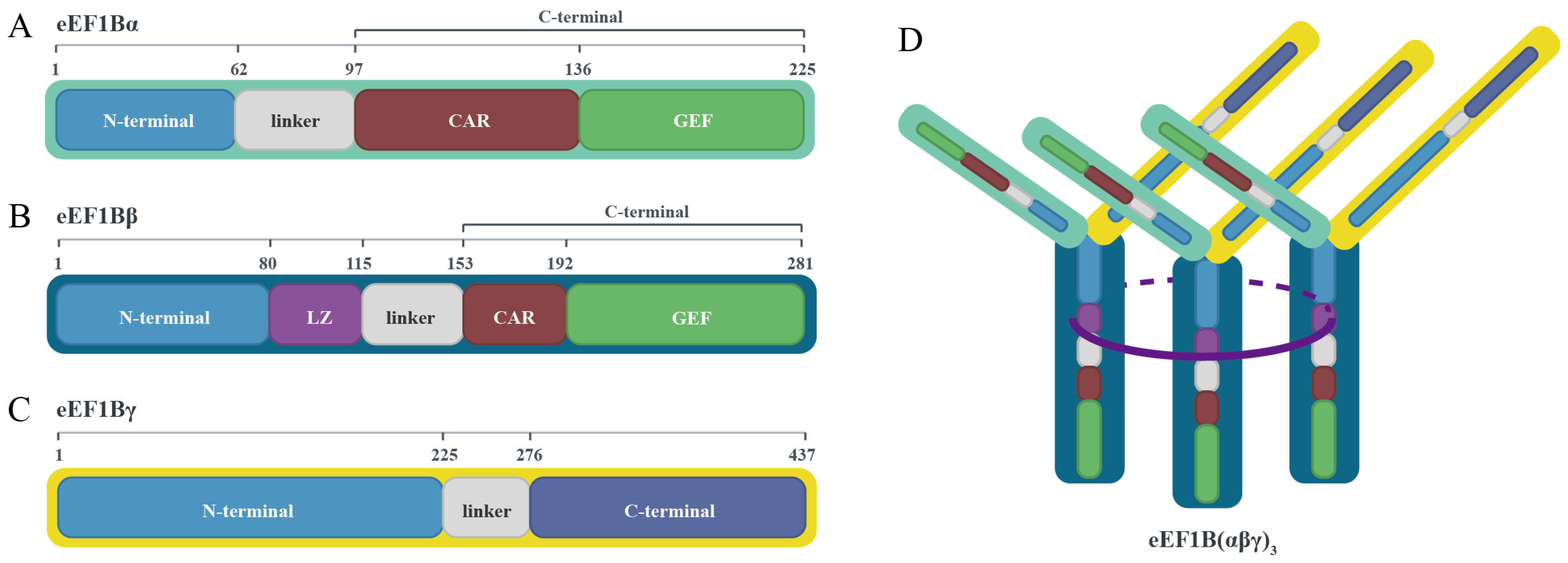

| Merrick’s Nomenclature [37] | Current Nomenclature [3,22,33,38,39] | Prior Nomenclature [24,25] |
|---|---|---|
| eEF1Bα | eEF1Bα/EEF1B2/eEF1B2 | eEF1β’(plant) |
| eEF1β (yeast and metazoan) | ||
| eEF1Bβ | eEF1Bβ (plant) eEF1Bδ (metazoan)/ EEF1D/eEF1D | eEF1β (plant) |
| eEF1δ (metazoan) | ||
| eEF1Bγ | eEF1Bγ/EEF1G/eEF1G | eEF1γ (yeast, plant and metazoan) |
| Virus | Effect Phase | Molecular Mechanism | References |
|---|---|---|---|
| TMV | Genome replication | Binds to RdRp, maintains polymerase catalytic activity | [65] |
| RSV | Genome replication; virus release | Directly interacts with nucleoprotein to stabilize replication complex; regulates actin stress fibers to promote viral budding and release | [66,67] |
| TBSV | Genome replication | Specifically binds to replication proteins p33 and p92pol, enhances stability of replication complexes, promotes efficient negative-strand RNA synthesis | [68,69] |
| Bacteriophage Qβ | Genome replication | Acts as an essential cofactor for phage RdRp complex, participates in the initiation phase of RNA synthesis | [70] |
| WNV | Genome replication | Specifically recognizes the 3’ terminal stem-loop structure (3’-SL) of viral genome to directly promote negative-strand RNA synthesis | [72,73] |
| TYMV | Genome replication | eEF1A·GTP binding to aminoacylated viral RNA inhibits negative-strand RNA synthesis; binding to 3’-TLS enhances translation efficiency | [74,75,76,77,78] |
| CSFV | Genome replication | Competitively occupies IRES to inhibit translation initiation efficiency; binds to NS5A protein interfering with replication complex assembly | [79] |
| AIV | Genome replication | Mediates binding to PB1/PB2 subunits via Ala206, disrupts PA–PB1 dimer stability, affects vRNP assembly | [80] |
| SARS-CoV-2 | Immune suppression | NSP12 hijacks eEF1A to regulate host mRNA translation efficiency, inhibits type I interferon production, promotes inflammatory cytokine expression | [90] |
| HIV-1 | Genome replication; assembly and release; immune suppression | Functionally binds to RT and 5’UTR, supporting viral reverse transcription; binds to Gag protein, enhancing viral particle formation efficiency; interacts with Nef protein inhibiting host cell apoptosis | [9,64,71,83,84,85,91] |
| Subunit | Virus | Effect Phase | Molecular Mechanism | References |
|---|---|---|---|---|
| eEF1B2 | NiV | Genome replication; VLP budding | Specifically binds to the nucleotides 81–100 region in the 5’UTR of M gene, enhances M protein mRNA translation efficiency and VLP budding | [93] |
| eEF1D | IAV | Genome replication | Interacts with IAV polymerase subunits (PB1, PB2, PA) and NP; inhibits NP interaction with importin α5 reducing nuclear import efficiency; weakens PB1 binding to RanBP5 hindering PA–PB1 heterodimer formation | [94] |
| eEF1D | HSV-1 | Genome replication | Viral kinase UL13 specifically phosphorylates Ser133 of eEF1D; ICP0 protein interacts with eEF1D | [95,96,97] |
| eEF1G | IAV | Genome replication | Interacts with IAV polymerase subunits (PB1, PB2, PA) and NP; significantly enhances translation efficiency of viral structural proteins (M1, NP) | [98,99] |
| eEF1G | FMDV | Genome replication; assembly and release | Interacts with non-structural protein 2B through C-terminal region (aa 208–437), drives cell membrane reorganization and vesicle formation | [100] |
| eEF1G | HIV-1 | Genome replication | Works synergistically with eEF1A on RT and IN, stabilizes the structure of RTC, ensures high efficiency of reverse transcription process | [64,91] |
| eEF1G | TBSV | Genome replication | Works synergistically with eEF1A by binding to viral RNA and regulating RdRp activity to jointly promote viral negative-strand RNA synthesis | [101] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Liu, K.; Wang, X.; Liu, X. Multidimensional Regulatory Mechanisms and Targeting Strategies of the eEF1 Family in RNA Virus Infection. Viruses 2025, 17, 682. https://doi.org/10.3390/v17050682
Wang X, Liu K, Wang X, Liu X. Multidimensional Regulatory Mechanisms and Targeting Strategies of the eEF1 Family in RNA Virus Infection. Viruses. 2025; 17(5):682. https://doi.org/10.3390/v17050682
Chicago/Turabian StyleWang, Xin, Kaituo Liu, Xiaoquan Wang, and Xiufan Liu. 2025. "Multidimensional Regulatory Mechanisms and Targeting Strategies of the eEF1 Family in RNA Virus Infection" Viruses 17, no. 5: 682. https://doi.org/10.3390/v17050682
APA StyleWang, X., Liu, K., Wang, X., & Liu, X. (2025). Multidimensional Regulatory Mechanisms and Targeting Strategies of the eEF1 Family in RNA Virus Infection. Viruses, 17(5), 682. https://doi.org/10.3390/v17050682






