Human T-Lymphotropic Virus (HTLV): Epidemiology, Genetic, Pathogenesis, and Future Challenges
Abstract
1. Introduction
2. Virological Characteristics of HTLVs and Infectious Cycle
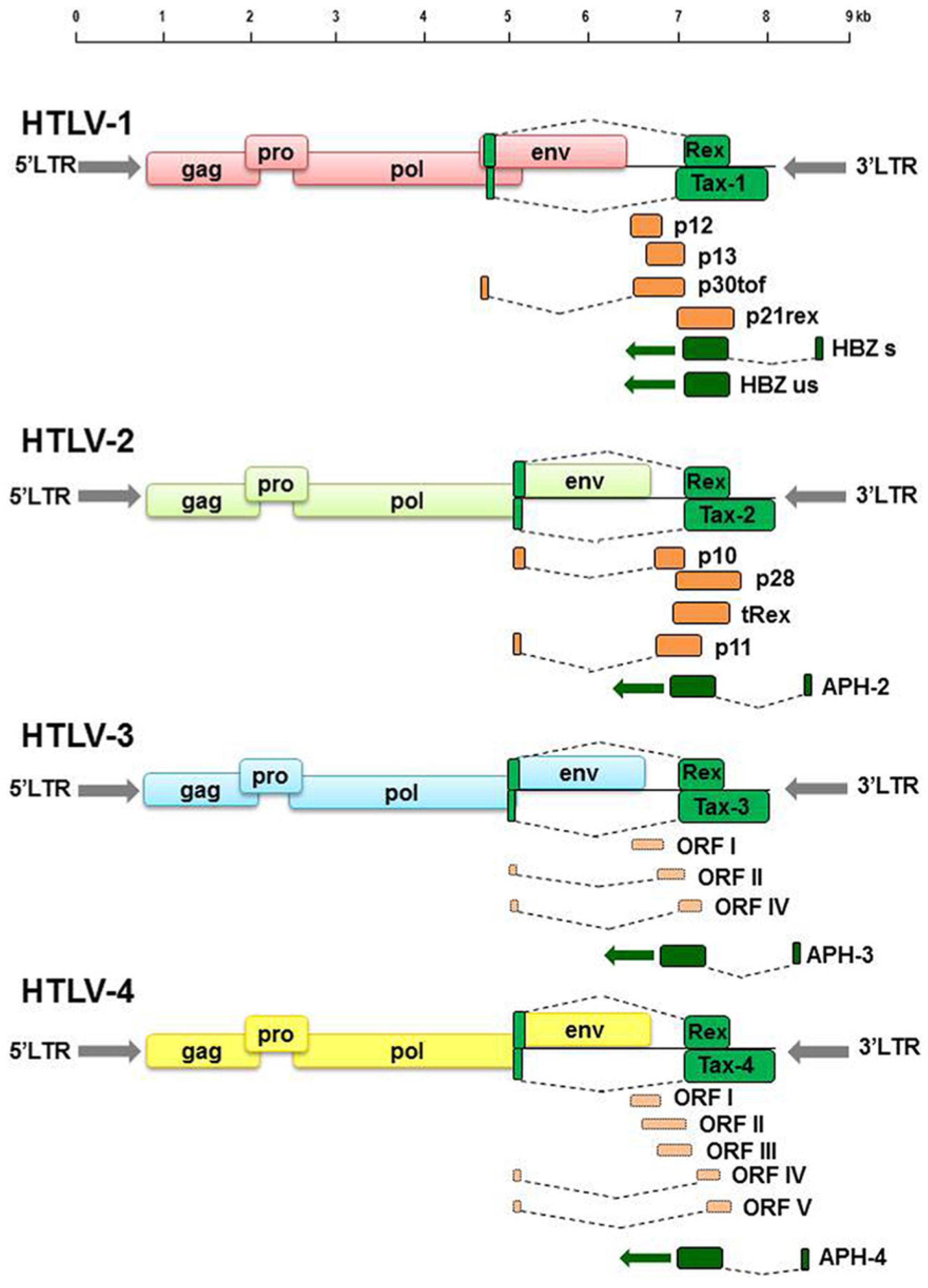
2.1. HTLV-1 and HTLV-2
2.2. HLTV-3 and HTLV-4
3. Pathogenesis and Pathophysiology
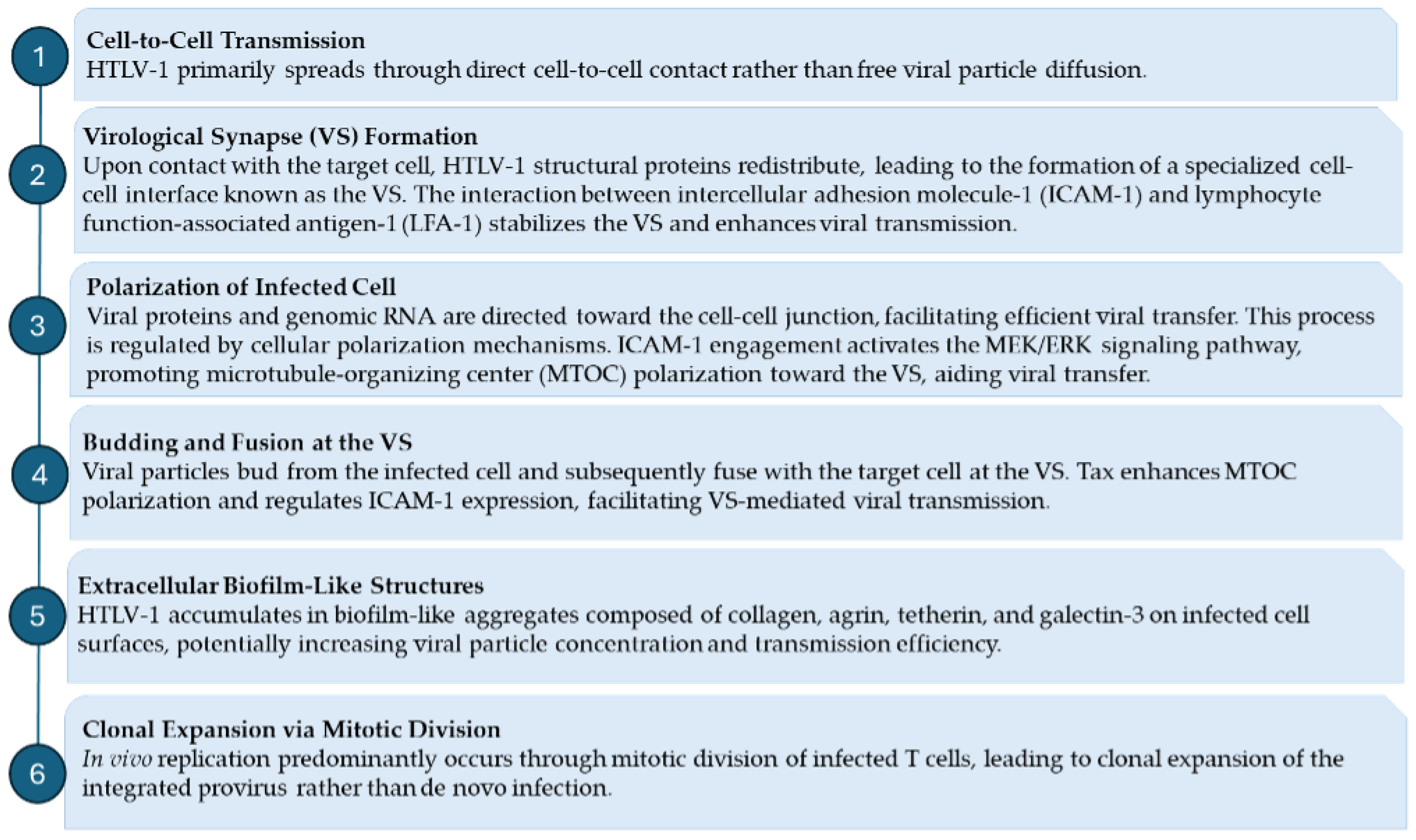
3.1. Mechanisms of HTLV-1 Infection and Persistence: Virological Synapse and Clonal Expansion
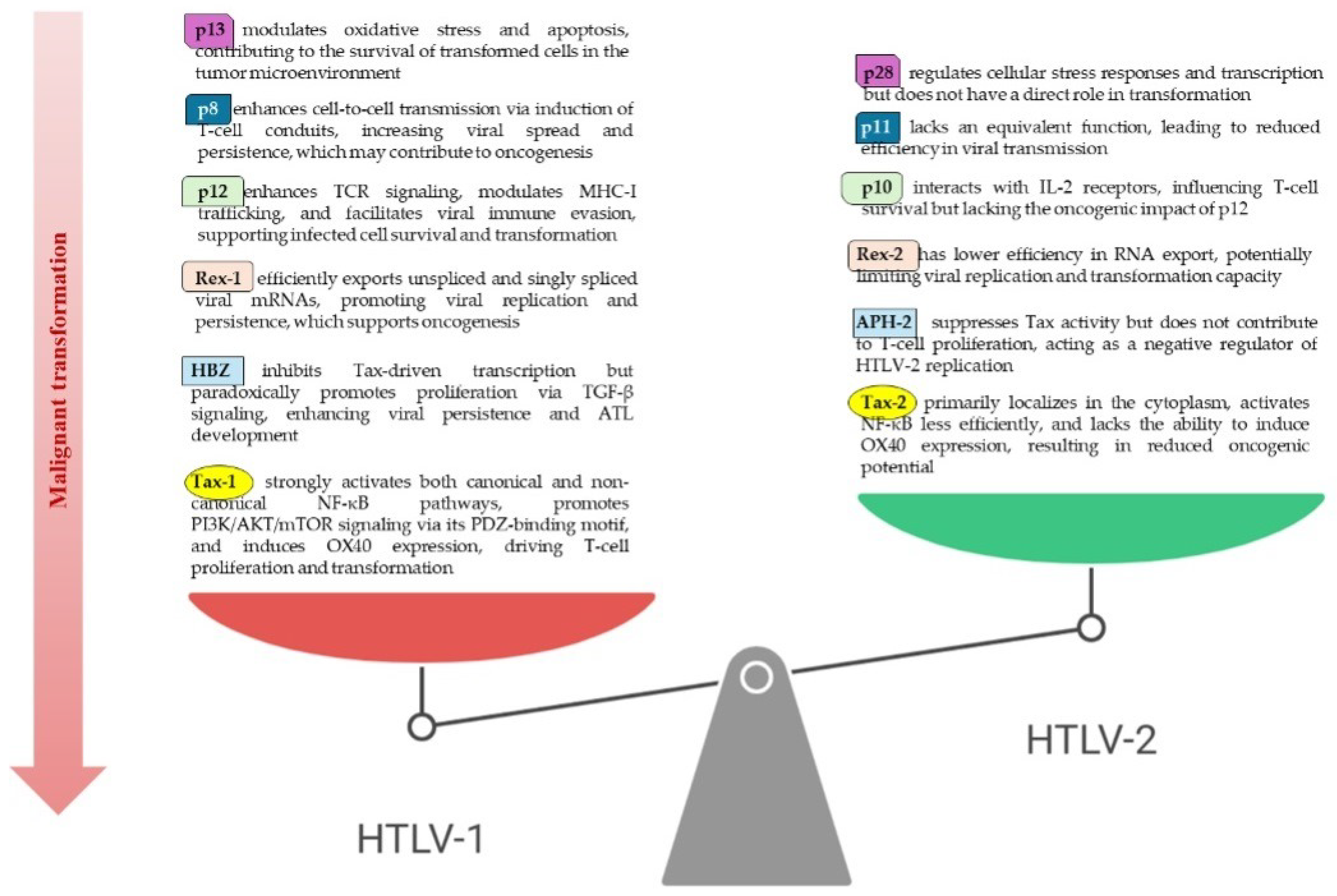
3.2. Differences in HTLV-1 and HTLV-2 Pathogenetic Profiles
4. Geographical Distribution and Epidemiological Insights of HTLV
Inter-Hosts Transmission Routes
5. Genetics and Genomics of HTLV
5.1. From Viral Architecture to Host Genome Impact
5.2. Beyond Viral Architecture: HTLV Integration as a Source of Genomic Instability
6. Diagnosis of HTLV Infections
Challenges and Future Directions in HTLV Diagnosis
7. Clinical Management of HTLV Infections
General Clinical Management Principles
8. Disease-Specific Management Strategies
8.1. Management of ATL
8.2. Management of HAM/TSP
8.3. Management of HTLV-1-Associated Inflammatory Disorders
- HTLV-1-associated uveitis: Chronic intraocular inflammation that may lead to vision impairment or blindness. Treatment includes topical, periocular, or systemic corticosteroids, with immunosuppressive agents (e.g., cyclosporine, methotrexate) reserved for refractory cases. Regular ophthalmologic evaluations are necessary to monitor disease activity and prevent complications [237,238,239,240].
8.4. Emerging Therapies and Future Directions
9. HIV/HTLV Coinfection: Implications for Disease Progression and Management
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Poiesz, B.J.; Ruscetti, F.W.; Gazdar, A.F.; Bunn, P.A.; Minna, J.D.; Gallo, R.C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA 1980, 77, 7415–7419. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, M.; Miyoshi, I.; Hinuma, Y. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc. Natl. Acad. Sci. USA 1982, 79, 2031–2035. [Google Scholar] [CrossRef]
- Calattini, S.; Chevalier, S.A.; Duprez, R.; Bassot, S.; Froment, A.; Mahieux, R.; Gessain, A. Discovery of a new human T-cell lymphotropic virus (HTLV-3) in Central Africa. Retrovirology 2005, 2, 30. [Google Scholar] [CrossRef]
- Mahieux, R.; Gessain, A. The human HTLV-3 and HTLV-4 retroviruses: New members of the HTLV family. Pathol. Biol. 2009, 57, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Legrand, N.; McGregor, S.; Bull, R.; Bajis, S.; Valencia, B.M.; Ronnachit, A.; Einsiedel, L.; Gessain, A.; Kaldor, J.; Martinello, M. Clinical and public health implications of human T-lymphotropic virus type 1 infection. Clin. Microbiol. Rev. 2022, 35, e00078-21. [Google Scholar] [CrossRef]
- Einsiedel, L.; Pham, H.; Talukder, M.R.; Taylor, K.; Wilson, K.; Kaldor, J.; Gessain, A.; Woodman, R. Very high prevalence of infection with the human T cell leukaemia virus type 1c in remote Australian Aboriginal communities: Results of a large cross-sectional community survey. PLoS Neglect. Trop. Dis. 2021, 15, e0009915. [Google Scholar] [CrossRef] [PubMed]
- Gessain, A.; Ramassamy, J.L.; Afonso, P.V.; Cassar, O. Geographic distribution, clinical epidemiology and genetic diversity of the human oncogenic retrovirus HTLV-1 in Africa, the world’s largest endemic area. Front. Immunol. 2023, 14, 1043600. [Google Scholar] [CrossRef]
- Sánchez-Núñez, J.P.; de Miguel-Balsa, E.; Soriano, V.; Lorenzo-Garrido, E.; Giménez-Richarte, A.; Otero-Rodriguez, S.; Celis-Salinas, J.C.; de Mendoza, C.; Casapia-Morales, M.; Ramos-Rincón, J.M. Prevalence of HTLV-1/2 infection in pregnant women in Central and South America and the Caribbean: A systematic review and meta-analysis. Int. J. Infect. Dis. 2024, 143, 107018. [Google Scholar] [CrossRef]
- Itabashi, K.; Miyazawa, T.; Uchimaru, K. How can we prevent mother-to-child transmission of HTLV-1? Int. J. Mol. Sci. 2023, 24, 6961. [Google Scholar] [CrossRef]
- Nunes, D.; Boa-Sorte, N.; Grassi, M.F.R.; Taylor, G.P.; Teixeira, M.G.; Barreto, M.L.; Dourado, I.; Galvão-Castro, B. HTLV-1 is predominantly sexually transmitted in Salvador, the city with the highest HTLV-1 prevalence in Brazil. PLoS ONE 2017, 12, e0171303. [Google Scholar] [CrossRef]
- Ramassamy, J.L.; Bilounga Ndongo, C.; Nnuka, P.; Antunes, M.; Le Mener, M.; Betsem a Betsem, E.; Njouom, R.; Cassar, O.; Fontanet, A.; Gessain, A. Epidemiological evidence of nosocomial and zoonotic transmission of human T-cell leukemia virus-1 in a large survey in a rural population of Central Africa. J. Infect. Dis. 2023, 227, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Bangham, C.R. Human T cell leukemia virus type 1: Persistence and pathogenesis. Annu. Rev. Immunol. 2018, 36, 43–71. [Google Scholar] [CrossRef] [PubMed]
- Igakura, T.; Stinchcombe, J.C.; Goon, P.K.; Taylor, G.P.; Weber, J.N.; Griffiths, G.M.; Tanaka, Y.; Osame, M.; Bangham, C.R. Spread of HTLV-I between lymphocytes by virus-induced polarization of the cytoskeleton. Science 2003, 299, 1713–1716. [Google Scholar] [CrossRef]
- Kalyanaraman, V.; Sarngadharan, M.; Robert-Guroff, M.; Miyoshi, I.; Blayney, D.; Golde, D.; Gallo, R.C. A new subtype of human T-cell leukemia virus (HTLV-II) associated with a T-cell variant of hairy cell leukemia. Science 1982, 218, 571–573. [Google Scholar] [CrossRef]
- Feuer, G.; Green, P.L. Comparative biology of human T-cell lymphotropic virus type 1 (HTLV-1) and HTLV-2. Oncogene 2005, 24, 5996–6004. [Google Scholar] [CrossRef]
- Hjelle, B.; Torrez-Martinez, N.; Mills, R.; Appenzeller, O.; Jahnke, R.; Alexander, S.; Ross, G. Chronic neurodegenerative disease associated with HTLV-II infection. Lancet 1992, 339, 645–646. [Google Scholar] [CrossRef]
- Marinho, T.A.; Okita, M.T.; Guimarães, R.A.; Zara, A.L.d.S.A.; Caetano, K.A.A.; Teles, S.A.; de Matos, M.A.D.; Carneiro, M.A.d.S.; Martins, R.M.B. The Global Prevalence of HTLV-1 and HTLV-2 Infections among Immigrants and Refugees—A Systematic Review and Meta-Analysis. Viruses 2024, 16, 1526. [Google Scholar] [CrossRef]
- Barbeau, B.; Peloponese, J.M.; Mesnard, J.M. Functional comparison of antisense proteins of HTLV-1 and HTLV-2 in viral pathogenesis. Front. Microbiol. 2013, 4, 226. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.P.; Al-Saleem, J.; Green, P.L. Comparative virology of HTLV-1 and HTLV-2. Retrovirology 2019, 16, 21. [Google Scholar] [CrossRef]
- Wolfe, N.D.; Heneine, W.; Carr, J.K.; Garcia, A.D.; Shanmugam, V.; Tamoufe, U.; Torimiro, J.N.; Prosser, A.T.; LeBreton, M.; Mpoudi-Ngole, E.; et al. Emergence of unique primate T-lymphotropic viruses among central African bushmeat hunters. Proc. Natl. Acad. Sci. USA 2005, 102, 7994–7999. [Google Scholar] [CrossRef]
- Calattini, S.; Chevalier, S.A.; Duprez, R.; Afonso, P.; Froment, A.; Gessain, A.; Mahieux, R. Human T-cell lymphotropic virus type 3: Complete nucleotide sequence and characterization of the human tax3 protein. J. Virol. 2006, 80, 9876–9888. [Google Scholar] [CrossRef] [PubMed]
- Duong, Y.T.; Jia, H.; Lust, J.A.; Garcia, A.D.; Tiffany, A.J.; Heneine, W.; Switzer, W.M. Absence of evidence of HTLV-3 and HTLV-4 in patients with large granular lymphocyte (LGL) leukemia. AIDS Res. Hum. Retroviruses 2008, 24, 1503–1505. [Google Scholar] [CrossRef]
- Thomas, A.; Perzova, R.; Abbott, L.; Benz, P.; Poiesz, M.J.; Dube, S.; Loughran, T.; Ferrer, J.; Sheremata, W.; Glaser, J.; et al. LGL leukemia and HTLV. AIDS Res. Hum. Retroviruses 2010, 26, 33–40. [Google Scholar] [CrossRef]
- Chevalier, S.A.; Durand, S.; Dasgupta, A.; Radonovich, M.; Cimarelli, A.; Brady, J.N.; Mahieux, R.; Pise-Masison, C.A. The transcription profile of Tax-3 is more similar to Tax-1 than Tax-2: Insights into HTLV-3 potential leukemogenic properties. PLoS ONE 2012, 7, e41003. [Google Scholar] [CrossRef]
- O’Donnell, J.S.; Jaberolansar, N.; Chappell, K.J. Human T-lymphotropic virus type 1 and antiretroviral therapy: Practical considerations for pre-exposure and post-exposure prophylaxis, transmission prevention, and mitigation of severe disease. Lancet Microbe 2024, 5, e400–e408. [Google Scholar] [CrossRef]
- Boostani, R.; Sadeghi, R.; Sabouri, A.; Ghabeli-Juibary, A. Human T-lymphotropic virus type I and breastfeeding; systematic review and meta-analysis of the literature. Iran. J. Neurol. 2018, 17, 174. [Google Scholar] [CrossRef] [PubMed]
- Roucoux, D.F.; Wang, B.; Smith, D.; Nass, C.C.; Smith, J.; Hutching, S.T.; Newman, B.; Lee, T.H.; Chafets, D.M.; Investigators, H.O.S. A prospective study of sexual transmission of human T lymphotropic virus (HTLV)–I and HTLV-II. J. Infect. Dis. 2005, 191, 1490–1497. [Google Scholar] [CrossRef]
- Hewitt, P.E.; Davison, K.; Howell, D.R.; Taylor, G.P. Human T-lymphotropic virus lookback in NHS B lood and T ransplant (E ngland) reveals the efficacy of leukoreduction. Transfusion 2013, 53, 2168–2175. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Public Health Impact and Implications for Future Actions: WHO Global Consultation on the Human T-Lymphotropic Virus Type 1, Tokyo, Japan, 13–15 November 2019; World Health Organization: Geneva, Switzerland, 2021.
- Hirons, A.; Khoury, G.; Purcell, D.F. Human T-cell lymphotropic virus type-1: A lifelong persistent infection, yet never truly silent. Lancet Infect. Dis. 2021, 21, e2–e10. [Google Scholar] [CrossRef]
- Kamoi, K.; Horiguchi, N.; Kurozumi-Karube, H.; Hamaguchi, I.; Yamano, Y.; Uchimaru, K.; Tojo, A.; Watanabe, T.; Ohno-Matsui, K. Horizontal transmission of HTLV-1 causing uveitis. Lancet Infect. Dis. 2021, 21, 578. [Google Scholar] [CrossRef]
- Yamamoto, N.; Matsumoto, T.; Koyanagi, Y.; Tanaka, Y.; Hinuma, Y. Unique cell lines harbouring both Epstein–Barr virus and adult T-cell leukaemia virus, established from leukaemia patients. Nature 1982, 299, 367–369. [Google Scholar] [CrossRef] [PubMed]
- Ho, D.D.; Rota, T.R.; Hirsch, M.S. Infection of human endothelial cells by human T-lymphotropic virus type I. Proc. Natl. Acad. Sci. USA 1984, 81, 7588–7590. [Google Scholar] [CrossRef] [PubMed]
- Longo, D.L.; Gelmann, E.P.; Cossman, J.; Young, R.A.; Gallo, R.C.; O’Brien, S.J.; Matis, L.A. Isolation of HTLV-transformed B-lymphocyte clone from a patient with HTLV-associated adult T-cell leukaemia. Nature 1984, 310, 505–506. [Google Scholar] [CrossRef] [PubMed]
- Ijichi, S.; Ramundo, M.; Takahashi, H.; Hall, W. In vivo cellular tropism of human T cell leukemia virus type II (HTLV-II). J. Exp. Med. 1992, 176, 293–296. [Google Scholar] [CrossRef]
- Rosenberg, A.R.; Delamarre, L.; Preira, A.; Dokhélar, M.C. Analysis of functional conservation in the surface and transmembrane glycoprotein subunits of human T-cell leukemia virus type 1 (HTLV-1) and HTLV-2. J. Virol. 1998, 72, 7609–7614. [Google Scholar] [CrossRef]
- Manel, N.; Kim, F.J.; Kinet, S.; Taylor, N.; Sitbon, M.; Battini, J.L. The ubiquitous glucose transporter GLUT-1 is a receptor for HTLV. Cell 2003, 115, 449–459. [Google Scholar] [CrossRef]
- Pinon, J.D.; Klasse, P.; Jassal, S.R.; Welson, S.; Weber, J.; Brighty, D.W.; Sattentau, Q.J. Human T-cell leukemia virus type 1 envelope glycoprotein gp46 interacts with cell surface heparan sulfate proteoglycans. J. Virol. 2003, 77, 9922–9930. [Google Scholar] [CrossRef]
- Jones, K.S.; Fugo, K.; Petrow-Sadowski, C.; Huang, Y.; Bertolette, D.C.; Lisinski, I.; Cushman, S.W.; Jacobson, S.; Ruscetti, F.W. Human T-cell leukemia virus type 1 (HTLV-1) and HTLV-2 use different receptor complexes to enter T cells. J. Virol. 2006, 80, 8291–8302. [Google Scholar] [CrossRef]
- Temin, H.M.; Mizutami, S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature 1970, 226, 1211–1213. [Google Scholar] [CrossRef]
- Baltimore, D. Viral RNA-dependent DNA polymerase: RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature 1970, 226, 1209–1211. [Google Scholar] [CrossRef]
- Kitamura, Y.; Lee, Y.; Coffin, J.M. Nonrandom integration of retroviral DNA in vitro: Effect of CpG methylation. Proc. Natl. Acad. Sci. USA 1992, 89, 5532–5536. [Google Scholar] [CrossRef]
- Doi, K.; Wu, X.; Taniguchi, Y.; Yasunaga, J.i.; Satou, Y.; Okayama, A.; Nosaka, K.; Matsuoka, M. Preferential selection of human T-cell leukemia virus type I provirus integration sites in leukemic versus carrier states. Blood 2005, 106, 1048–1053. [Google Scholar] [CrossRef]
- Holman, A.G.; Coffin, J.M. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proc. Natl. Acad. Sci. USA 2005, 102, 6103–6107. [Google Scholar] [CrossRef]
- Derse, D.; Crise, B.; Li, Y.; Princler, G.; Lum, N.; Stewart, C.; McGrath, C.F.; Hughes, S.H.; Munroe, D.J.; Wu, X. Human T-cell leukemia virus type 1 integration target sites in the human genome: Comparison with those of other retroviruses. J. Virol. 2007, 81, 6731–6741. [Google Scholar] [CrossRef] [PubMed]
- Niederer, H.A.; Laydon, D.J.; Melamed, A.; Elemans, M.; Asquith, B.; Matsuoka, M.; Bangham, C.R. HTLV-1 proviral integration sites differ between asymptomatic carriers and patients with HAM/TSP. Virol. J. 2014, 11, 172. [Google Scholar] [CrossRef] [PubMed]
- Kashanchi, F.; Brady, J.N. Transcriptional and post-transcriptional gene regulation of HTLV-1. Oncogene 2005, 24, 5938–5951. [Google Scholar] [CrossRef] [PubMed]
- Baranger, A.M.; Palmer, C.R.; Hamm, M.K.; Giebler, H.A.; Brauweiler, A.; Nyborg, J.K.; Schepartz, A. Mechanism of DNA-binding enhancement by the human T-cell leukaemia virus transactivator Tax. Nature 1995, 376, 606–608. [Google Scholar] [CrossRef]
- Hidaka, M.; Inoue, J.; Yoshida, M.; Seiki, M. Post-transcriptional regulator (rex) of HTLV-1 initiates expression of viral structural proteins but suppresses expression of regulatory proteins. EMBO J. 1988, 7, 519–523. [Google Scholar] [CrossRef]
- Butsch, M.; Boris-Lawrie, K. Destiny of unspliced retroviral RNA: Ribosome and/or virion? J. Virol. 2002, 76, 3089–3094. [Google Scholar] [CrossRef]
- Fogarty, K.H.; Chen, Y.; Grigsby, I.F.; Macdonald, P.J.; Smith, E.M.; Johnson, J.L.; Rawson, J.M.; Mansky, L.M.; Mueller, J.D. Characterization of cytoplasmic Gag-gag interactions by dual-color z-scan fluorescence fluctuation spectroscopy. Biophys. J. 2011, 100, 1587–1595. [Google Scholar] [CrossRef]
- Konvalinka, J.; Kräusslich, H.G.; Müller, B. Retroviral proteases and their roles in virion maturation. Virology 2015, 479, 403–417. [Google Scholar] [CrossRef]
- Panfil, A.R.; Dissinger, N.J.; Howard, C.M.; Murphy, B.M.; Landes, K.; Fernandez, S.A.; Green, P.L. Functional comparison of HBZ and the related APH-2 protein provides insight into human T-cell leukemia virus type 1 pathogenesis. J. Virol. 2016, 90, 3760–3772. [Google Scholar] [CrossRef] [PubMed]
- Panfil, A.R.; Al-Saleem, J.; Howard, C.M.; Shkriabai, N.; Kvaratskhelia, M.; Green, P.L. Stability of the HTLV-1 antisense-derived protein, HBZ, is regulated by the E3 ubiquitin-protein ligase, UBR5. Front. Microbiol. 2018, 9, 80. [Google Scholar] [CrossRef] [PubMed]
- Mahieux, R.; Gessain, A. HTLV-3/STLV-3 and HTLV-4 viruses: Discovery, epidemiology, serology and molecular aspects. Viruses 2011, 3, 1074–1090. [Google Scholar] [CrossRef] [PubMed]
- Gessain, A.; Rua, R.; Betsem, E.; Turpin, J.; Mahieux, R. HTLV-3/4 and simian foamy retroviruses in humans: Discovery, epidemiology, cross-species transmission and molecular virology. Virology 2013, 435, 187–199. [Google Scholar] [CrossRef]
- Romanelli, M.G.; Diani, E.; Bergamo, E.; Casoli, C.; Ciminale, V.; Bex, F.; Bertazzoni, U. Highlights on distinctive structural and functional properties of HTLV Tax proteins. Front. Microbiol. 2013, 4, 271. [Google Scholar] [CrossRef]
- Zuo, X.; Zhou, R.; Yang, S.; Ma, G. HTLV-1 persistent infection and ATLL oncogenesis. J. Med. Virol. 2023, 95, e28424. [Google Scholar] [CrossRef]
- Chiale, C.; Marchese, A.M.; Robek, M.D. Innate immunity and HBV persistence. Curr. Opin. Virol. 2021, 49, 13–20. [Google Scholar] [CrossRef]
- Pavia, G.; Quirino, A.; Marascio, N.; Veneziano, C.; Longhini, F.; Bruni, A.; Garofalo, E.; Pantanella, M.; Manno, M.; Gigliotti, S.; et al. Persistence of SARS-CoV-2 infection and viral intra-and inter-host evolution in COVID-19 hospitalized patients. J. Med. Virol. 2024, 96, e29708. [Google Scholar] [CrossRef]
- Marascio, N.; Mazzitelli, M.; Pavia, G.; Giancotti, A.; Barreca, G.S.; Costa, C.; Pisani, V.; Greco, G.; Serapide, F.; Trecarichi, E.M.; et al. Clinical, virological characteristics, and outcomes of treatment with sofosbuvir/ledipasvir in two pediatric patients infected by HCV genotype 4. Cells 2019, 8, 416. [Google Scholar] [CrossRef]
- Marascio, N.; Pavia, G.; Romeo, I.; Talarico, C.; Di Salvo, S.; Reale, M.; Marano, V.; Barreca, G.S.; Fabiani, F.; Perrotti, N.; et al. Real-life 3D therapy failure: Analysis of NS5A 93H RAS plus 108 K polymorphism in complex with ombitasvir by molecular modeling. J. Med. Virol. 2018, 90, 1257–1263. [Google Scholar] [CrossRef]
- Marascio, N.; Pavia, G.; Strazzulla, A.; Dierckx, T.; Cuypers, L.; Vrancken, B.; Barreca, G.S.; Mirante, T.; Malanga, D.; Oliveira, D.M.; et al. Detection of natural resistance-associated substitutions by ion semiconductor technology in HCV1b positive, direct-acting antiviral agents-naïve patients. Int. J. Mol. Sci. 2016, 17, 1416. [Google Scholar] [CrossRef] [PubMed]
- Romeo, I.; Marascio, N.; Pavia, G.; Talarico, C.; Costa, G.; Alcaro, S.; Artese, A.; Torti, C.; Liberto, M.C.; Focà, A. Structural Modeling of New Polymorphism Clusters of HCV Polymerase Isolated from Direct-Acting Antiviral Naïve Patients: Focus on Dasabuvir and Setrobuvir Binding Affinity. ChemistrySelect 2018, 3, 6009–6017. [Google Scholar] [CrossRef]
- Lairmore, M.D.; Haines, R.; Anupam, R. Mechanisms of human T-lymphotropic virus type 1 transmission and disease. Curr. Opin. Virol. 2012, 2, 474–481. [Google Scholar] [CrossRef] [PubMed]
- Forlani, G.; Shallak, M.; Accolla, R.S.; Romanelli, M.G. HTLV-1 infection and pathogenesis: New insights from cellular and animal models. Int. J. Mol. Sci. 2021, 22, 8001. [Google Scholar] [CrossRef] [PubMed]
- Gross, C.; Thoma-Kress, A.K. Molecular mechanisms of HTLV-1 cell-to-cell transmission. Viruses 2016, 8, 74. [Google Scholar] [CrossRef]
- Fan, N.; Gavalchin, J.; Paul, B.; Wells, K.; Lane, M.; Poiesz, B. Infection of peripheral blood mononuclear cells and cell lines by cell-free human T-cell lymphoma/leukemia virus type I. J. Clin. Microbiol. 1992, 30, 905–910. [Google Scholar] [CrossRef]
- Majorovits, E.; Nejmeddine, M.; Tanaka, Y.; Taylor, G.P.; Fuller, S.D.; Bangham, C.R. Human T-lymphotropic virus-1 visualized at the virological synapse by electron tomography. PLoS ONE 2008, 3, e2251. [Google Scholar] [CrossRef]
- Nejmeddine, M.; Negi, V.S.; Mukherjee, S.; Tanaka, Y.; Orth, K.; Taylor, G.P.; Bangham, C.R. HTLV-1–Tax and ICAM-1 act on T-cell signal pathways to polarize the microtubule-organizing center at the virological synapse. Blood J. Am. Soc. Hematol. 2009, 114, 1016–1025. [Google Scholar] [CrossRef]
- Qualley, D.F.; Stewart-Maynard, K.M.; Wang, F.; Mitra, M.; Gorelick, R.J.; Rouzina, I.; Williams, M.C.; Musier-Forsyth, K. C-terminal domain modulates the nucleic acid chaperone activity of human T-cell leukemia virus type 1 nucleocapsid protein via an electrostatic mechanism. J. Biol. Chem. 2010, 285, 295–307. [Google Scholar] [CrossRef]
- Nejmeddine, M.; Barnard, A.L.; Tanaka, Y.; Taylor, G.P.; Bangham, C.R. Human T-lymphotropic virus, type 1, tax protein triggers microtubule reorientation in the virological synapse. J. Biol. Chem. 2005, 280, 29653–29660. [Google Scholar] [CrossRef]
- Fukudome, K.; Furuse, M.; Fukuhara, N.; Orita, S.; Imai, T.; Takagi, S.; Nagira, M.; Hinuma, Y.; Yoshie, O. Strong induction of ICAM-1 in human T cells transformed by human T-cell-leukemia virus type 1 and depression of ICAM-1 or LFA-1 in adult T-cell-leukemia-derived cell lines. Int. J. Cancer 1992, 52, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, S.A.; Turpin, J.; Cachat, A.; Afonso, P.V.; Gessain, A.; Brady, J.N.; Pise-Masison, C.A.; Mahieux, R. Gem-induced cytoskeleton remodeling increases cellular migration of HTLV-1-infected cells, formation of infected-to-target T-cell conjugates and viral transmission. PLoS Pathog. 2014, 10, e1003917. [Google Scholar] [CrossRef] [PubMed]
- Pais-Correia, A.M.; Sachse, M.; Guadagnini, S.; Robbiati, V.; Lasserre, R.; Gessain, A.; Gout, O.; Alcover, A.; Thoulouze, M.I. Biofilm-like extracellular viral assemblies mediate HTLV-1 cell-to-cell transmission at virological synapses. Nat. Med. 2010, 16, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Gessain, A.; Gallo, R.C.; Franchini, G. Low degree of human T-cell leukemia/lymphoma virus type I genetic drift in vivo as a means of monitoring viral transmission and movement of ancient human populations. J. Virol. 1992, 66, 2288–2295. [Google Scholar] [CrossRef]
- Van Dooren, S.; Pybus, O.G.; Salemi, M.; Liu, H.F.; Goubau, P.; Remondegui, C.; Talarmin, A.; Gotuzzo, E.; Alcantara, L.C.J.; Galvão-Castro, B.; et al. The low evolutionary rate of human T-cell lymphotropic virus type-1 confirmed by analysis of vertical transmission chains. Mol. Biol. Evol. 2004, 21, 603–611. [Google Scholar] [CrossRef]
- Ohshima, K.; Ohgami, A.; Matsuoka, M.; Etoh, K.; Utsunomiya, A.; Makino, T.; Ishiguro, M.; Suzumiya, J.; Kikuchi, M. Random integration of HTLV-I provirus; increasing chromosomal instability. Cancer Lett. 1998, 132, 203–212. [Google Scholar] [CrossRef]
- Wattel, E.; Vartanian, J.P.; Pannetier, C.; Wain-Hobson, S. Clonal expansion of human T-cell leukemia virus type I-infected cells in asymptomatic and symptomatic carriers without malignancy. J. Virol. 1995, 69, 2863–2868. [Google Scholar] [CrossRef]
- Etoh, K.i.; Tamiya, S.; Yamaguchi, K.; Okayama, A.; Tsubouchi, H.; Ideta, T.; Mueller, N.; Takatsuki, K.; Matsuoka, M. Persistent clonal proliferation of human T-lymphotropic virus type I-infected cells in vivo. Cancer Res. 1997, 57, 4862–4867. [Google Scholar]
- Shirinian, M.; Kfoury, Y.; Dassouki, Z.; El-Hajj, H.; Bazarbachi, A. Tax-1 and Tax-2 similarities and differences: Focus on post-translational modifications and NF-κB activation. Front. Microbiol. 2013, 4, 231. [Google Scholar] [CrossRef]
- Jin, D.Y.; Jeang, K.T. HTLV-I Tax self-association in optimal trans-activation function. Nucleic Acids Res. 1997, 25, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Boxus, M.; Twizere, J.C.; Legros, S.; Dewulf, J.F.; Kettmann, R.; Willems, L. The HTLV-1 tax interactome. Retrovirology 2008, 5, 76. [Google Scholar] [CrossRef]
- Cherian, M.A.; Baydoun, H.H.; Al-Saleem, J.; Shkriabai, N.; Kvaratskhelia, M.; Green, P.; Ratner, L. Akt pathway activation by human T-cell leukemia virus type 1 Tax oncoprotein. J. Biol. Chem. 2015, 290, 26270–26281. [Google Scholar] [CrossRef] [PubMed]
- Meertens, L.; Chevalier, S.; Weil, R.; Gessain, A.; Mahieux, R. A 10-amino acid domain within human T-cell leukemia virus type 1 and type 2 tax protein sequences is responsible for their divergent subcellular distribution. J. Biol. Chem. 2004, 279, 43307–43320. [Google Scholar] [CrossRef]
- Journo, C.; Filipe, J.; About, F.; Chevalier, S.A.; Afonso, P.V.; Brady, J.N.; Flynn, D.; Tangy, F.; Israel, A.; Vidalain, P.O.; et al. NRP/optineurin cooperates with TAX1BP1 to potentiate the activation of NF-κB by human T-lymphotropic virus type 1 tax protein. PLoS Pathog. 2009, 5, e1000521. [Google Scholar] [CrossRef]
- Journo, C.; Bonnet, A.; Favre-Bonvin, A.; Turpin, J.; Vinera, J.; Côté, E.; Chevalier, S.A.; Kfoury, Y.; Bazarbachi, A.; Pique, C.; et al. Human T cell leukemia virus type 2 tax-mediated NF-κB activation involves a mechanism independent of Tax conjugation to ubiquitin and SUMO. J. Virol. 2013, 87, 1123–1136. [Google Scholar] [CrossRef]
- Shibata, Y.; Tokunaga, F.; Goto, E.; Komatsu, G.; Gohda, J.; Saeki, Y.; Tanaka, K.; Takahashi, H.; Sawasaki, T.; Inoue, S.; et al. HTLV-1 tax induces formation of the active macromolecular IKK complex by generating Lys63-and Met1-linked hybrid polyubiquitin chains. PLoS Pathog. 2017, 13, e1006162. [Google Scholar] [CrossRef]
- Motai, Y.; Takahashi, M.; Takachi, T.; Higuchi, M.; Hara, T.; Mizuguchi, M.; Aoyagi, Y.; Terai, S.; Tanaka, Y.; Fujii, M. Human T-cell leukemia virus type 1 (HTLV-1) Tax1 oncoprotein but not HTLV-2 Tax2 induces the expression of OX40 ligand by interacting with p52/p100 and RelB. Virus Genes 2016, 52, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Turci, M.; Lodewick, J.; Di Gennaro, G.; Rinaldi, A.S.; Marin, O.; Diani, E.; Sampaio, C.; Bex, F.; Bertazzoni, U.; Romanelli, M.G. Ubiquitination and sumoylation of the HTLV-2 Tax-2B protein regulate its NF-κB activity: A comparative study with the HTLV-1 Tax-1 protein. Retrovirology 2012, 9, 102. [Google Scholar] [CrossRef]
- Ariumi, Y.; Kaida, A.; Lin, J.Y.; Hirota, M.; Masui, O.; Yamaoka, S.; Taya, Y.; Shimotohno, K. HTLV-1 tax oncoprotein represses the p53-mediated trans-activation function through coactivator CBP sequestration. Oncogene 2000, 19, 1491–1499. [Google Scholar] [CrossRef]
- Nicot, C. HTLV-I Tax-mediated inactivation of cell cycle checkpoints and DNA repair pathways contribute to cellular transformation: “A random mutagenesis model”. J. Cancer Sci. 2015, 2. [Google Scholar] [CrossRef]
- Gaudray, G.; Gachon, F.; Basbous, J.; Biard-Piechaczyk, M.; Devaux, C.; Mesnard, J.M. The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription. J. Virol. 2002, 76, 12813–12822. [Google Scholar] [CrossRef]
- Halin, M.; Douceron, E.; Clerc, I.; Journo, C.; Ko, N.L.; Landry, S.; Murphy, E.L.; Gessain, A.; Lemasson, I.; Mesnard, J.M.; et al. Human T-cell leukemia virus type 2 produces a spliced antisense transcript encoding a protein that lacks a classic bZIP domain but still inhibits Tax2-mediated transcription. Blood J. Am. Soc. Hematol. 2009, 114, 2427–2438. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Kannian, P.; Dissinger, N.; Haines, R.; Niewiesk, S.; Green, P.L. Human T-cell leukemia virus type 2 antisense viral protein 2 is dispensable for in vitro immortalization but functions to repress early virus replication in vivo. J. Virol. 2012, 86, 8412–8421. [Google Scholar] [CrossRef]
- Hakata, Y.; Yamada, M.; Shida, H. Rat CRM1 is responsible for the poor activity of human T-cell leukemia virus type 1 Rex protein in rat cells. J. Virol. 2001, 75, 11515–11525. [Google Scholar] [CrossRef] [PubMed]
- Ciminale, V.; Zotti, L.; D’Agostino, D.M.; Chieco-Bianchi, L. Inhibition of human T-cell leukemia virus type 2 Rex function by truncated forms of Rex encoded in alternatively spliced mRNAs. J. Virol. 1997, 71, 2810–2818. [Google Scholar] [CrossRef] [PubMed]
- Younis, I.; Khair, L.; Dundr, M.; Lairmore, M.D.; Franchini, G.; Green, P.L. Repression of human T-cell leukemia virus type 1 and type 2 replication by a viral mRNA-encoded posttranscriptional regulator. J. Virol. 2004, 78, 11077–11083. [Google Scholar] [CrossRef]
- Hiraragi, H.; Kim, S.J.; Phipps, A.J.; Silic-Benussi, M.; Ciminale, V.; Ratner, L.; Green, P.L.; Lairmore, M.D. Human T-lymphotropic virus type 1 mitochondrion-localizing protein p13II is required for viral infectivity in vivo. J. Virol. 2006, 80, 3469–3476. [Google Scholar] [CrossRef]
- Valeri, V.W.; Hryniewicz, A.; Andresen, V.; Jones, K.; Fenizia, C.; Bialuk, I.; Chung, H.K.; Fukumoto, R.; Parks, R.W.; Ferrari, M.G.; et al. Requirement of the human T-cell leukemia virus p12 and p30 products for infectivity of human dendritic cells and macaques but not rabbits. Blood J. Am. Soc. Hematol. 2010, 116, 3809–3817. [Google Scholar] [CrossRef]
- Cimarelli, A.; Duclos, C.A.; Gessain, A.; Casoli, C.; Bertazzoni, U. Clonal expansion of human T-cell leukemia virus type II in patients with high proviral load. Virology 1996, 223, 362–364. [Google Scholar] [CrossRef]
- Eusebio-Ponce, E.; Anguita, E.; Paulino-Ramirez, R.; Candel, F.J. HTLV-1 infection: An emerging risk. Pathogenesis, epidemiology, diagnosis and associated diseases. Rev. Esp. Quimioter. 2019, 32, 485. [Google Scholar]
- Gessain, A.; Cassar, O. Epidemiological aspects and world distribution of HTLV-1 infection. Front. Microbiol. 2012, 3, 388. [Google Scholar] [CrossRef] [PubMed]
- Brito, W.R.d.S.; Cardoso-Costa, G.d.L.; Roland Junior, L.M.; Pereira, K.A.S.; Lopes, F.T.; Santos, B.C.d.; de Lima, A.C.R.; Abreu, I.N.; Lima, C.N.C.; Lima, S.S.; et al. Prevalence and risk factors for HTLV-1/2 infection in quilombo remnant communities living in the Brazilian Amazon. Front. Public Health 2022, 10, 871865. [Google Scholar] [CrossRef]
- Cook, L.B.; Taylor, G.P. HTLV-1 and HTLV-2 prevalence in the United States. J. Infect. Dis. 2014, 209, 486–487. [Google Scholar] [CrossRef] [PubMed]
- Ishak, R.; de Oliveira Guimarães Ishak, M.; Abreu, I.N.; Machado, L.F.A.; Lima, S.S.; Queiroz, M.A.F.; Cayres-Vallinoto, I.M.; Guerreiro, J.F.; Vallinoto, A.C.R. Long-term prevalence follow-up (1967–2022) of HTLV-2 among vulnerable indigenous populations in the Amazon region of Brazil. Front. Microbiol. 2023, 14, 1217134. [Google Scholar] [CrossRef] [PubMed]
- Zunt, J.R.; Tapia, K.; Thiede, H.; Lee, R.; Hagan, H. HTLV-2 infection in injection drug users in King County, Washington. Scand. J. Infect. Dis. 2006, 38, 654–663. [Google Scholar] [CrossRef]
- Murphy, E.L.; Fridey, J.; Smith, J.; Engstrom, J.; Sacher, R.; Miller, K.; Gibble, J.; Stevens, J.; Thomson, R.; Hansma, D.; et al. HTLV-associated myelopathy in a cohort of HTLV-I and HTLV-II-infected blood donors. Neurology 1997, 48, 315–320. [Google Scholar] [CrossRef]
- Ishak, R.; Guimarães Ishak, M.d.O.; Azevedo, V.N.; Machado, L.F.A.; Vallinoto, I.M.C.; Queiroz, M.A.F.; Costa, G.d.L.C.; Guerreiro, J.F.; Vallinoto, A.C.R. HTLV in South America: Origins of a silent ancient human infection. Virus Evol. 2020, 6, veaa053. [Google Scholar] [CrossRef]
- Djuicy, D.D.; Mouinga-Ondémé, A.; Cassar, O.; Ramassamy, J.L.; Idam Mamimandjiami, A.; Bikangui, R.; Fontanet, A.; Gessain, A. Risk factors for HTLV-1 infection in Central Africa: A rural population-based survey in Gabon. PLoS Neglect. Trop. Dis. 2018, 12, e0006832. [Google Scholar] [CrossRef]
- Gonçalves, D.U.; Proietti, F.A.; Ribas, J.G.R.; Araújo, M.G.; Pinheiro, S.R.; Guedes, A.C.; Carneiro-Proietti, A.B.F. Epidemiology, treatment, and prevention of human T-cell leukemia virus type 1-associated diseases. Clin. Microbiol. Rev. 2010, 23, 577–589. [Google Scholar] [CrossRef]
- Wiktor, S.Z.; Pate, E.J.; Murphy, E.L.; Palker, T.J.; Champegnie, E.; Ramlal, A.; Cranston, B.; Hanchard, B.; Blattner, W.A. Mother-to-child transmission of human T-cell lymphotropic virus type I (HTLV-I) in Jamaica: Association with antibodies to envelope glycoprotein (gp46) epitopes. JAIDS J. Acquir. Immune Defic. Syndr. 1993, 6, 1162–1167. [Google Scholar]
- Takahashi, K.; Takezaki, T.; Oki, T.; Kawakami, K.; Yashiki, S.; Fujiyoshi, T.; Usuku, K.; GROUP, M.T.C.T.S.; Mueller, N.; Osame, M.; et al. Inhibitory effect of maternal antibody on mother-to-child transmission of human T-lymphotropic virus type I. Int. J. Cancer 1991, 49, 673–677. [Google Scholar] [CrossRef] [PubMed]
- Nyambi, P.N.; Ville, Y.; Louwagie, J.; Bedjabaga, I.; Glowaczower, E.; Peeters, M.; Kerouedan, D.; Dazza, M.C.; Larouzé, B.; van der Groen, G.; et al. Mother-to-child transmission of human T-cell lymphotropic virus types I and II (HTLV-I/II) in Gabon: A prospective follow-up of 4 years. JAIDS J. Acquir. Immune Defic. Syndr. 1996, 12, 187–192. [Google Scholar] [CrossRef]
- Li, H.C.; Biggar, R.J.; Miley, W.J.; Maloney, E.M.; Cranston, B.; Hanchard, B.; Hisada, M. Provirus load in breast milk and risk of mother-to-child transmission of human T lymphotropic virus type I. J. Infect. Dis. 2004, 190, 1275–1278. [Google Scholar] [CrossRef] [PubMed]
- Martin-Latil, S.; Gnädig, N.F.; Mallet, A.; Desdouits, M.; Guivel-Benhassine, F.; Jeannin, P.; Prevost, M.C.; Schwartz, O.; Gessain, A.; Ozden, S.; et al. Transcytosis of HTLV-1 across a tight human epithelial barrier and infection of subepithelial dendritic cells. Blood J. Am. Soc. Hematol. 2012, 120, 572–580. [Google Scholar] [CrossRef]
- Murphy, E. Infection with human T-lymphotropic virus types-1 and-2 (HTLV-1 and-2): Implications for blood transfusion safety. Transfus. Clin. Biol. 2016, 23, 13–19. [Google Scholar] [CrossRef]
- Khabbaz, R.F.; Onorato, I.M.; Cannon, R.O.; Hartley, T.M.; Roberts, B.; Hosein, B.; Kaplan, J.E. Seroprevalence of HTLV-I and HTLV-II among intravenous drug users and persons in clinics for sexually transmitted diseases. N. Engl. J. Med. 1992, 326, 375–380. [Google Scholar] [CrossRef]
- Filippone, C.; Betsem, E.; Tortevoye, P.; Cassar, O.; Bassot, S.; Froment, A.; Fontanet, A.; Gessain, A. A severe bite from a nonhuman primate is a major risk factor for HTLV-1 infection in hunters from Central Africa. Clin. Infect. Dis. 2015, 60, 1667–1676. [Google Scholar] [CrossRef]
- Kazanji, M.; Mouinga-Ondémé, A.; Lekana-Douki-Etenna, S.; Caron, M.; Makuwa, M.; Mahieux, R.; Gessain, A. Origin of HTLV-1 in hunters of nonhuman primates in Central Africa. J. Infect. Dis. 2015, 211, 361–365. [Google Scholar] [CrossRef]
- LeBreton, M.; Switzer, W.M.; Djoko, C.F.; Gillis, A.; Jia, H.; Sturgeon, M.M.; Shankar, A.; Zheng, H.; Nkeunen, G.; Tamoufe, U.; et al. A gorilla reservoir for human T-lymphotropic virus type 4. Emerg. Microbes Infect. 2014, 3, 1–12. [Google Scholar] [CrossRef]
- Melamed, A.; Laydon, D.J.; Gillet, N.A.; Tanaka, Y.; Taylor, G.P.; Bangham, C.R. Genome-wide determinants of proviral targeting, clonal abundance and expression in natural HTLV-1 infection. PLoS Pathog. 2013, 9, e1003271. [Google Scholar] [CrossRef] [PubMed]
- Lupiáñez, D.G.; Kraft, K.; Heinrich, V.; Krawitz, P.; Brancati, F.; Klopocki, E.; Horn, D.; Kayserili, H.; Opitz, J.M.; Laxova, R.; et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 2015, 161, 1012–1025. [Google Scholar] [CrossRef] [PubMed]
- Krivtsov, A.; Armstrong, S. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer 2007, 7, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Satou, Y.; Miyazato, P.; Ishihara, K.; Yaguchi, H.; Melamed, A.; Miura, M.; Fukuda, A.; Nosaka, K.; Watanabe, T.; Rowan, A.G.; et al. The retrovirus HTLV-1 inserts an ectopic CTCF-binding site into the human genome. Proc. Natl. Acad. Sci. USA 2016, 113, 3054–3059. [Google Scholar] [CrossRef]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory activities of transposable elements: From conflicts to benefits. Nat. Rev. Genet. 2016, 18, 71–86. [Google Scholar] [CrossRef]
- Gallo, R.; Willems, L. Proviral latency and the reprogramming of host gene expression in HTLV-1 infection. Front. Microbiol. 2021, 12, 657050. [Google Scholar]
- Firouzi, S.; Farmanbar, A.; Nakai, K.; Iwanaga, M.; Uchimaru, K.; Utsunomiya, A.; Suzuki, Y.; Watanabe, T. Clonality of HTLV-1–infected T cells as a risk indicator for development of adult T-cell leukemia. Blood Adv. 2017, 1, 1195–1205. [Google Scholar] [CrossRef]
- Ratner, L. Epigenetic Regulation of Human T-Cell Leukemia Virus Gene Expression. Microorganisms 2022, 10, 84. [Google Scholar] [CrossRef]
- Matsuoka, M.; Green, P.L. The HBZ gene, a key player in HTLV-1 pathogenesis. Retrovirology 2009, 6, 71. [Google Scholar] [CrossRef]
- Matsuoka, M.; Jeang, K.T. Human T-cell leukemia virus type 1 (HTLV-1) and leukemic transformation: Viral infectivity, Tax, HBZ and therapy. Oncogene 2011, 30, 1379–1389. [Google Scholar] [CrossRef]
- Sugata, K.; Yasunaga, J.; Kinosada, H.; Mitobe, Y.; Furuta, R.; Mahgoub, M.; Onishi, C.; Nakashima, K.; Ohshima, K.; Matsuoka, M. HTLV-1 Viral Factor HBZ Induces CCR4 to Promote T-cell Migration and Proliferation. Cancer Res. 2016, 76, 5068–5079. [Google Scholar] [CrossRef]
- Panfil, A.R.; Green, P.L.; Yoder, K.E. CRISPR Genome Editing Applied to the Pathogenic Retrovirus HTLV-1. Front. Cell Infect. Microbiol. 2020, 10, 580371. [Google Scholar] [CrossRef]
- Leonardi, F.e.a. Deep learning applied to classification of HTLV subtypes. Front. Genet. 2020, 11, 487. [Google Scholar]
- Yamagishi, M.; Kubokawa, M.; Kuze, Y.; Suzuki, A.; Yokomizo, A.; Kobayashi, S.; Nakashima, M.; Makiyama, J.; Iwanaga, M.; Fukuda, T.; et al. Chronological genome and single-cell transcriptome integration characterizes the evolutionary process of adult T cell leukemia-lymphoma. Nat. Commun. 2021, 12, 4821. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Jia, J.; Jeong, H.H.; Zhao, Z. Deep learning for detecting and elucidating human T-cell leukemia virus type 1 integration in the human genome. Patterns (NY) 2023, 4, 100674. [Google Scholar] [CrossRef]
- Melamed, A.; Yaguchi, H.; Miura, M.; Witkover, A.; Fitzgerald, T.W.; Birney, E.; Bangham, C.R. The human leukemia virus HTLV-1 alters the structure and transcription of host chromatin in cis. Elife 2018, 7, e36245. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Rao, S.; Crispino, J.D.; Ntziachristos, P. Determinants and role of chromatin organization in acute leukemia. Leukemia 2020, 34(10), 2561–2575. [Google Scholar] [CrossRef] [PubMed]
- Yaguchi, H.; Melamed, A.; Ramanayake, S.; Kiik, H.; Witkover, A.; Bangham, C.R.M. The impact of HTLV-1 expression on the 3D structure and expression of host chromatin. PLoS Pathog. 2024, 20(3), e1011716. [Google Scholar] [CrossRef]
- Shichijo, T.; Yasunaga, J.I. Stratagems of HTLV-1 for persistent infection and the resultant oncogenesis: Immune evasion and clonal expansion. Leuk. Res. 2025, 152, 107680. [Google Scholar] [CrossRef]
- Chen, M.; Huang, X.; Wang, C.; Wang, S.; Jia, L.; Li, L. Endogenous retroviral solo-LTRs in human genome. Front. Genet. 2024, 15, 1358078. [Google Scholar] [CrossRef]
- Smith, S.; Seth, J.; Midkiff, A.; Stahl, R.; Syu, Y.-C.; Shkriabai, N.; Kvaratskhelia, M.; Musier-Forsyth, K.; Jain, P.; Green, P.L.; et al. The Pleiotropic Effects of YBX1 on HTLV-1 Transcription. Int. J. Mol. Sci. 2023, 24, 13119. [Google Scholar] [CrossRef]
- Baydoun, H.H.; Bai, X.T.; Shelton, S.; Nicot, C. HTLV-I Tax Increases Genetic Instability by Inducing DNA Double Strand Breaks during DNA Replication and Switching Repair to NHEJ. PLoS ONE 2012, 7, e42226. [Google Scholar] [CrossRef]
- Danovski, G.; Panova, G.; Keister, B.; Georgiev, G.; Atemin, A.; Uzunova, S.; Stamatov, R.; Kanev, P.B.; Aleksandrov, R.; Blagoev, K.B.; et al. Diffusion of activated ATM explains γH2AX and MDC1 spread beyond the DNA damage site. iScience 2024, 27, 110826. [Google Scholar] [CrossRef] [PubMed]
- Rowan, A.G.; Dillon, R.; Witkover, A.; Melamed, A.; Demontis, M.A.; Gillet, N.A.; Mun, L.J.; Bangham, C.R.M.; Cook, L.B.; Fields, P.A.; et al. Evolution of retrovirus-infected premalignant T-cell clones prior to adult T-cell leukemia/lymphoma diagnosis. Blood 2020, 135, 2023–2032. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Vercruyssen, M.; Cook, L. HTLV-1-related adult T-cell leukemia/lymphoma: Insights in early detection and management. Curr. Opin. Oncol. 2022, 34, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Jalaeikhoo, H.; Soleymani, M.; Rajaeinejad, M.; Keyhani, M. Prevalence of human T-lymphotropic virus type 1 (HTLV-1) infection in patients with hematologic disorders and non-hematologic malignancies in a tertiary referral hospital. Arch. Iran. Med. 2017, 20, 224–228. [Google Scholar]
- Salehi, M.; Mostafavi, S.K.S.; Ghasemian, A.; Gholami, M.; Kazemi-Vardanjani, A.; Rahimi, M.K. Seroepidemiology of HTLV-1 and HTLV-2 infection in Neyshabur city, North-Eastern Iran, during 2010–2014. Iran. Biomed. J. 2017, 21, 57. [Google Scholar] [CrossRef]
- Sabino, E.C.; Zrein, M.; Taborda, C.P.; Otani, M.M.; Ribeiro-Dos-Santos, G.; Sáez-Alquézar, A. Evaluation of the INNO-LIA HTLV I/II assay for confirmation of human T-cell leukemia virus-reactive sera in blood bank donations. J. Clin. Microbiol. 1999, 37, 1324–1328. [Google Scholar] [CrossRef]
- Rosadas, C.; Caterino-de Araujo, A.; Taylor, G.P. Specificity of HTLV screening tests and its impact on health care program costs: The perspective of antenatal screening in Brazil. Rev. Soc. Bras. Med. Trop. 2021, 54, e0853-2020. [Google Scholar] [CrossRef]
- da Silva Brito, V.; Santos, F.L.N.; Gonçalves, N.L.S.; Araujo, T.H.A.; Nascimento, D.S.V.; Pereira, F.M.; Boa-Sorte, N.C.A.; Grassi, M.F.R.; Caterino-de Araujo, A.; Galvão-Castro, B. Performance of commercially available serological screening tests for human T-cell lymphotropic virus infection in Brazil. J. Clin. Microbiol. 2018, 56, 10–1128. [Google Scholar] [CrossRef]
- Hjelle, B.; Wilson, C.; Cyrus, S.; Bradshaw, P.; Lo, J.; Schammel, C.; Wiltbank, T.; Alexander, S. Human T-cell leukemia virus type II infection frequently goes undetected in contemporary US blood donors. Blood 1993, 81, 1641–1644. [Google Scholar] [CrossRef]
- Abrams, A.; Akahata, Y.; Jacobson, S. The prevalence and significance of HTLV-I/II seroindeterminate Western blot patterns. Viruses 2011, 3, 1320–1331. [Google Scholar] [CrossRef]
- Medeiros, A.C.; Vidal, L.R.; Von Linsingen, R.; Ferin, A.N.; Bessani Strapasson, T.; de Almeida, S.M.; Raboni, S.M.; Carvalho, N.S.; Bordignon Nogueira, M. Confirmatory molecular method for HTLV-1/2 infection in high-risk pregnant women. J. Med. Virol. 2018, 90, 998–1001. [Google Scholar] [CrossRef] [PubMed]
- Moreno, C.; Balangero, M.; Barbás, M.G.; Cudolá, A.; Gallego, S. Diagnóstico serológico de HTLV-1/2: Combinación de técnicas de tamizaje para definir el estatus serológico en donantes de sangre. Rev. Argent. Microbiol. 2013, 45, 165–168. [Google Scholar] [CrossRef] [PubMed]
- Fani, M.; Rezayi, M.; Meshkat, Z.; Rezaee, S.A.; Makvandi, M.; Abouzari-Lotf, E.; Ferns, G.A. Current approaches for detection of human T-lymphotropic virus Type 1: A systematic review. J. Cell. Physiol. 2019, 234, 12433–12441. [Google Scholar] [CrossRef] [PubMed]
- Cassar, O.; Gessain, A. Serological and molecular methods to study epidemiological aspects of human T-cell lymphotropic virus type 1 infection. In Human T-Lymphotropic Viruses: Methods and Protocols; Humana Press: New York, NY, USA, 2017; pp. 3–24. [Google Scholar]
- Ohshima, K.; Hashimoto, K.; Izumo, S.; Suzumiya, J.; Kikuchi, M. Detection of human T lymphotrophic virus type I (HTLV-I) DNA and mRNA in individual cells by polymerase chain reaction (PCR) in situ hybridization (ISH) and reverse transcription (RT)-PCR ISH. Hematol. Oncol. 1996, 14, 91–100. [Google Scholar] [CrossRef]
- de Oliveira Andrade, F.; Cucco, M.S.; Borba, M.M.N.; Neto, R.C.; Gois, L.L.; de Almeida Rego, F.F.; Santos, L.A.; Barreto, F.K. An overview of sequencing technology platforms applied to HTLV-1 studies: A systematic review. Arch. Virol. 2021, 166, 3037–3048. [Google Scholar] [CrossRef]
- Rahimian, M.; Panahi, B. Next generation sequencing-based transcriptome data mining for virus identification and characterization: Review on recent progress and prospect. J. Clin. Virol. Plus 2024, 4, 100194. [Google Scholar] [CrossRef]
- Kamali, P.; Zandi, M.; Ghasemzadeh-Moghaddam, H.; Fani, M. Comparison between various biosensor methods for human T-lymphotropic virus-1 (HTLV-1) detection. Mol. Biol. Rep. 2022, 49, 1513–1517. [Google Scholar] [CrossRef]
- Vigneshvar, S.; Sudhakumari, C.; Senthilkumaran, B.; Prakash, H. Recent advances in biosensor technology for potential applications—An overview. Front. Bioeng. Biotechnol. 2016, 4, 11. [Google Scholar] [CrossRef]
- Karunakaran, C.; Rajkumar, R.; Bhargava, K. Introduction to biosensors. In Biosensors and Bioelectronics; Elsevier: Amsterdam, The Netherlands, 2015; pp. 1–68. [Google Scholar]
- Perumal, V.; Hashim, U. Advances in biosensors: Principle, architecture and applications. J. Appl. Biomed. 2014, 12, 1–15. [Google Scholar] [CrossRef]
- Salek-Maghsoudi, A.; Vakhshiteh, F.; Torabi, R.; Hassani, S.; Ganjali, M.R.; Norouzi, P.; Hosseini, M.; Abdollahi, M. Recent advances in biosensor technology in assessment of early diabetes biomarkers. Biosens. Bioelectron. 2018, 99, 122–135. [Google Scholar] [CrossRef]
- Silveira, C.M.; Monteiro, T.; Almeida, M.G. Biosensing with paper-based miniaturized printed electrodes–a modern trend. Biosensors 2016, 6, 51. [Google Scholar] [CrossRef]
- Santos, F.A.; Catão, C.L.S.; Martins, J.P.; Pessoa, U.H.S.; Sousa, I.V.; Melo, J.S.; Souza, G.L.; Araújo, N.D.; Magalhães-Gama, F.; Abrahim, C.M.d.M.; et al. Performance of immunological assays for universal and differential diagnosis of HTLV-1/2 infection in candidates for blood donations from the Brazilian Amazon. PLoS ONE 2024, 19, e0298710. [Google Scholar] [CrossRef]
- Matavele Chissumba, R.; Silva-Barbosa, S.D.; Augusto, Â.; Maueia, C.; Mabunda, N.; Gudo, E.S.; Bhatt, N.; Jani, I.; Savino, W. CD4+ CD25 High Treg cells in HIV/HTLV co-infected patients with neuropathy: High expression of Alpha4 integrin and lower expression of Foxp3 transcription factor. BMC Immunol. 2015, 16, 52. [Google Scholar] [CrossRef] [PubMed]
- Freitas, N.L.; Gomes, Y.C.P.; Souza, F.d.S.; Torres, R.C.; Echevarria-Lima, J.; Leite, A.C.C.B.; Lima, M.A.S.D.; Araújo, A.Q.C.; Silva, M.T.T.; Espíndola, O.d.M. Lessons from the cerebrospinal fluid analysis of HTLV-1-infected individuals: Biomarkers of inflammation for HAM/TSP development. Viruses 2022, 14, 2146. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Rana, P.S.; Bawa, S. Hybrid machine learning models for predicting types of Human T-cell Lymphotropic Virus. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 18, 1524–1534. [Google Scholar] [CrossRef]
- Wang, T.T.; Hirons, A.; Doerflinger, M.; Morris, K.V.; Ledger, S.; Purcell, D.F.; Kelleher, A.D.; Ahlenstiel, C.L. Current state of therapeutics for HTLV-1. Viruses 2024, 16, 1616. [Google Scholar] [CrossRef]
- El Hajj, H.; Hermine, O.; Bazarbachi, A. Therapeutic advances for the management of adult T cell leukemia: Where do we stand? Leuk. Res. 2024, 147, 107598. [Google Scholar] [CrossRef]
- Puccioni-Sohler, M.; Poton, A.R.; Cabral-Castro, M.J.; Yamano, Y.; Taylor, G.; Casseb, J. Human T lymphotropic virus 1-associated myelopathy: Overview of human T cell lymphotropic virus-1/2 tests and potential biomarkers. AIDS Res. Hum. Retroviruses 2022, 38, 924–932. [Google Scholar] [CrossRef]
- Iwanaga, M.; Watanabe, T.; Utsunomiya, A.; Okayama, A.; Uchimaru, K.; Koh, K.R.; Ogata, M.; Kikuchi, H.; Sagara, Y.; Uozumi, K.; et al. Human T-cell leukemia virus type I (HTLV-1) proviral load and disease progression in asymptomatic HTLV-1 carriers: A nationwide prospective study in Japan. Blood J. Am. Soc. Hematol. 2010, 116, 1211–1219. [Google Scholar] [CrossRef] [PubMed]
- Hodson, A.; Crichton, S.; Montoto, S.; Mir, N.; Matutes, E.; Cwynarski, K.; Kumaran, T.; Ardeshna, K.M.; Pagliuca, A.; Taylor, G.P.; et al. Use of zidovudine and interferon alfa with chemotherapy improves survival in both acute and lymphoma subtypes of adult T-cell leukemia/lymphoma. J. Clin. Oncol. 2011, 29, 4696–4701. [Google Scholar] [CrossRef] [PubMed]
- Taylor, G.P.; Goon, P.; Furukawa, Y.; Green, H.; Barfield, A.; Mosley, A.; Nose, H.; Babiker, A.; Rudge, P.; Usuku, K.; et al. Zidovudine plus lamivudine in human T-lymphotropic virus type-I-associated myelopathy: A randomised trial. Retrovirology 2006, 3, 63. [Google Scholar] [CrossRef]
- Taylor, G.P.; Hall, S.E.; Navarrete, S.; Michie, C.A.; Davis, R.; Witkover, A.D.; Rossor, M.; Nowak, M.A.; Rudge, P.; Matutes, E.; et al. Effect of lamivudine on human T-cell leukemia virus type 1 (HTLV-1) DNA copy number, T-cell phenotype, and anti-tax cytotoxic T-cell frequency in patients with HTLV-1-associated myelopathy. J. Virol. 1999, 73, 10289–10295. [Google Scholar] [CrossRef]
- Macchi, B.; Balestrieri, E.; Ascolani, A.; Hilburn, S.; Martin, F.; Mastino, A.; Taylor, G.P. Susceptibility of primary HTLV-1 isolates from patients with HTLV-1-associated myelopathy to reverse transcriptase inhibitors. Viruses 2011, 3, 469. [Google Scholar] [CrossRef] [PubMed]
- Trevino, A.; Parra, P.; Bar-Magen, T.; Garrido, C.; de Mendoza, C.; Soriano, V. Antiviral effect of raltegravir on HTLV-1 carriers. J. Antimicrob. Chemother. 2012, 67, 218–221. [Google Scholar] [CrossRef]
- Abad-Fernández, M.; Cabrera, C.; García, E.; Vallejo, A. Transient increment of HTLV-2 proviral load in HIV-1-co-infected patients during treatment intensification with raltegravir. J. Clin. Virol. 2014, 59, 204–207. [Google Scholar] [CrossRef]
- Enose-Akahata, Y.; Billioux, B.J.; Azodi, S.; Dwyer, J.; Vellucci, A.; Ngouth, N.; Nozuma, S.; Massoud, R.; Cortese, I.; Ohayon, J.; et al. Clinical trial of raltegravir, an integrase inhibitor, in HAM/TSP. Ann. Clin. Transl. Neurol. 2021, 8, 1970–1985. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, T.; Marconi, C.; Montaño-Castellón, I.; Deminco, F.; Brites, C. A Systematical Review on ART Use in HTLV Infection: Clinical, Virological, and Immunological Outcomes. Pathogens 2024, 13, 721. [Google Scholar] [CrossRef]
- Ducasa, N.; Domínguez, D.; Benencio, P.; Alfie, L.; Etcheves, P.; Scarton, G.; Biglione, M.; Caputo, M. Low-cost and simple PCR process for access to molecular diagnosis of HTLV-1/2 in low-resource countries. Acta Trop. 2024, 260, 107395. [Google Scholar] [CrossRef]
- Dangana, A.; Abdullahi, I.N.; Billyrose, O.M.A.; Emeribe, A.U.; Abu, J.M.; Anka, A.U.; Animasaun, O.S.; Ghamba, P.E. Sero-epidemiology of human T-cell lymphotropic viruses-1 and -2 infection among pregnant women attending Abuja Teaching Hospital, Nigeria. Hum. Antibodies 2021, 29, 101–108. [Google Scholar] [CrossRef]
- Sandler, S.G.; Fang, C.T.; Williams, A.E. Human T-cell lymphotropic virus type I and II in transfusion medicine. Transfus. Med. Rev. 1991, 5, 93–107. [Google Scholar] [CrossRef] [PubMed]
- de Mendoza, C.; Bautista, J.M.; Pérez-Benavente, S.; Kwawu, R.; Fobil, J.; Soriano, V.; Díez, A. Screening for retroviruses and hepatitis viruses using dried blood spots reveals a high prevalence of occult hepatitis B in Ghana. Ther. Adv. Infect. Dis. 2019, 6, 2049936119851464. [Google Scholar] [CrossRef] [PubMed]
- Altaf, F.; Qureshi, Z.A.; Moore, S.; Golek, T.M.; Chawala, A. A Textbook Case of Human T-lymphotropic Virus-1 (HTLV-1)-Induced Adult T-cell Leukemia Treated With Cyclophosphamide, Hydroxydaunorubicin, Oncovin, and Prednisone/Prednisolone (CHOP). Cureus 2023, 15, e49169. [Google Scholar] [CrossRef] [PubMed]
- Saeed, H.; Sandoval-Sus, J.; Castillo-Tokumori, F.; Dong, N.; Pullukkara, J.J.; Boisclair, S.; Brahim, A.; Walker, D.; Bridgellal, S.; Zhang, L.; et al. Poor Outcome of Adult T-Cell Leukemia/Lymphoma with Current Available Therapy: An Experience of Two Centers. Oncol. Res. Treat. 2023, 46, 459–465. [Google Scholar] [CrossRef]
- Taylor, G.P.; Matsuoka, M. Natural history of adult T-cell leukemia/lymphoma and approaches to therapy. Oncogene 2005, 24, 6047–6057. [Google Scholar] [CrossRef]
- Lin, Y.; Zhang, R.D.; Chen, R.L.; Chen, J.; Wu, Y.; Chen, Q. Clinical Study of Chemotherapy Combined with Antivirals for Adult T-cell Leukemia/Lymphoma. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2022, 30, 1407–1414. [Google Scholar]
- Katsuya, H. Current and emerging therapeutic strategies in adult T-cell leukemia–lymphoma. Int. J. Hematol. 2023, 117, 512–522. [Google Scholar] [CrossRef]
- Ishida, T.; Joh, T.; Uike, N.; Yamamoto, K.; Utsunomiya, A.; Yoshida, S.; Saburi, Y.; Miyamoto, T.; Takemoto, S.; Suzushima, H.; et al. Defucosylated anti-CCR4 monoclonal antibody (KW-0761) for relapsed adult T-cell leukemia-lymphoma: A multicenter phase II study. J. Clin. Oncol. 2012, 30, 837–842. [Google Scholar] [CrossRef]
- Ishida, T.; Jo, T.; Takemoto, S.; Suzushima, H.; Uozumi, K.; Yamamoto, K.; Uike, N.; Saburi, Y.; Nosaka, K.; Utsunomiya, A.; et al. Dose-intensified chemotherapy alone or in combination with mogamulizumab in newly diagnosed aggressive adult T-cell leukaemia-lymphoma: A randomized phase II study. Br. J. Haematol. 2015, 169, 672–682. [Google Scholar] [CrossRef]
- Sakamoto, Y.; Ishida, T.; Masaki, A.; Murase, T.; Yonekura, K.; Tashiro, Y.; Tokunaga, M.; Utsunomiya, A.; Ito, A.; Kusumoto, S.; et al. CCR4 mutations associated with superior outcome of adult T-cell leukemia/lymphoma under mogamulizumab treatment. Blood J. Am. Soc. Hematol. 2018, 132, 758–761. [Google Scholar] [CrossRef]
- Ishida, T.; Jo, T.; Takemoto, S.; Suzushima, H.; Suehiro, Y.; Choi, I.; Yoshimitsu, M.; Saburi, Y.; Nosaka, K.; Utsunomiya, A.; et al. Follow-up of a randomised phase II study of chemotherapy alone or in combination with mogamulizumab in newly diagnosed aggressive adult T-cell leukaemia-lymphoma: Impact on allogeneic haematopoietic stem cell transplantation. Br. J. Haematol. 2019, 184, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, N.; Mori, S.; Kiyotani, K.; Ota, Y.; Gotoh, O.; Kusumoto, S.; Nakano, N.; Suehiro, Y.; Ito, A.; Choi, I.; et al. Genomic determinants impacting the clinical outcome of mogamulizumab treatment for adult T-cell leukemia/lymphoma. Haematologica 2022, 107, 2418. [Google Scholar] [CrossRef]
- Shichijo, T.; Nosaka, K.; Tatetsu, H.; Higuchi, Y.; Endo, S.; Inoue, Y.; Toyoda, K.; Kikukawa, Y.; Kawakita, T.; Yasunaga, J.I.; et al. Beneficial impact of first-line mogamulizumab-containing chemotherapy in adult T-cell leukaemia-lymphoma. Br. J. Haematol. 2022, 198, 983–987. [Google Scholar] [CrossRef] [PubMed]
- Fuji, S.; Inoue, Y.; Utsunomiya, A.; Moriuchi, Y.; Uchimaru, K.; Choi, I.; Otsuka, E.; Henzan, H.; Kato, K.; Tomoyose, T.; et al. Pretransplantation anti-CCR4 antibody mogamulizumab against adult T-cell leukemia/lymphoma is associated with significantly increased risks of severe and corticosteroid-refractory graft-versus-host disease, nonrelapse mortality, and overall mortality. J. Clin. Oncol. 2016, 34, 3426–3433. [Google Scholar] [CrossRef]
- Sakamoto, H.; Itonaga, H.; Sawayama, Y.; Furumoto, T.; Fujioka, M.; Chiwata, M.; Toriyama, E.; Kasai, S.; Nakashima, J.; Horai, M.; et al. Treatment with mogamulizumab or lenalidomide for relapsed adult T-cell leukemia/lymphoma after allogeneic hematopoietic stem cell transplantation: The Nagasaki transplant group experience. Hematol. Oncol. 2020, 38, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y.; Endo, S.; Matsuno, N.; Kikukawa, Y.; Shichijo, T.; Koga, K.; Takaki, A.; Iwanaga, K.; Nishimura, N.; Fuji, S.; et al. Safety of mogamulizumab for relapsed ATL after allogeneic hematopoietic cell transplantation. Bone Marrow Transplant. 2019, 54, 338–342. [Google Scholar] [CrossRef]
- Ishida, T.; Fujiwara, H.; Nosaka, K.; Taira, N.; Abe, Y.; Imaizumi, Y.; Moriuchi, Y.; Jo, T.; Ishizawa, K.; Tobinai, K.; et al. Multicenter phase II study of lenalidomide in relapsed or recurrent adult T-cell leukemia/lymphoma: ATLL-002. J. Clin. Oncol. 2016, 34, 4086–4093. [Google Scholar] [CrossRef]
- Tanaka, T.; Inamoto, Y.; Ito, A.; Watanabe, M.; Takeda, W.; Aoki, J.; Kim, S.W.; Fukuda, T. Lenalidomide treatment for recurrent adult T-cell leukemia/lymphoma after allogeneic hematopoietic cell transplantation. Hematol. Oncol. 2023, 41, 389–395. [Google Scholar] [CrossRef]
- Hourigan, C.S.; Forde, P.M.; Ambinder, R.F.; Gladstone, D.E. Bortezomib salvage therapy in refractory acute adult T-cell leukemia/lymphoma. Leuk. Lymphoma 2013, 54, 2563–2564. [Google Scholar] [CrossRef]
- Ratner, L.; Rauch, D.; Abel, H.; Caruso, B.; Noy, A.; Barta, S.; Parekh, S.; Ramos, J.; Ambinder, R.; Phillips, A.; et al. Dose-adjusted EPOCH chemotherapy with bortezomib and raltegravir for human T-cell leukemia virus-associated adult T-cell leukemia lymphoma. Blood Cancer J. 2016, 6, e408. [Google Scholar] [CrossRef] [PubMed]
- Fuji, S.; Yamaguchi, T.; Inoue, Y.; Utsunomiya, A.; Moriuchi, Y.; Owatari, S.; Miyagi, T.; Sawayama, Y.; Otsuka, E.; Yoshida, S.I.; et al. VCAP-AMP-VECP as a preferable induction chemotherapy in transplant-eligible patients with aggressive adult T-cell leukemia-lymphoma: A propensity score analysis. Bone Marrow Transplant. 2019, 54, 1399–1405. [Google Scholar] [CrossRef]
- Fuji, S.; Fujiwara, H.; Nakano, N.; Wake, A.; Inoue, Y.; Fukuda, T.; Hidaka, M.; Moriuchi, Y.; Miyamoto, T.; Uike, N.; et al. Early application of related SCT might improve clinical outcome in adult T-cell leukemia/lymphoma. Bone Marrow Transplant. 2016, 51, 205–211. [Google Scholar] [CrossRef]
- Choi, I.; Tanosaki, R.; Uike, N.; Utsunomiya, A.; Tomonaga, M.; Harada, M.; Yamanaka, T.; Kannagi, M.; Okamura, J. Long-term outcomes after hematopoietic SCT for adult T-cell leukemia/lymphoma: Results of prospective trials. Bone Marrow Transplant. 2011, 46, 116–118. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Hishizawa, M.; Kato, K.; Tanosaki, R.; Fukuda, T.; Taniguchi, S.; Eto, T.; Takatsuka, Y.; Miyazaki, Y.; Moriuchi, Y.; et al. Allogeneic hematopoietic stem cell transplantation for adult T-cell leukemia-lymphoma with special emphasis on preconditioning regimen: A nationwide retrospective study. Blood J. Am. Soc. Hematol. 2012, 120, 1734–1741. [Google Scholar] [CrossRef]
- Inoue, Y.; Nakano, N.; Fuji, S.; Eto, T.; Kawakita, T.; Suehiro, Y.; Miyamoto, T.; Sawayama, Y.; Uchida, N.; Kondo, T.; et al. Impact of conditioning intensity and regimen on transplant outcomes in patients with adult T-cell leukemia-lymphoma. Bone Marrow Transplant. 2021, 56, 2964–2974. [Google Scholar] [CrossRef] [PubMed]
- Ito, A.; Nakano, N.; Tanaka, T.; Fuji, S.; Makiyama, J.; Inoue, Y.; Choi, I.; Nakamae, H.; Nagafuji, K.; Takase, K.; et al. Improved survival of patients with aggressive ATL by increased use of allo-HCT: A prospective observational study. Blood Adv. 2021, 5, 4156–4166. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, T.; Itonaga, H.; Moriuchi, Y.; Yoshida, S.; Taguchi, J.; Imaizumi, Y.; Imanishi, D.; Tsushima, H.; Sawayama, Y.; Matsuo, E.; et al. Feasibility of cord blood transplantation in chemosensitive adult T-cell leukemia/lymphoma: A retrospective analysis of the Nagasaki Transplantation Network. Int. J. Hematol. 2013, 97, 485–490. [Google Scholar] [CrossRef]
- Kato, K.; Choi, I.; Wake, A.; Uike, N.; Taniguchi, S.; Moriuchi, Y.; Miyazaki, Y.; Nakamae, H.; Oku, E.; Murata, M.; et al. Treatment of patients with adult T cell leukemia/lymphoma with cord blood transplantation: A Japanese nationwide retrospective survey. Biol. Blood Marrow Transplant. 2014, 20, 1968–1974. [Google Scholar] [CrossRef]
- Tanaka, T.; Nakamae, H.; Ito, A.; Fuji, S.; Hirose, A.; Eto, T.; Henzan, H.; Takase, K.; Yamasaki, S.; Makiyama, J.; et al. A phase I/II multicenter trial of HLA-haploidentical PBSCT with PTCy for aggressive adult T cell leukemia/lymphoma. Transplant. Cell. Ther. 2021, 27, 928.e1–928.e7. [Google Scholar] [CrossRef]
- Yamauchi, J.; Tanabe, K.; Sato, T.; Nakagawa, M.; Matsuura, E.; Tsuboi, Y.; Tamaki, K.; Sakima, H.; Ishihara, S.; Ohta, Y.; et al. Efficacy of corticosteroid therapy for HTLV-1-associated myelopathy: A randomized controlled trial (HAMLET-P). Viruses 2022, 14, 136. [Google Scholar] [CrossRef]
- Araújo, A.d.Q.; Leite, A.C.; Dultra, S.V.; Andrada-Serpa, M.J. Progression of neurological disability in HTLV-I-associated myelopathy/tropical spastic paraparesis (HAM/TSP). J. Neurol. Sci. 1995, 129, 147–151. [Google Scholar] [CrossRef] [PubMed]
- Yamano, Y.; Sato, T. Clinical pathophysiology of human T-lymphotropic virus-type 1-associated myelopathy/tropical spastic paraparesis. Front. Microbiol. 2012, 3, 389. [Google Scholar] [CrossRef]
- Izumo, S.; Goto, I.; Itoyama, Y.; Okajima, T.; Watanabe, S.; Kuroda, Y.; Araki, S.; Mori, M.; Nagataki, M.; Matsukura, S.; et al. Interferon-alpha is effective in HTLV-I-associated myelopathy: A multicenter, randomized, double-blind, controlled trial. Neurology 1996, 46, 1016–1021. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, M.; Nakahara, K.; Maruyama, Y.; Kawabata, M.; Higuchi, I.; Kubota, H.; Izumo, S.; Arimura, K.; Osame, M. Therapeutic trials in 200 patients with HTLV-Iassociated myelopathy/tropical spastic paraparesis. J. Neurovirol. 1996, 2, 345–355. [Google Scholar] [CrossRef]
- Martin, F.; Castro, H.; Gabriel, C.; Adonis, A.; Fedina, A.; Harrison, L.; Brodnicki, L.; Demontis, M.A.; Babiker, A.G.; Weber, J.N.; et al. Ciclosporin A proof of concept study in patients with active, progressive HTLV-1 associated myelopathy/tropical spastic paraparesis. PLoS Neglect. Trop. Dis. 2012, 6, e1675. [Google Scholar] [CrossRef]
- Izumi, Y.; Kojima, H.; Koga, Y.; Yokota, K.; Mori, H.; Ohno, T.; Miyashita, T.; Ito, M.; Motomura, M.; Mine, M.; et al. Successful treatment of HTLV-1-related overlap syndrome using tacrolimus. Intern. Med. 2011, 50, 1849–1853. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Yagishita, N.; Tamaki, K.; Inoue, E.; Hasegawa, D.; Nagasaka, M.; Suzuki, H.; Araya, N.; Coler-Reilly, A.; Hasegawa, Y.; et al. Proposal of classification criteria for HTLV-1-associated myelopathy/tropical spastic paraparesis disease activity. Front. Microbiol. 2018, 9, 1651. [Google Scholar] [CrossRef]
- Sato, T.; Coler-Reilly, A.L.; Yagishita, N.; Araya, N.; Inoue, E.; Furuta, R.; Watanabe, T.; Uchimaru, K.; Matsuoka, M.; Matsumoto, N.; et al. Mogamulizumab (Anti-CCR4) in HTLV-1–associated myelopathy. N. Engl. J. Med. 2018, 378, 529–538. [Google Scholar] [CrossRef]
- Agarwal, S.; Patel, T.; Shah, N.; Patel, B.M. Comparative study of therapeutic response to baclofen vs tolperisone in spasticity. Biomed. Pharmacother. 2017, 87, 628–635. [Google Scholar] [CrossRef]
- Groves, L.; Shellenberger, M.; Davis, C. Tizanidine treatment of spasticity: A meta-analysis of controlled, double-blind, comparative studies with baclofen and diazepam. Adv. Ther. 1998, 15, 241–251. [Google Scholar]
- Abdelmageed, S.; Horak, V.J.; Mossner, J.; Wang, R.; Krater, T.; Raskin, J.S. Safety and efficacy of intrathecal baclofen trials for the treatment of hypertonia: A retrospective cohort study. J. Neurosurg. Pediatr. 2023, 33, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Levy, J.; Karam, P.; Forestier, A.; Loze, J.Y.; Bensmail, D. Botulinum toxin use in patients with post-stroke spasticity: A nationwide retrospective study from France. Front. Neurol. 2023, 14, 1245228. [Google Scholar] [CrossRef] [PubMed]
- Troisgros, O.; Barnay, J.L.; Darbon-Naghibzadeh, F.; Olive, P.; René-Corail, P. Retrospective clinic and urodynamic study in the neurogenic bladder dysfunction caused by human T cell lymphotrophic virus type 1 associated myelopathy/tropical spastic paraparesis (HAM/TSP). Neurourol. Urodynamics 2017, 36, 449–452. [Google Scholar] [CrossRef]
- Ananda, I.G.Y.P.; Surya, R.N.H.; Surya, P.A.; Putratama, A.; Andhika, D.P. Efficacy and safety of solifenacin for overactive bladder: An updated systematic review and meta-analysis. Urol. Ann. 2025, 17, 2–8. [Google Scholar] [CrossRef]
- Iijima, N.; Yamauchi, J.; Yagishita, N.; Araya, N.; Aratani, S.; Tanabe, K.; Sato, T.; Takata, A.; Yamano, Y. Clinical course of neurogenic bladder dysfunction in human T-cell leukemia virus type-1-associated myelopathy/tropical spastic paraparesis: A nationwide registry study in Japan. Orphanet J. Rare Dis. 2021, 16, 355. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, J.; Abrams, P. Alpha-adrenoceptor antagonists in neurogenic lower urinary tract dysfunction. Urology 1999, 53, 21–28. [Google Scholar] [CrossRef]
- Mota, R.d.S.; Macêdo, M.C.; Corradini, S.; Patricío, N.A.; Baptista, A.F.; Sá, K.N. The effect of home exercise on the posture and mobility of people with HAM/TSP: A randomized clinical trial. Arq. Neuro-Psiquiatr. 2020, 78, 149–157. [Google Scholar] [CrossRef]
- Lim, Y.M.; Kim, S.K.; Yoo, D.H.; Kim, H. Effects of assistive technology-based occupational therapy on community-dwelling people recovering from stroke. Assist. Technol. 2022, 34, 273–280. [Google Scholar] [CrossRef]
- Kosse, N.M.; Dutmer, A.L.; Dasenbrock, L.; Bauer, J.M.; Lamoth, C.J. Effectiveness and feasibility of early physical rehabilitation programs for geriatric hospitalized patients: A systematic review. BMC Geriatr. 2013, 13, 107. [Google Scholar] [CrossRef]
- Gascón, M.R.P.; Mellão, M.d.A.; Mello, S.H.; Negrão, R.M.; Casseb, J.; Oliveira, A.C.P.d. The impact of urinary incontinence on the quality of life and on the sexuality of patients with HAM/TSP. Braz. J. Infect. Dis. 2018, 22, 288–293. [Google Scholar] [CrossRef]
- Boa-Sorte, N.; Galvão-Castro, A.V.; Borba, D.; Lima, R.B.N.d.C.; Galvão-Castro, B. HAM/TSP and major depression: The role of age. Braz. J. Infect. Dis. 2015, 19, 314–318. [Google Scholar] [CrossRef] [PubMed]
- Souza, L.S.; Lins-Silva, D.H.; Dorea-Bandeira, I.; Barouh, J.L.; Tolentino, A.; Bandeira, I.D.; Quarantini, L.C. Prevalence and factors associated with depression and anxiety in people living with HTLV-1: A systematic review with meta-analysis and meta-regression. Gen. Hosp. Psychiatry 2021, 73, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Terada, Y.; Miyata, K.; Shoji, N.; Mochizuki, M. Human T-cell Leukemia Virus Type 1 (HTLV-1)-induced Uveitis. Ocul. Immunol. Inflamm. 2023, 31, 1416–1424. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Kamoi, K.; Ando, N.; Kurozumi-Karube, H.; Ohno-Matsui, K. Mechanism of secondary glaucoma development in HTLV-1 uveitis. Front. Microbiol. 2022, 13, 738742. [Google Scholar] [CrossRef] [PubMed]
- Siverio-Llosa, C.; Silva-Ocas, I.; Gálvez-Olórtegui, T.; Arana-Kaik, G. Clinical course of HTLV-1 infection associated intermediate uveitis. Arch. Soc. Esp. Oftalmol. (Engl. Ed.) 2022, 97, 386–390. [Google Scholar] [CrossRef]
- Kamoi, K.; Watanabe, T.; Uchimaru, K.; Okayama, A.; Kato, S.; Kawamata, T.; Kurozumi-Karube, H.; Horiguchi, N.; Zong, Y.; Yamano, Y.; et al. Updates on HTLV-1 uveitis. Viruses 2022, 14, 794. [Google Scholar] [CrossRef] [PubMed]
- Smadja, D.; Bellance, R.; Cabre, P.; Arfi, S.; Vernant, J.C. Clinical characteristics of HTLV-1 associated dermato-polymyositis. Acta Neurol. Scand. 1995, 92, 206–212. [Google Scholar] [CrossRef]
- Martin, F.; Taylor, G.P.; Jacobson, S. Inflammatory manifestations of HTLV-1 and their therapeutic options. Expert Rev. Clin. Immunol. 2014, 10, 1531–1546. [Google Scholar] [CrossRef]
- Araujo, A.Q.; Wedemann, D. HTLV-1 associated neurological complex. What is hidden below the water? Aids Rev. 2019, 21, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Dias, Á.R.N.; Falcão, L.F.M.; Quaresma, J.A.S. An overview of human T-lymphotropic virus Type 1 lung injury. Front. Immunol. 2022, 13, 914498. [Google Scholar] [CrossRef] [PubMed]
- Einsiedel, L.; Chiong, F.; Jersmann, H.; Taylor, G.P. Human T-cell leukaemia virus type 1 associated pulmonary disease: Clinical and pathological features of an under-recognised complication of HTLV-1 infection. Retrovirology 2021, 18, 1. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, A.; Takeda, S.; Kariya, R.; Matsuda, K.; Urano, E.; Okada, S.; Komano, J. A novel therapeutic molecule against HTLV-1 infection targeting provirus. Leukemia 2013, 27, 1621–1627. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, Y.; Hong, W.; Huang, M.; Wu, M.; Zhao, X. Applications of genome editing technology in the targeted therapy of human diseases: Mechanisms, advances and prospects. Signal Transduct. Target. Ther. 2020, 5, 1. [Google Scholar] [CrossRef]
- Zheng, R.; Zhu, X.; Xiao, Y. Advances in CAR-T-cell therapy in T-cell malignancies. J. Hematol. Oncol. 2024, 17, 49. [Google Scholar] [CrossRef]
- Nicolás, D.; Ambrosioni, J.; Paredes, R.; Marcos, M.Á.; Manzardo, C.; Moreno, A.; Miró, J.M. Infection with human retroviruses other than HIV-1: HIV-2, HTLV-1, HTLV-2, HTLV-3 and HTLV-4. Expert Rev. Anti-Infect. Ther. 2015, 13, 947–963. [Google Scholar] [CrossRef]
- Das, C.; Kundu, C.N. Decoding the molecular complexity of viruses in human cancer: Insights into host cell infection, oncogenesis, and therapeutic prospects. Crit. Rev. Microbiol. 2025, 1–24. [Google Scholar] [CrossRef]
- Iino, T.; Hasegawa, A.; Matsutani, T.; Akashi, K.; Kannagi, M.; Suehiro, Y. Elimination of residual adult T-cell leukaemia clones by Tax-targeted dendritic cell vaccine. EJHaem 2025, 6, e1072. [Google Scholar] [CrossRef]
- Nakamura-Hoshi, M.; Ishii, H.; Nomura, T.; Nishizawa, M.; Hau, T.T.T.; Kuse, N.; Okazaki, M.; Ainai, A.; Suzuki, T.; Hasegawa, H.; et al. Prophylactic vaccination inducing anti-Env antibodies can result in protection against HTLV-1 challenge in macaques. Mol. Ther. 2024, 32, 2328–2339. [Google Scholar] [CrossRef]
- Akbarin, M.M.; Rafatpanah, H.; Soleimanpour, S.; Amini, A.A.; Arian, A.; Mosavat, A.; Rezaee, S.A. TAX and HBZ: HFc ɣ 1 proteins as targets for passive immunotherapy. Iran. J. Basic Med Sci. 2022, 25, 586. [Google Scholar]
- Ticona, E.; Huaman, M.A.; Yanque, O.; Zunt, J.R. HIV and HTLV-1 coinfection: The need to initiate antiretroviral therapy. J. Int. Assoc. Provid. AIDS Care (JIAPAC) 2013, 12, 373–374. [Google Scholar] [CrossRef] [PubMed]
- Montano-Castellon, I.; Marconi, C.S.C.; Saffe, C.; Brites, C. Clinical and laboratory outcomes in HIV-1 and HTLV-1/2 coinfection: A systematic review. Front. Public Health 2022, 10, 820727. [Google Scholar] [CrossRef] [PubMed]
- Pilotti, E.; Bianchi, M.V.; De Maria, A.; Bozzano, F.; Romanelli, M.G.; Bertazzoni, U.; Casoli, C. HTLV-1/-2 and HIV-1 co-infections: Retroviral interference on host immune status. Front. Microbiol. 2013, 4, 372. [Google Scholar] [CrossRef] [PubMed]
- Casoli, C.; Pilotti, E.; Bertazzoni, U. Molecular and cellular interactions of HIV-1/HTLV coinfection and impact on AIDS progression. Aids Rev. 2007, 9, 140–149. [Google Scholar] [PubMed]
- Abad-Fernández, M.; Hernández-Walias, F.J.; Ruiz de León, M.J.; Vivancos, M.J.; Pérez-Elías, M.J.; Moreno, A.; Casado, J.L.; Quereda, C.; Dronda, F.; Moreno, S.; et al. HTLV-2 Enhances CD8+ T Cell-Mediated HIV-1 Inhibition and Reduces HIV-1 Integrated Proviral Load in People Living with HIV-1. Viruses 2022, 14, 2472. [Google Scholar] [CrossRef]
| HTLV Type | Region | Key Countries/Areas | Prevalence Range (General Population/Specific Groups) | Data Collection Period |
|---|---|---|---|---|
| HTLV-1 | Southwestern Japan | Kyushu, Okinawa | Blood donors: 1% (Hokkaido) to >6% (Kyushu, Okinawa) | 2006–2016 |
| Sub-Saharan Africa | Gabon, DRC, Nigeria, Ghana, Guinea-Bissau | Adults: 0.3–3%; Older women (Gabon/DRC): 10–25%; Pregnant women (West Africa): 0.2–7.7% | early 2000s–2010s | |
| South America | Peru, Colombia, French Guiana, Brazil | Blood donors (Brazil): 0.04–1% | 2000s–2010s | |
| Caribbean Area | Jamaica, Haiti | Jamaica (mean): 6.1%; Pregnant women (Haiti): 2.2–4.2% | 1990s–2000s | |
| Middle East | Iran (Mashad region) | Adults: 0.77–3% | 2003–2011 | |
| Australo-Melanesia | Central Australia, PNG, Solomon Islands | Aboriginal Australians: up to 44%; Tribes: 1.2–3% | 1990s–2000s | |
| Southeastern USA | Prevalence in blood donors, regional variations | 2007–2015 | ||
| HTLV-2 | Indigenous populations of the Americas | Brazil (Amazon), Panama, USA | Kayapó: up to 41.2%; Native American tribes: up to 13%; Mexico: 0.23% | 2000s–2010s |
| People who inject drugs (PWID) | North America, Europe | Estimated prevalence: 20% (USA) | 1990s–2010s | |
| Some Indigenous people in Africa | Cameroon, DRC (Pygmy populations) | Detected in Pygmy populations | 2000s | |
| USA | Blood donors: HTLV-2 more common than HTLV-1; overall prevalence: 0.016% | 2007–2015 |
| HTLV Type | Mode of Transmission | At-Risk Populations |
|---|---|---|
| HTLV-1 | Mother-to-child transmission | Infants breastfed for prolonged periods by HTLV-1 positive mothers |
| Sexual transmission | Sex partners of HTLV-1 infected individuals, particularly female partners of infected males | |
| Contaminated blood products | Recipients of unscreened blood transfusions or organ transplants | |
| Parenteral transmission | Intravenous drug users sharing needles | |
| Nosocomial transmission | Individuals undergoing medical procedures with inadequately sterilized equipment (suggested in Central Africa) | |
| HTLV-1 | Zoonotic transmission | Individuals with close contact to infected non-human primates, e.g., through bites (Central Africa) |
| General Risk (Endemic Areas) | Residents of highly endemic regions, women (increased prevalence with age), specific ethnic groups (e.g., Aboriginal Australians) | |
| HTLV-2 | Mother-to-child transmission | Infants breastfed by HTLV-2 positive mothers |
| Sexual transmission | Sex partners of HTLV-2 infected individuals | |
| Contaminated blood products | Recipients of unscreened blood transfusions or organ transplants | |
| Parenteral transmission | Intravenous drug users sharing needles | |
| General Risk (Endemic Areas) | Indigenous populations of the Americas, PWID, some Indigenous populations in Africa |
| HTLV Type | Diagnostic Method | Test Type | Interpretation | Clinical Relevance |
|---|---|---|---|---|
| HTLV-1 | Serology (Screening) | ELISA | Detects anti-HTLV-1/2 antibodies; requires confirmation due to cross-reactivity | First-line screening for HTLV infection |
| Confirmatory Serology | Western Blot (WB)/Line Immunoassay (LIA) | Differentiates HTLV-1 from HTLV-2 based on specific viral protein bands | Confirms infection; may yield indeterminate results | |
| Molecular Testing | PCR | Detects HTLV-1 proviral DNA in PBMCs | Essential for confirming serology and diagnosing asymptomatic carriers | |
| Proviral Load Quantification | qPCR | Measures HTLV-1 proviral DNA levels | High proviral load associated with ATL and HAM/TSP | |
| Flow Cytometry | CCR4+ CD4+ T-cell analysis | Evaluates CCR4 expression in ATL cells | Used for prognosis and treatment decisions in ATL | |
| HTLV-2 | Serology (Screening) | ELISA | Detects HTLV-1/2 antibodies; requires differentiation from HTLV-1 | Initial screening test |
| Confirmatory Serology | Western Blot (WB)/Line Immunoassay (LIA) | Differentiates HTLV-2 from HTLV-1 by detecting specific viral proteins | Confirms HTLV-2 infection but may yield indeterminate results | |
| Molecular Testing | PCR | Detects HTLV-2 proviral DNA in PBMCs | Useful for confirmation in serologically indeterminate cases | |
| Proviral Load Quantification | qPCR | Measures HTLV-2 proviral DNA levels | HTLV-2 has lower pathogenicity; routine monitoring is not usually required | |
| HTLV-3/HTLV-4 | Serology | ELISA/Western Blot | Limited availability; assays still under development | Rarely tested in clinical settings due to uncertain pathogenicity |
| Molecular Testing | PCR/Next-Generation Sequencing (NGS) | Identifies HTLV-3/HTLV-4-specific genetic sequences | Used for epidemiological research, not routine diagnosis |
| HTLV Type | Associated Diseases | Diagnostic Methods | Main Symptoms and Clinical Features |
|---|---|---|---|
| HTLV-1 | 1. Malignancies: - ATL: Aggressive CD4+ T-cell malignancy with subtypes (acute, lymphoma, chronic, smoldering). 2. Neuroinflammatory Diseases: - HAM/TSP: Chronic inflammatory demyelinating disorder affecting the spinal cord. 3. Inflammatory & Autoimmune Conditions: - HTLV-1 associated uveitis (HAU): T-cell-mediated intraocular inflammation. - HTLV-1 associated polymyositis: Chronic inflammatory muscle disorder. - HTLV-1 associated arthritis: Immune-mediated joint inflammation. - HTLV-1 associated alveolitis: Interstitial lung disease due to lymphocytic infiltration. - Chronic infectious dermatitis (CID): Severe, recurrent skin infections, particularly in children. | 1. Serological testing: - Enzyme-linked immunosorbent assay (ELISA): Detects anti-HTLV-1 antibodies. - Western Blot: Confirms seropositivity and distinguishes from HTLV-2. 2. Molecular testing: - Polymerase Chain Reaction (PCR): Detects and quantifies HTLV-1 proviral DNA in peripheral blood mononuclear cells (PBMCs). - Southern Blot Analysis: Determines clonal integration in ATL. 3. Histopathology & Imaging: - Bone Marrow Biopsy & Flow Cytometry (for ATL): Detects leukemic infiltration and abnormal CD4+/CD25+ T-cell expansion. - MRI of the Spinal Cord (for HAM/TSP): Shows atrophy, demyelination, and inflammatory changes in the thoracic spinal cord. - Slit-Lamp Biomicroscopy (for HAU): Reveals inflammatory cells in the anterior chamber and vitritis. | 1. ATL (Leukemia/Lymphoma Subtypes): - Generalized lymphadenopathy, hepatosplenomegaly, skin lesions (erythematous plaques, nodules), hypercalcemia-induced nephropathy, lytic bone lesions, opportunistic infections. 2. HAM/TSP (Neurological Syndrome): - Progressive lower limb spasticity and weakness, hyperreflexia, sensory deficits, urinary urgency/incontinence, erectile dysfunction, lumbar pain. 3. HTLV-1 Associated Uveitis: - Blurred vision, floaters, photophobia, eye pain, granulomatous anterior uveitis, optic nerve involvement. 4. Polymyositis/Arthritis/Alveolitis: - Muscle weakness, elevated creatine kinase (CK), interstitial lung fibrosis, chronic inflammatory arthritis with morning stiffness. 5. Chronic Infectious Dermatitis (CID): - Persistent eczematous rash, secondary bacterial/fungal infections. |
| HTLV-2 | Possible but Unconfirmed Disease Associations: - HTLV-2 is not definitively linked to specific malignancies or inflammatory disorders, but it has been implicated in neurological dysfunctions similar to HAM/TSP. - Some studies suggest increased susceptibility to opportunistic infections (e.g., bacterial pneumonia, urinary tract infections). - Possible link to neurodegenerative diseases, but conclusive evidence is lacking. | 1. Serological & molecular testing: - ELISA & Western Blot: Differentiates HTLV-1 from HTLV-2. - PCR: Confirms proviral DNA integration of HTLV-2. 2. Neurological & Immune Function Assessments: - Electromyography (EMG) & Nerve Conduction Studies (NCS): Detects subclinical neuropathies. - Cerebrospinal Fluid (CSF) Analysis: Occasionally shows pleocytosis and elevated IgG index. | 1. Neurological Symptoms (HAM/TSP-like Syndrome): - Gait disturbances, muscle weakness, spasticity, bladder dysfunction, but with a slower and less aggressive progression than HTLV-1-associated HAM/TSP. 2. Immune Dysregulation & Infections: - Recurrent bacterial and viral infections due to immune alterations. - Mild cognitive impairment reported in some cases. |
| HTLV-3 | No confirmed pathogenicity: - HTLV-3 shares genetic similarities with HTLV-1, but no associated diseases have been identified. - Limited epidemiological data due to its recent discovery in Cameroon. | Research-based diagnostic methods: - PCR: Used for detecting HTLV-3 proviral DNA. - No commercially available serological tests. | No known symptoms or clinical manifestations: - Further research is needed to determine its pathogenic potential. |
| HTLV-4 | No known associated diseases: - HTLV-4 was identified in a small number of asymptomatic individuals in Central Africa. - Its clinical significance is currently unknown. | Research-Based Diagnostic Methods: - PCR: Used for viral identification in experimental settings. - No available serological testing. | No known symptoms or disease associations. |
| Therapeutic Area | Strategy | Mechanism and Considerations |
|---|---|---|
| Gene-Editing Therapies | CRISPR-Cas9, ZFNs | Selective excision of HTLV-1 proviral DNA, prevention of oncogenic transformation. Challenges include delivery efficiency and off-target effects. |
| Immunotherapy | PD-1/PD-L1 inhibitors (nivolumab, pembrolizumab), CAR-T therapy | Enhances immune response against ATL but requires optimization to mitigate immune-related toxicity. CAR-T therapy is being developed to target CD4+ ATL cells. |
| Targeted Therapies for ATL | Mogamulizumab (anti-CCR4), lenalidomide, bortezomib | Depletes infected leukemic cells, modulates immune responses, and disrupts NF-B-mediated survival pathways. |
| HAM/TSP Treatment | JAK-STAT inhibitors, IL-15 blockade, neuroprotective agents (minocycline, rapamycin) | Reduces neuroinflammation, slows disease progression, and protects against neuronal damage. |
| Antiviral Therapies | Azacytidine, conventional antiretroviral therapy | Limited efficacy of ART due to HTLV clonal persistence. Azacytidine shows promise in reactivating and eliminating infected cells. |
| HTLV-3 & HTLV-4 Surveillance | Epidemiological studies, genomic sequencing | Investigating potential disease associations and monitoring transmission dynamics in endemic regions. |
| Vaccine Development | mRNA-based vaccines, VLP-based vaccines, therapeutic vaccines (Tax/HBZ-based) | Aims to prevent HTLV-1 infection or enhance immune responses in infected individuals. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Branda, F.; Romano, C.; Pavia, G.; Bilotta, V.; Locci, C.; Azzena, I.; Deplano, I.; Pascale, N.; Perra, M.; Giovanetti, M.; et al. Human T-Lymphotropic Virus (HTLV): Epidemiology, Genetic, Pathogenesis, and Future Challenges. Viruses 2025, 17, 664. https://doi.org/10.3390/v17050664
Branda F, Romano C, Pavia G, Bilotta V, Locci C, Azzena I, Deplano I, Pascale N, Perra M, Giovanetti M, et al. Human T-Lymphotropic Virus (HTLV): Epidemiology, Genetic, Pathogenesis, and Future Challenges. Viruses. 2025; 17(5):664. https://doi.org/10.3390/v17050664
Chicago/Turabian StyleBranda, Francesco, Chiara Romano, Grazia Pavia, Viola Bilotta, Chiara Locci, Ilenia Azzena, Ilaria Deplano, Noemi Pascale, Maria Perra, Marta Giovanetti, and et al. 2025. "Human T-Lymphotropic Virus (HTLV): Epidemiology, Genetic, Pathogenesis, and Future Challenges" Viruses 17, no. 5: 664. https://doi.org/10.3390/v17050664
APA StyleBranda, F., Romano, C., Pavia, G., Bilotta, V., Locci, C., Azzena, I., Deplano, I., Pascale, N., Perra, M., Giovanetti, M., Ciccozzi, A., De Vito, A., Quirino, A., Marascio, N., Matera, G., Madeddu, G., Casu, M., Sanna, D., Ceccarelli, G., ... Scarpa, F. (2025). Human T-Lymphotropic Virus (HTLV): Epidemiology, Genetic, Pathogenesis, and Future Challenges. Viruses, 17(5), 664. https://doi.org/10.3390/v17050664



















