Abstract
Handling cultured isolates and clinical, environmental, or wildlife surveillance samples containing Risk Group 3 and 4 pathogens presents considerable biosafety challenges in minimizing human exposure during processing and transport. Safe handling typically requires high- or maximum-containment facilities, demanding substantial logistical planning and resources. We evaluated PrimeStore Molecular Transport Medium (PS-MTM), a guanidine-based solution created to kill pathogens and preserve nucleic acids at ambient temperatures, for inactivating Crimean-Congo hemorrhagic fever, eastern equine encephalitis, Ebola, Hendra, Japanese encephalitis, Lassa, Marburg, Nipah, Rift Valley fever, and West Nile viruses. To mimic diagnostic conditions, human whole blood spiked with any of these viruses was incubated with PS-MTM for 20-, 30-, or 60-min. Samples with titers up to 107 PFU/mL exposed to PS-MTM at all time points resulted in complete loss of infectivity judged by plaque assays. A 30-min incubation provided a 50% safety margin over the minimum inactivation time and was used for quantification with the tissue culture infectious dose (TCID50) assay, enabling evaluation of PS-MTM’s activity for viruses that do or do not produce well-defined plaques. Results confirmed that PS-MTM inactivated all tested viruses at titers up to 107 TCID50/mL, underscoring its reliability for enhancing biosafety in diagnostics, outbreak management, and surveillance.
1. Introduction
Risk Group 3 and 4 pathogens are associated with severe and often fatal diseases in humans and other animals. These pathogens pose significant biological threats, and many have the potential to cause pandemics. Even under controlled conditions, handling diagnostic samples containing these pathogens presents considerable biosafety risks, particularly during processing and transportation [1]. Safe handling of samples typically requires specialized high- (biosafety level 3 [BSL-3]) or maximum-containment (BSL-4) laboratories [2]. However, establishing and maintaining these facilities requires substantial logistical planning and financial resources, the latter of which can be challenging to sustain even in well-resourced regions [3]. Additionally, many diagnostic workflows rely on maintaining a cold chain from the point of sample collection to the laboratory to preserve sample integrity and to ensure accurate downstream analysis [4]. Implementing and sustaining such systems can be logistically demanding in well-resourced settings and present additional challenges in resource-limited regions. These complexities are particularly evident during active surveillance studies, which are critical for monitoring pathogen circulation in endemic and non-endemic areas [5].
PrimeStore Molecular Transport Medium (PS-MTM), the first Class II microbial nucleic acid storage and stabilization device cleared by the United States (U.S.) Food and Drug Administration (FDA) [6] offers a promising solution to these challenges. This medium is primarily composed of the chaotropic agent guanidine thiocyanate, which denatures proteins, lyses cells and virus particles and inactivates DNases and RNases to prevent nucleic acid degradation [7,8]. PS-MTM inactivates bacteria (e.g., Bacillus subtilis, Escherichia coli, Mycobacterium tuberculosis, Pseudomonas aeruginosa, and Staphylococcus aureus), fungi (e.g., Candida albicans, Aspergillus brasiliensis), and viruses (e.g., eastern equine encephalitis virus [EEEV]/Sindbis virus [SINV] chimera, human adenovirus 5, influenza A virus [FLUAV] H3N2 and H5N1, monkeypox virus [MPXV], and severe acute respiratory syndrome coronavirus 2 [SARS-CoV-2]) at ambient temperatures [8,9,10,11]. PS-MTM has been widely used for the collection, storage, and transport of samples in animal and clinical surveillance studies [12,13,14]. For example, the U.S. Walter Reed Army Institute of Research has used PS-MTM for genomic surveillance of SARS-CoV-2 in NATO/U.S. military camps in Afghanistan, including during the withdrawal of U.S. forces, a period marked by logistical disruptions and the collapse of the cold chain [13]. Similarly, the U.S. Department of Agriculture has used PS-MTM for surveillance of highly pathogenic avian FLUAV in wildlife [15]. Additionally, PS-MTM effectively stabilizes nucleic acids at room temperature, facilitating their use in molecular assays, including PCR and sequencing [8,13,15,16,17].
Although PS-MTM has been shown to be active against some viruses, the inactivation of most viruses listed on the National Institute of Allergy and Infectious Diseases (NIAID) Biodefense Pathogen List [18] remains unassessed [8,9,15]. Pandemic threats from high-consequence viruses (e.g., Crimean-Congo hemorrhagic fever virus [CCHFV], Ebola virus [EBOV], Hendra virus [HeV], Japanese encephalitis virus [JeV], Lassa virus [LASV], Marburg virus [MARV], Nipah virus [NiV], Rift Valley fever virus [RVFV], and West Nile virus [WNV]) remain a major concern. These pathogens are often endemic in resource-limited regions, where finite diagnostic, surveillance, cold chain, and containment infrastructure exacerbate their impact [5,19,20]. Coupled with high case-fatality rates, human-to-human transmission, and emerging zoonotic risks, these challenges present significant barriers to effective disease control. This underscores the importance of approaches that increase diagnostic accuracy, biosafety, and biosecurity for high-consequence pathogens, even in resource-limited settings.
In this study, we evaluated the effectiveness of PS-MTM in inactivating high-consequence pathogens from the virus families Arenaviridae (LASV), Filoviridae (EBOV, MARV, Reston virus [RESTV]), Flaviviridae (JeV, WNV), Nairoviridae (CCHFV), Paramyxoviridae (HeV, NiV), Phenuiviridae (RVFV), and Togaviridae (EEEV) using virus stocks available at the Integrated Research Facility at Fort Detrick (IRF-Frederick). We assessed the activity of PS-MTM in reducing viral infectivity using plaque assays and tissue culture infectious dose assays.
2. Materials and Methods
All work with infectious viruses was conducted in a maximum containment (BSL-4) laboratory.
2.1. Cells and Viruses
2.1.1. Cells
Grivet (Chlorocebus aethiops (Linnaeus, 1758)) epithelial kidney Vero E6 cells (Catalog No. NR-596, BEI Resources, Manassas, VA, USA) and golden hamster (Mesocricetus auratus (Waterhouse, 1839)) kidney BHK-21 fibroblasts (Catalog No. CCL-10, American Culture Collection [ATCC], Manassas, VA, USA [ATCC]) were cultured at 37 °C with 5% carbon dioxide (CO2), whereas human adrenal cortex epithelial SW-13 cells (Catalog No. CCL-105, ATCC) were cultured at 37 °C without extraneous CO2. Vero E6 cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM; Catalog No. 11995040, Gibco, Waltham, MA, USA), BHK-21 cells in Eagle’s Minimum Essential Media (EMEM; Catalog No. 30-2003, ATCC), and SW-13 cells in Leibovitz’s L-15 medium (Catalog No. 11415064, Gibco). All media were supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS; Catalog No. F4135, Millipore Sigma, Burlington, MA, USA) without antibiotics (in accordance with institutional standard operating procedures to maintain aseptic conditions and enable contamination monitoring). Cells were seeded in T25 flasks or T75 flasks for passaging in tissue culture, 6-well plates for plaque assays, and 96-well plates for tissue culture infectious dose (TCID50) assays. Cells were incubated for 24 h prior to use.
2.1.2. Viruses
Crimean-Congo hemorrhagic fever virus isolate IbAr10200 (CCHFV; Internal Reference No. IRF0414) was obtained from the World Reference Collection of Emerging Viruses and Arboviruses (WRCEVA) at the University of Texas Medical Branch at Galveston (UTMB), Galveston, TX, USA. Eastern equine encephalitis virus isolate FL93-939 (EEEV; Internal Reference No. IRF0539), isolated in 1993 from a pool of black-tailed mosquitoes (Culiseta melanura (Coquillett, 1902)) in Florida, USA, was obtained from BEI Resources (Item No. NR-41567). Ebola virus variant Makona isolate C05 (EBOV; Internal Reference No. IRF0137), isolated in 2014 from the blood of a patient [21], was provided by the Public Health Agency of Canada (PHAC), Ottawa, ON, Canada [21]. Reston virus strain Philippines89-AZ-1435 (RESTV; Internal Reference No. IRF0195) was provided by the U.S. Army Medical Research Institute of Infectious Diseases (USAMRIID; Fort Detrick, Frederick, MD, USA). Hendra virus (HeV; Internal Reference No. IRF0359) was provided by USAMRIID. Japanese encephalitis virus isolate Nakayama (JEV), originally isolated in 1935 from the cerebrospinal fluid of a 6-year-old patient, was obtained from BEI Resources (Item No. NR-92). Lassa virus isolate SN61 (LASV; Internal Reference No. IRF0562) was provided by the National Microbiology Laboratory (NML), Winnipeg, Manitoba, Canada. Marburg virus variant Mt. Elgon isolate Musoke (MARV; Internal Reference No. IRF0296) was provided by USAMRIID. Nipah virus Bangladesh isolate 810398 (NiV-B, Internal Reference No. IRF0284), isolated in 2004 from an oropharyngeal swab from a patient in Bangladesh [22], was provided by the Special Pathogens Branch of the U.S. Centers for Disease Control and Prevention (CDC), Atlanta, GA, USA. NiV Malaysia (NiV-M; Internal Reference No. IRF0523) was provided by USAMRIID. Rift Valley fever virus isolate ZH501 (RVFV) was obtained from BEI Resources (Item No. NR-37378). Venezuelan equine encephalitis TC83 virus acquired from BEI Resources (NR-63). West Nile virus isolate 385-99 (WNV) was obtained from WRCEVA.
Table A1 lists all viruses and cell lines used for infection or virus propagation.
2.2. Virus Inactivation with PS-MTM
To better simulate diagnostic conditions, the inactivation process was evaluated on human whole blood spiked with test virus at titers higher or comparable to those found in clinical samples [23,24,25,26,27]. Whole blood from healthy human donors (BioIVT, Westbury, NY, USA) was spiked with CCHFV, EBOV, RESTV, EEEV, HeV, JEV, LASV, MARV, NiV-B, NiV-M, RVFV, or WNV. Non-spiked whole blood and non-spiked whole blood treated with PS-MTM served as negative controls. Samples were treated with PS-MTM (Catalog No. LH-PS-MTM-12, Longhorn Vaccines and Diagnostics, Bethesda, MD, USA) following the manufacturer’s instructions. One part of spiked whole blood or negative control was mixed with three parts of PS-MTM and incubated at room temperature for 20-, 30-, or 60-min (Scheme 1). For each condition, samples derived from the same source material were inactivated in triplicate for the plaque assay and in duplicate for the TCID50 assay.
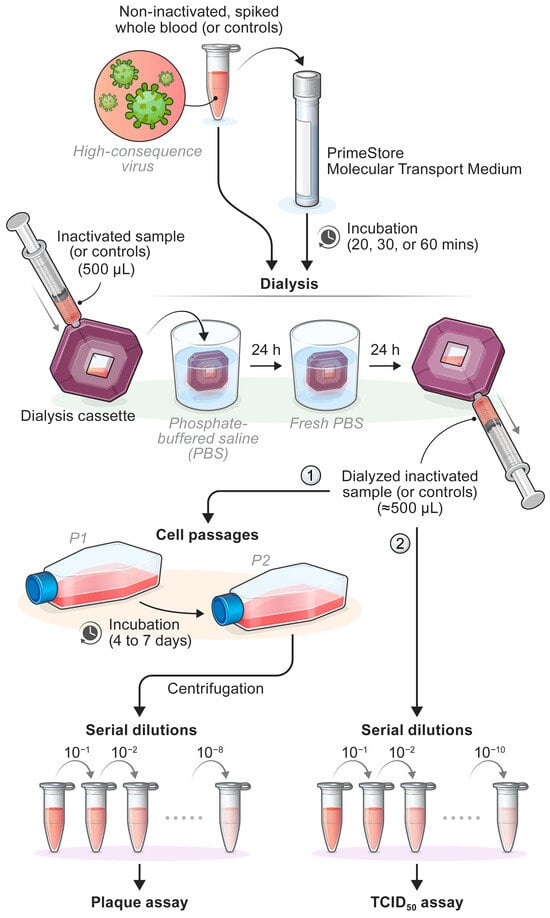
Scheme 1.
Inactivation Procedure for High-Consequence Pathogens using PrimeStore Molecular Transport Media (PS-MTM). ① For safety testing, dialyzed samples were subjected to two blind serial passages in naïve Vero E6 cells prior to serial dilution and plaque assay. ② For TCID50 assays, serial passaging was not performed. Samples were immediately serial-diluted and assessed.
2.3. Evaluation of PS-MTM Removal by Dialysis
To assess virus loss during the removal of the inactivation reagent, virus stocks from representative virus families were dialyzed prior to inactivation with PS-MTM. Testing was performed using VEEV (Togaviridae), RVFV (Phenuiviridae), EBOV (Filoviridae), LASV (Arenaviridae), HeV (Paramyxoviridae), and WNV (Flaviviridae). For each virus, three 500-µL aliquots were dialyzed in phosphate-buffered saline (PBS) using Slide-A-Lyzer dialysis cassettes with a molecular weight cutoff of 20 kDa (Catalog No. 66005, Thermo Fisher Scientific, Waltham, MA, USA). Dialysis was performed at room temperature for 24 h, after which PBS was replaced with fresh buffer, and dialysis continued for an additional 24 h. The final volume (≈500 µL) from each replicate was collected and used to inoculate cells for plaque assays to evaluate viral titers after dialysis. Undialyzed aliquots from each virus were used to inoculate cells for plaque assays for viral titer comparison. Comparisons of paired data (pre-dialysis vs. post-dialysis) were performed using the Wilcoxon matched-pairs signed-rank test (two-tailed) in GraphPad Prism (version 10.2.0 for Windows, GraphPad Software, Boston, MA, USA).
2.4. Sample Dialysis and Serial Passaging for Safety Testing
After evaluating potential virus loss during dialysis, inactivated samples, the positive control (non-inactivated spiked whole blood), and PS-MTM alone were dialyzed in PBS to remove residual detergent and minimize cytotoxicity. As described above, each 500 µL sample was dialyzed using Slide-A-Lyzer dialysis cassettes with a molecular weight cutoff of 20 kDa. Dialysis was performed at room temperature for 24 h, followed by PBS replacement and an additional 24 h dialysis. The final volume (≈500 µL) was collected and used to inoculate cells for the validation of chemical inactivation via plaque assay or TCID50 assay (Scheme 1).
For safety testing by plaque assay, dialyzed samples underwent two blind serial passages on naïve cultured Vero E6 cells. In contrast, serial passaging was not required for the TCID50 assay. Cells were incubated for 4 (EEEV-FL93-939, HeV, RVFV-ZH501, and WNV-385-99), 6 (LASV), or 7 d (EBOV). Following the first passage, the cell culture supernatants and monolayers from each T25 flask were collected and clarified by centrifugation. The entire clarified supernatants were then transferred to fresh T75 flasks containing Vero E6 cells for the second passage. These were incubated under the same conditions, and the resulting clarified supernatants (900 µL) were used for plaque assays (Scheme 1).
2.5. Plaque Assays
Vero E6 cells were seeded in 6-well plates 24 h before use and subsequently transferred to the BSL-4 laboratory. A 2.5% Avicel 591 overlay was prepared by mixing Avicel PH-101 cellulose resin (Catalog No. 11365-1KG, Millipore Sigma) with 2× Temin’s Modified Eagle Medium (Catalog No. 11935046, Gibco, Waltham, MA, USA), supplemented with 4% heat-inactivated FBS (v/v) and 2% penicillin-streptomycin (w/v) in a 1:1 ratio to achieve a final 1X concentration. Samples were serially diluted 10-fold in DMEM supplemented with 2% heat-inactivated FBS (v/v) and 1.06% penicillin-streptomycin (w/v) with a dilution range of 10−1–10−8 (Scheme 1). Cell culture supernatants from the 6-well plates were removed, and 300 μL of diluted samples were added to wells in triplicate. Plates were incubated at 37 °C with 5% CO2 for 1 h and gently rocked every 15 min. After incubation, 2 mL of Avicel 591 overlay were added to each well, and plates were incubated for 4 (EEEV, HeV, RVFV, and WNV), 6 (LASV), or 7 d (EBOV). After incubation, overlays were discarded, and cells were stained for 30 min with a 0.2% crystal violet solution (2% Gentian Violet [Catalog No. 3233, Ricca Chemical Company, Arlington, TX, USA] prepared in 10% neutral buffered formalin [Catalog No. 16004-126, VWR, Radnor, PA, USA]). Plaques were visually counted immediately after staining, and their numbers were recorded.
2.6. TCID50 Assays
Vero E6, BHK-21, and SW-13 cells were seeded in 96-well plates 24 h before use and subsequently transferred to the BSL-4 laboratory. Ten-fold serial dilutions of the inactivated samples, as well as positive and negative controls, were prepared using a cell culture medium (as described in the Section 2.1), supplemented with 2% heat-inactivated FBS (v/v). Dilutions (range of 10−1–10−10) were added to wells in triplicate (100 µL per well) for each technical replicate (Scheme 1). Non-spiked whole blood and non-spiked whole blood treated with PS-MTM were included on each plate.
Plates were incubated at 37 °C with 5% CO2 for 1 h and gently rocked every 15 min; SW-13 cells were incubated at 37 °C without extraneous CO2. After incubation, 100 μL of DMEM, supplemented with 2% heat-inactivated FBS, were added to wells. Then, plates were incubated for 4 (JEV, NiV-B, and NiV-M), 5 (HeV), 6 (RVFV), 7 (CCHFV), 10 (EBOV and MARV), or 13 d (RESTV).
Following incubation, cells were stained with 100 μL of 0.4% crystal violet solution for 30 min at room temperature. Plates were then washed twice with deionized water and air dried. TCID50s were calculated using the Reed–Munch method [28].
2.7. Statistical Analysis
Statistical comparisons of control and treatment groups were not performed due to the absence of detectable variability in plaque assay or TCID50 values for treatment groups, which consistently did not result in cytopathic effects (CPE) or infectivity.
3. Results
3.1. Plaque Assay Confirms That PS-MTM Effectively Inactivates High-Consequence Pathogens After Incubation for 20, 30, and 60 Min
To determine the time required for PS-MTM to fully inactivate high-consequence pathogens and evaluate its effectiveness in reducing viral infectivity, representative viruses from the Arenaviridae, Filoviridae, Flaviviridae, Paramyxoviridae, Phenuiviridae, and Togaviridae families were treated with PS-MTM for 20-, 30- and 60-min, and viral infectivity was subsequently assessed using plaque assays. Inactivation was evaluated in the presence of whole blood to better simulate diagnostic sample conditions. Prior to each inactivation, the initial viral concentration was determined to establish the maximum starting titer (Table 1). Virus-spiked whole blood samples were then treated with PS-MTM, whereas non-spiked whole blood negative controls and spiked whole blood positive controls were left untreated. All samples and controls were dialyzed before being assessed for infectivity by plaque assay.

Table 1.
Maximum starting viral titer as determined by plaque assay.
All tested samples were fully inactivated following PS-MTM treatment. No viable virus was detected at 20-, 30-, or 60-min post-inactivation for any of the viruses tested (Table 2). For the positive controls, CPE was evident; however, for LASV-, no CPE was observed, despite detectable infectivity by plaque assay.

Table 2.
Viral titers of PS-MTM-inactivated samples and controls obtained by plaque assay.
3.2. Dialysis Does Not Significantly Reduce Virus Titer
To determine whether dialysis affected viral recovery, high-titer virus stocks from each representative family were dialyzed in PBS prior to inactivation. Plaque assays performed on pre- and post-dialysis samples showed minimal reduction in virus titers, indicating that the dialysis procedure itself did not contribute to a significant loss of viral infectivity (Table A2). Moreover, no cytopathic effects were observed in wells inoculated with dialyzed PS-MTM controls, indicating minimal residual toxicity (Table 2). These findings support the validity of subsequent inactivation assays conducted after dialysis.
3.3. TCID50 Assay Confirms Complete Inactivation of High-Consequence Pathogens by PS-MTM
For viruses, such as CCHFV that do not produce observable plaques [29] or HeV and NiV that may produce poorly defined plaques due to syncytia formation [30], TCID50 assays were used as an alternative method to quantify infectious virus particles. Furthermore, TCID50 assays were used with other viruses at the IRF-Frederick known to form well-defined plaques to comprehensively evaluate its utility in titrating high-consequence pathogens (EBOV, RESTV, MARV, JEV, RVFV). Although a plaque assay was successfully performed on LASV, the TCID50 assay was not able to be used for this virus due to the absence and/or inconsistency of CPE, which is required as an endpoint for the TCID50 assay. Thus, we relied on the plaque assay to quantify the viral infectivity of LASV.
The ability of PS-MTM to fully reduce viral infectivity was evaluated after a 30-min incubation. This duration was chosen based on previous studies demonstrating that a 30-min incubation effectively inactivated a representative subset of viruses and provided a 50% safety margin over the minimum inactivation time determined by plaque assay (Table 2). Although not all viruses were tested at 20 and 60 min, the 30-min incubation was selected to standardize the protocol and ensure consistency across experiments.
For these subsequent experiments, inactivation was assessed directly by TCID50 assay following dialysis rather than by serial passaging and plaque assay, which had already been used to confirm complete inactivation for representative viruses from each viral family. An exception was CCHFV, the representative member of the Nairoviridae family, for which serial passaging was not performed.
Results from the TCID50 assay showed that PS-MTM fully inactivated CCHFV, EBOV, RESTV, HeV, JEV, MARV, NiV-B, NiV-M, and RVFV in whole blood within 30 min (Figure 1). Post-inactivation, no CPE was observed, and virus titers were reduced by 5–7 log units. Although the observed inactivation of CCHFV supports PS-MTM’s broader efficacy, further validation using serial passaging is needed to fully confirm inactivation for nairovirids.
Figure 1.
Inactivation of High-Consequence Pathogens by PrimeStore Molecular Transport Media (PS-MTM) at 30 min. Bar graphs represent the mean log10 TCID50/mL from duplicates, with the error bars indicating the 95% confidence intervals. CCHFV, Crimean-Congo hemorrhagic fever virus; EBOV, Ebola virus; HeV, Hendra virus; JEV, Japanese encephalitis virus; MARV, Marburg virus; NiV-B, Nipah virus Bangladesh; NiV-M, Nipah virus Malaysia; RESTV, Reston virus; RVFV, Rift Valley fever virus; TCID50, tissue culture infectious dose.
4. Discussion
Handling high-consequence pathogens in a BSL-2 environment requires an inactivation step to ensure biosafety [31]. Several methods have been described for enveloped viruses, including heat inactivation, ultraviolet radiation, lipid solvents, and guanidine-based lysis solutions [32,33,34,35,36,37]. The choice of method depends on whether the goal is to preserve protein structure (e.g., for serological assays) or to maintain nucleic acid integrity (e.g., for molecular assays). PS-MTM is a guanidine-thiocyanate-based solution with ethanol and N-lauroylsarcosine sodium salt, specifically designed to inactivate pathogens while stabilizing nucleic acids [7]. Traditional guanidine-thiocyanate-based solutions, such as AVL buffer (Qiagen) and TRIzol LS reagent (Invitrogen), are widely used for pathogen inactivation and RNA extraction but differ in long-term stabilization [32]. AVL buffer, particularly when combined with ethanol, effectively inactivates pathogens but is not suitable for long-term storage, requiring refrigeration or freezing (−20 or −80 °C) to prevent RNA degradation [38,39]. Similarly, TRIzol LS is not designed for long-term stability at ambient temperature and requires frozen storage [40]. In contrast, PS-MTM stabilizes RNA and DNA at ambient temperatures for up to 30 d, thereby eliminating the need for refrigeration or freezing [15,41,42].
This study demonstrates that PS-MTM effectively inactivates high-consequence representative pathogens from the virus families Arenaviridae, Filoviridae, Flaviviridae, Nairoviridae, Paramyxoviridae, Phenuiviridae, and Togaviridae, within 30 min of incubation in whole blood. Inactivation was confirmed at the highest tested titers (104–107 TCID50/mL or PFU/mL) by TCID50 and plaque assays. These titers were comparable to or exceed those reported in clinical samples for some of the pathogens, such as EBOV (up to 1 × 107 PFU/mL in serum) [23,24], LASV (up to 2.5 × 105 PFU/mL in cerebrospinal fluid after serial passage) [27], MARV (up to 1 × 107 TCID50/mL in whole blood and bodily fluids, based on cycle threshold (Ct) conversions) [25], and RVFV (up to 1 × 106 TCID50/mL in whole blood) [26]. These findings highlight PS-MTM’s ability to inactivate viruses even at concentrations reflective of severe infections. For some pathogens, such as EEEV, HeV, JEV, and NiV, data on viral loads in human clinical samples are scarce, limiting direct comparisons. Most available information comes from in vitro and/or in vivo experimental infection studies [43,44,45,46,47], whereas, for JEV, diagnostics primarily relies on detecting virus-specific antibodies or RNA rather than quantifying viral titers [48,49]. For CCHFV, viral loads are typically reported in copies per mL rather than infectious titers, with values reaching up to 1.3 × 1010 copies per mL in sera, depending on disease severity [50,51]. Since only a fraction of virus particles are replication-competent, these values likely overestimate infectious titers, making direct comparisons challenging [52]. Nonetheless, the infectious titers measured in this study likely represent biologically relevant levels, reinforcing the utility of PS-MTM in diagnosis and surveillance. Future studies could incorporate low-titer spiked controls to empirically determine assay sensitivity.
In addition to demonstrating effective viral inactivation, an important consideration for chemical agents such as PS-MTM is their potential cytotoxicity, which can interfere with the accurate assessment of infectivity. PS-MTM contains chaotropic agents that are cytotoxic to cells if not diluted or adequately removed [8,15]. To reduce cytotoxicity prior to infectivity assays, we used a two-step dialysis procedure in which virus-containing samples were placed in dialysis cassettes and submerged in PBS. Consequently, low molecular weight compounds, such as residual inactivation reagents, could diffuse out through the membrane while virus particles were retained. The absence of CPE in the dialyzed whole-blood and PS-MTM controls, combined with the ability to detect viruses in non-inactivated and pre-dialyzed samples, supports the effectiveness of this purification method. Previous studies have demonstrated that cytotoxic components can be effectively removed using methods such as dialysis, dilution of the inactivating reagent, extensive washing steps, or size-exclusion spin column filtration [10,15,36,53,54,55,56]. Herein, the dialysis method used aligns with these studies and with safety testing procedures at our facility. To expand PS-MTM use for diagnostic or research applications, future studies could directly compare purification methods and quantify residual cytotoxicity. Additionally, although our current experimental design confirmed the removal of cytotoxicity and the ability of cells to support virus replication post dialysis, we did not include a control in which infectious virus was spiked into PS-MTM-treated whole blood samples after dialysis. This control would help confirm that residual PS-MTM components, even after purification, did not interfere with virus entry or replication. Including this type of control in future experiments would further strengthen confidence in assay sensitivity.
Overall, this study demonstrates PS-MTM’s ability to inactivate high-consequence pathogens, but several areas warrant further investigation to fully understand its potential and optimize its use. For example, future studies should assess PS-MTM’s inactivation efficacy against additional Risk Group 3 and 4 priority pathogens, including bacteria, fungi, and toxins. Additionally, the inclusion of more diverse sample matrices, such as cerebrospinal fluid, oral swabs, seminal fluid, or tissue homogenates, would provide greater insight into its versatility.
One key area for future research is the evaluation of PS-MTM’s potency. Determining the minimum inactivation concentration (MIC) of PS-MTM or the concentration required to inactivate 50% of viral infectivity (IC50) in samples with higher viral titers would provide valuable insights into the efficiency of PS-MTM under varying conditions. This is particularly critical for optimizing the sample-to-PS-MTM ratio in scenarios in which precious or cultured samples contain limited copies of target genetic material that may fall below detectable levels if diluted. At the current 1:3 ratio, the target material may be considerably diluted. Furthermore, the identification of the maximum viral concentration that can be completely inactivated at the recommended sample-to-PS-MTM ratio and exposure time would strengthen its applicability in both clinical and research settings.
Another important focus should be the assessment of how long preserved nucleic acids maintain integrity at ambient temperatures for sequencing or other advanced applications. Additionally, the stability of host genetic material in PS-MTM-treated samples presents an important avenue for exploration. Investigating whether host nucleic acids remain intact could enable the use of inactivated samples for RNA sequencing and facilitate the identification of host-specific genomic markers associated with specific pathologies. This would expand the potential applications of PS-MTM beyond pathogen detection to include studies on host-pathogen interactions and disease mechanisms. These studies would not only establish PS-MTM’s broad applicability but also optimize its usage across diverse diagnostic and research workflows.
5. Conclusions
The effective inactivation of representative high-consequence pathogens from the Arenaviridae, Filoviridae, Flaviviridae, Nairoviridae, Paramyxoviridae, Phenuiviridae, and Togaviridae families by PS-MTM has significant implications for public health, biosafety, diagnostics, and research. By mitigating the risk of accidental exposure during the handling and transportation of high-risk biological samples, PS-MTM enhances laboratory safety and facilitates safer diagnostic workflows. This capability is particularly beneficial in field studies and resource-limited settings, where high- or maximum-containment facilities may not be accessible. Furthermore, PS-MTM contributes to advancing global surveillance of high-consequence pathogens, strengthening pandemic preparedness and supporting infectious disease research. Enabling safer diagnostic practices also accelerates the development of countermeasures against potential bioterrorism threats and improves outbreak response capabilities.
Author Contributions
J.H.K., G.F., I.M.B., P.C. and J.T. conceived the study and provided project organization and supervision of the work. P.C., G.K., D.F., N.M. and K.A. contributed to the design of and executed the experiments, and B.S.-H., P.C. and M.B. performed data analyses. B.S.-H. was primarily responsible for drafting the manuscript, while M.B. contributed to writing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded in part through a Laulima Government Solutions, LLC, prime contract with the National Institute of Allergy and Infectious Diseases (Contract No. HHSN272201800013C). B.S.-H., P.C., G.K., M.B., D.F., N.M., K.A. and I.M.B. performed this work as employees or affiliates of Laulima Government Solutions, LLC. J.H.K. performed this work as an employee of Tunnell Government Services (TGS), a subcontractor of Laulima Government Solutions, LLC, under Contract No. HHSN272201800013C. The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the U.S. Department of Health and Human Services or of the institutions and companies affiliated with the authors, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Institutional Review Board Statement
Work with infectious viruses at the NIH NIAID DCR IRF-Frederick was approved by the Institutional Biosafety Committee (IBC), code REQ0033892. The related pathogen registration numbers are: Alphaviruses: 24-BMC-147; Arenaviruses: 24-BMC-211; Bunyaviruses: 25-BMC-040; Filoviruses: 4905; Flaviviruses: 5797; Paramyxoviruses: 24-BMC-131.
Informed Consent Statement
Human whole blood samples were obtained from BioIVT with appropriate ethical approval and informed consent for research use. Samples were de-identified before distribution.
Data Availability Statement
All raw data are available upon request.
Acknowledgments
The authors thank Longhorn Vaccines and Diagnostics, LLC, for providing PrimeStore MTM. The authors also acknowledge Anya Crane (National Institute of Allergy and Infectious Disease) for editorial revisions of the manuscript.
Conflicts of Interest
G.F. has a financial (ownership) interest in, and J.T. is employed by, Longhorn Vaccines and Diagnostics, LLC, which developed and markets PS-MTM, the subject of this study. Neither participated in the experimental procedures, data collection or data analysis. All other authors have disclosed any potential conflicts, and the study design, data interpretation and conclusions were conducted with scientific integrity and without influence from financial or proprietary considerations.
Abbreviations
The following abbreviations are used in this manuscript:
| ATCC | American Type Culture Collection |
| BSL | biosafety level |
| CCHFV | Crimean-Congo hemorrhagic fever virus |
| CDC | Centers for Disease Control and Prevention |
| CO2 | carbon dioxide |
| CPE | cytopathic effect |
| Ct | cycle threshold |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| EBOV | Ebola virus |
| EEEV | Eastern equine encephalitis virus |
| EMEM | Eagle’s Minimum Essential Media |
| FBS | fetal bovine serum |
| FDA | Food and Drug Administration |
| FLUAV | influenza A virus |
| HeV | Hendra virus |
| IC50 | concentration required to inactivate 50% of viral infectivity |
| IRF-Frederick | Integrated Research Facility at Fort Detrick |
| JEV | Japanese encephalitis virus |
| LASV | Lassa virus |
| MARV | Marburg virus |
| MIC | minimum inactivation concentration |
| MPXV | monkeypox virus |
| MRSA | methicillin-resistant Staphylococcus aureus |
| NIAID | National Institute of Allergy and Infectious Diseases |
| NiV | Nipah virus |
| NiV-B | Nipah virus Bangladesh |
| NiV-M | Nipah virus Malaysia |
| NML | National Microbiology Laboratory |
| PBS | phosphate-buffered saline |
| PHAC | Public Health Agency of Canada |
| qPCR | real-time polymerase chain reaction |
| RESTV | Reston virus |
| RVFV | Rift Valley fever virus |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus 2 |
| SINV | Sindbis virus |
| TCID50 | tissue culture infectious dose |
| U.S. | United States |
| USAMRIID | U.S. Army Medical Research Institute of Infectious Diseases |
| USDA | U.S. Department of Agriculture |
| UTMB | University of Texas Medical Branch at Galveston |
| WNV | West Nile virus |
| WRCEVA | World Reference Collection of Emerging Viruses and Arboviruses |
Appendix A

Table A1.
List of viruses used in the study and the cell lines used for infection or propagation.
Table A1.
List of viruses used in the study and the cell lines used for infection or propagation.
| Virus | Strain/Variant/Isolate | Cell Line | Reference GenBank Accession Number |
|---|---|---|---|
| CCHFV | IbAr10200 | SW-13 | PQ463984, PQ463983, PQ463982 |
| EBOV | Makona-C05 | Vero E6 | KX000400 |
| RESTV | Philippines89-AZ-1435 | Vero E6 | KY008770 |
| EEEV | FL93-939 | Vero E6 | EF151502 |
| HeV | unnamed | Vero E6 | AF017149 |
| JEV | Nakayama | BHK-21 | EF571853 |
| LASV | SN61 | Vero E6 | MZ169798, MZ169799 |
| MARV | Musoke | Vero E6 | AY430365 |
| NiV-M | IRF523 | Vero E6 | PQ463988 |
| NiV-B | 810398 | Vero E6 | MK673564 |
| RVFV | ZH501 | Vero E6 | PQ463985, PQ463986, PQ463986 |
| WNV | 385-99 | Vero E6 | AY842931 |
CCHFV, Crimean-Congo hemorrhagic fever virus; EBOV, Ebola virus; EEEV, eastern equine encephalitis virus; HeV, Hendra virus; JEV, Japanese encephalitis virus; LASV, Lassa virus; MARV, Marburg virus; NiV-B, Nipah virus Bangladesh; NiV-M, Nipah virus Malaysia; RESTV, Reston virus; RVFV, Rift Valley fever virus; WNV, West Nile virus.

Table A2.
Viral titer loss during inactivation reagent removal.
Table A2.
Viral titer loss during inactivation reagent removal.
| Samples | Average Titer (Log10 PFU/mL) | Reduction from Initial Titer (Log10 PFU/mL) | Sum of Signed Ranks (p-Value) 1 |
|---|---|---|---|
| EBOV initial titration | 7.32 | N/A | N/A |
| EBOV after dialysis 1 | 7.25 | 0.07 | −6.00 (p = 0.25) |
| EBOV after dialysis 2 | 7.23 | 0.09 | −6.00 (p = 0.25) |
| EBOV after dialysis 3 | 7.24 | 0.08 | −6.00 (p = 0.25) |
| HeV initial titration | 6.95 | N/A | N/A |
| HeV after dialysis 1 | 7.01 | −0.06 * | 6.00 (p = 0.25) |
| HeV after dialysis 2 | 6.98 | −0.03 * | 3.00 (p = 0.50) |
| HeV after dialysis 3 | 6.90 | 0.05 | −4.00 (p = 0.50) |
| LASV initial titration | 6.92 | N/A | N/A |
| LASV after dialysis 1 | 6.89 | 0.03 | −1.00 (p > 0.99) |
| LASV after dialysis 2 | 6.82 | 0.10 | −6.00 (p = 0.25) |
| LASV after dialysis 3 | 6.72 | 0.20 | −6.00 (p = 0.25) |
| RVFV initial titration | 6.81 | N/A | N/A |
| RVFV after dialysis 1 | 6.58 | 0.23 | −6.00 (p = 0.25) |
| RVFV after dialysis 2 | 6.61 | 0.20 | −6.00 (p = 0.25) |
| RVFV after dialysis 3 | 6.59 | 0.22 | −6.00 (p = 0.25) |
| VEEV initial titration | 8.87 | N/A | N/A |
| VEEV after dialysis 1 | 8.85 | 0.02 | −4.00 (p = 0.50) |
| VEEV after dialysis 2 | 8.78 | 0.09 | −6.00 (p = 0.25) |
| VEEV after dialysis 3 | 8.83 | 0.03 | −2.00 (p = 0.75) |
| WNV initial titration | 7.96 | N/A | N/A |
| WNV after dialysis 1 | 7.98 | −0.02 * | 1.00 (p > 0.99) |
| WNV after dialysis 2 | 7.83 | 0.13 | −6.00 (p = 0.25) |
| WNV after dialysis 3 | 7.82 | 0.14 | −6.00 (p = 0.25) |
* Negative reduction indicates an increase in titer. 1 Wilcoxon matched-pairs signed rank test statistic and two-tailed p-value are reported. Each group was compared to the initial viral titers prior to dialysis. EBOV, Ebola virus; HeV, Hendra virus; LASV, Lassa virus; RVFV, Rift Valley fever virus; VEEV, Venezuelan equine encephalitis virus; WNV, West Nile virus.
References
- Ravnholdt, A.R.; Ratnesar-Shumate, S.A.; Santarpia, J.L. A review of accidental aerosol generation in laboratories and laboratory-associated infections. Appl. Biosaf. 2024. [Google Scholar] [CrossRef]
- U.S. Department of Health and Human Services; Public Health Service; Centers for Disease Control and Prevention; National Institutes of Health. Biosafety in Microbiological and Biomedical Laboratories (BMBL) 6th Edition. 2020. Available online: https://www.cdc.gov/labs/pdf/SF__19_308133-A_BMBL6_00-BOOK-WEB-final-3.pdf (accessed on 16 February 2025).
- Yeh, K.B.; Tabynov, K.; Parekh, F.K.; Mombo, I.; Parker, K.; Tabynov, K.; Bradrick, S.S.; Tseng, A.S.; Yang, J.-R.; Gardiner, L.; et al. Significance of high-containment biological laboratories performing work during the COVID-19 pandemic: Biosafety level-3 and -4 labs. Front. Bioeng. Biotechnol. 2021, 9, 720315. [Google Scholar] [CrossRef]
- Dsa, O.C.; Kadni, T.S.; N, S. From cold chain to ambient temperature: Transport of viral specimens—A review. Ann. Med. 2023, 55, 2257711. [Google Scholar] [CrossRef]
- Mukadi-Bamuleka, D.; Mambu-Mbika, F.; de Weggheleire, A.; Edidi-Atani, F.; Bulabula-Penge, J.; Mfumu, M.M.K.; Legand, A.; Nkuba-Ndaye, A.; N’kasar, Y.T.T.; Mbala-Kingebeni, P.; et al. Efficiency of field laboratories for Ebola virus disease outbreak during chronic insecurity, eastern Democratic Republic of the Congo, 2018–2020. Emerg. Infect. Dis. 2023, 29, e221025. [Google Scholar] [CrossRef]
- Longhorn Vaccines and Diagnostics. FDA De Novo Clearance: The Original FDA-Cleared Inactivation Media. Available online: https://www.lhnvd.com/product-development (accessed on 16 February 2025).
- Longhorn Vaccines and Diagnostics. Guidelines for the Safe and Appropriate Use of PrimeStore® MTM. Available online: https://www.lhnvd.com/safety-information (accessed on 16 February 2025).
- Daum, L.T.; Worthy, S.A.; Yim, K.C.; Nogueras, M.; Schuman, R.F.; Choi, Y.W.; Fischer, G.W. A clinical specimen collection and transport medium for molecular diagnostic and genomic applications. Epidemiol. Infect. 2011, 139, 1764–1773. [Google Scholar] [CrossRef] [PubMed]
- UK Health Security Agency. Monkeypox Virus Inactivation Testing Report: PrimeStore Molecular Transport Medium. 2022. Available online: https://assets.publishing.service.gov.uk/media/630289058fa8f5387723df9e/HCM-MPx-002-v3_PrimeStore_Molecular_Transport_Medium.pdf (accessed on 16 February 2025).
- Welch, S.R.; Davies, K.A.; Buczkowski, H.; Hettiarachchi, N.; Green, N.; Arnold, U.; Jones, M.; Hannah, M.J.; Evans, R.; Burton, C.; et al. Analysis of inactivation of SARS-CoV-2 by specimen transport media, nucleic acid extraction reagents, detergents, and fixatives. J. Clin. Microbiol. 2020, 58, e01713–e01720. [Google Scholar] [CrossRef] [PubMed]
- Daum, L.T.; Choi, Y.; Worthy, S.A.; Rodriguez, J.D.; Chambers, J.P.; Fischer, G.W. A molecular transport medium for collection, inactivation, transport, and detection of Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 2014, 18, 847–849. [Google Scholar] [CrossRef]
- Schlaudecker, E.P.; Heck, J.P.; MacIntyre, E.T.; Martinez, R.; Dodd, C.N.; McNeal, M.M.; Staat, M.A.; Heck, J.E.; Steinhoff, M.C. Comparison of a new transport medium with universal transport medium at a tropical field site. Diagn. Microbiol. Infect. Dis. 2014, 80, 107–110. [Google Scholar] [CrossRef]
- Maljkovic Berry, I.; Hang, J.; Fung, C.; Yang, Y.; Chibucos, M.; Pollio, A.; Gandhi, J.; Li, T.; Conte, M.A.; Lidl, G.M.; et al. Genomic surveillance of SARS-CoV-2 in US military compounds in Afghanistan reveals multiple introductions and outbreaks of Alpha and Delta variants. BMC Genom. 2022, 23, 513. [Google Scholar] [CrossRef]
- Daum, L.T.; Peters, R.P.; Fourie, P.B.; Jonkman, K.; Worthy, S.A.; Rodriguez, J.D.; Ismail, N.A.; Omar, S.V.; Fischer, G.W. Molecular detection of Mycobacterium tuberculosis from sputum transported in PrimeStore® from rural settings. Int. J. Tuberc. Lung Dis. 2015, 19, 552–557. [Google Scholar] [CrossRef][Green Version]
- Welch, J.L.; Shrestha, R.; Hutchings, H.; Pal, N.; Levings, R.; Robbe-Austerman, S.; Palinski, R.; Shanmuganatham, K.K. Inactivation of highly transmissible livestock and avian viruses including influenza A and Newcastle disease virus for molecular diagnostics. Front. Vet. Sci. 2024, 11, 1304022. [Google Scholar] [CrossRef] [PubMed]
- Zeineldin, M.M.; Lehman, K.; Camp, P.; Farrell, D.; Thacker, T.C. Diagnostic evaluation of the IS1081-targeted real-time PCR for detection of Mycobacterium bovis DNA in bovine milk samples. Pathogens 2023, 12, 972. [Google Scholar] [CrossRef]
- Pikalo, J.; Deutschmann, P.; Fischer, M.; Roszyk, H.; Beer, M.; Blome, S. African swine fever laboratory diagnosis—Lessons learned from recent animal trials. Pathogens 2021, 10, 177. [Google Scholar] [CrossRef]
- National Institute of Allergy and Infectious Diseases. NIAID Biodefense Pathogens. 2024. Available online: https://www.niaid.nih.gov/research/niaid-biodefense-pathogens (accessed on 16 February 2025).
- Kennedy, S.B.; Dogba, J.B.; Wasunna, C.L.; Sahr, P.; Eastman, C.B.; Bolay, F.K.; Mason, G.T.; Kieh, M.W.S. Pre-Ebola virus disease laboratory system and related challenges in Liberia. Afr. J. Lab. Med. 2016, 5, 508. [Google Scholar] [CrossRef] [PubMed]
- Shannon, F.Q., 2nd; Bawo, L.L.; Crump, J.A.; Sharples, K.; Egan, R.; Hill, P.C. Evaluation of Ebola virus disease surveillance system capability to promptly detect a new outbreak in Liberia. BMJ Glob. Health 2023, 8, e012369. [Google Scholar] [CrossRef]
- Baize, S.; Pannetier, D.; Oestereich, L.; Rieger, T.; Koivogui, L.; Magassouba, N.F.; Soropogui, B.; Sow, M.S.; Keïta, S.; de Clerck, H.; et al. Emergence of Zaire Ebola virus disease in Guinea. N. Engl. J. Med. 2014, 371, 1418–1425. [Google Scholar] [CrossRef]
- Harcourt, B.H.; Lowe, L.; Tamin, A.; Liu, X.; Bankamp, B.; Bowden, N.; Rollin, P.E.; Comer, J.A.; Ksiazek, T.G.; Hossain, M.J.; et al. Genetic characterization of Nipah virus, Bangladesh, 2004. Emerg. Infect. Dis. 2005, 11, 1594–1597. [Google Scholar] [CrossRef]
- Towner, J.S.; Rollin, P.E.; Bausch, D.G.; Sanchez, A.; Crary, S.M.; Vincent, M.; Lee, W.F.; Spiropoulou, C.F.; Ksiazek, T.G.; Lukwiya, M.; et al. Rapid diagnosis of Ebola hemorrhagic fever by reverse transcription-PCR in an outbreak setting and assessment of patient viral load as a predictor of outcome. J. Virol. 2004, 78, 4330–4341. [Google Scholar] [CrossRef] [PubMed]
- Cross, R.W.; Boisen, M.L.; Millett, M.M.; Nelson, D.S.; Oottamasathien, D.; Hartnett, J.N.; Jones, A.B.; Goba, A.; Momoh, M.; Fullah, M.; et al. Analytical validation of the ReEBOV antigen rapid test for point-of-care diagnosis of Ebola virus infection. J. Infect. Dis. 2016, 214 (Suppl. 3), S210–S217. [Google Scholar] [CrossRef]
- Kurosaki, Y.; Grolla, A.; Fukuma, A.; Feldmann, H.; Yasuda, J. Development and evaluation of a simple assay for Marburg virus detection using a reverse transcription-loop-mediated isothermal amplification method. J. Clin. Microbiol. 2010, 48, 2330–2336. [Google Scholar] [CrossRef]
- de St. Maurice, A.; Harmon, J.; Nyakarahuka, L.; Balinandi, S.; Tumusiime, A.; Kyondo, J.; Mulei, S.; Namutebi, A.; Knust, B.; Shoemaker, T.; et al. Rift Valley fever viral load correlates with the human inflammatory response and coagulation pathway abnormalities in humans with hemorrhagic manifestations. PLoS Negl. Trop. Dis. 2018, 12, e0006460. [Google Scholar] [CrossRef] [PubMed]
- Günther, S.; Weisner, B.; Roth, A.; Grewing, T.; Asper, M.; Drosten, C.; Emmerich, P.; Petersen, J.; Wilczek, M.; Schmitz, H. Lassa fever encephalopathy: Lassa virus in cerebrospinal fluid but not in serum. J. Infect. Dis. 2001, 184, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Li, H.; Smith, G.; Goolia, M.; Marszal, P.; Pickering, B.S. Comparative characterization of Crimean-Congo hemorrhagic fever virus cell culture systems with application to propagation and titration methods. Virol. J. 2023, 20, 128. [Google Scholar] [CrossRef]
- Gamble, A.; Yeo, Y.Y.; Butler, A.A.; Tang, H.; Snedden, C.E.; Mason, C.T.; Buchholz, D.W.; Bingham, J.; Aguilar, H.C.; Lloyd-Smith, J.O. Drivers and distribution of henipavirus-induced syncytia: What do we know? Viruses 2021, 13, 1755. [Google Scholar] [CrossRef]
- Pan American Health Organization; World Health Organization. General Procedures for Inactivation of Potentially Infectious Samples with Ebola Virus and Other Highly Pathogenic Viral Agents. 2014. Available online: https://www3.paho.org/hq/dmdocuments/2014/2014-cha-procedures-inactivation-ebola.pdf (accessed on 16 February 2025).
- Elveborg, S.; Monteil, V.M.; Mirazimi, A. Methods of inactivation of highly pathogenic viruses for molecular, serology or vaccine development purposes. Pathogens 2022, 11, 271. [Google Scholar] [CrossRef]
- Haddock, E.; Feldmann, F.; Feldmann, H. Effective chemical inactivation of Ebola virus. Emerg. Infect. Dis. 2016, 22, 1292–1294. [Google Scholar] [CrossRef]
- Hume, A.J.; Ames, J.; Rennick, L.J.; Duprex, W.P.; Marzi, A.; Tonkiss, J.; Mühlberger, E. Inactivation of RNA viruses by gamma irradiation: A study on mitigating factors. Viruses 2016, 8, 204. [Google Scholar] [CrossRef]
- Ngo, K.A.; Jones, S.A.; Church, T.M.; Fuschino, M.E.; George, K.S.; Lamson, D.M.; Maffei, J.; Kramer, L.D.; Ciota, A.T. Unreliable inactivation of viruses by commonly used lysis buffers. Appl. Biosaf. 2017, 22, 56–59. [Google Scholar] [CrossRef]
- Olejnik, J.; Hume, A.J.; Ross, S.J.; Scoon, W.A.; Seitz, S.; White, M.R.; Slutzky, B.; Yun, N.E.; Mühlberger, E. Art of the kill: Designing and testing viral inactivation procedures for highly pathogenic negative sense RNA viruses. Pathogens 2023, 12, 952. [Google Scholar] [CrossRef]
- Auerswald, H.; Yann, S.; Dul, S.; In, S.; Dussart, P.; Martin, N.J.; Karlsson, E.A.; Garcia-Rivera, J.A. Assessment of inactivation procedures for SARS-CoV-2. J. Gen. Virol. 2021, 102, 001539. [Google Scholar] [CrossRef] [PubMed]
- Smither, S.J.; Weller, S.A.; Phelps, A.; Eastaugh, L.; Ngugi, S.; O’Brien, L.M.; Steward, J.; Lonsdale, S.G.; Lever, M.S. Buffer AVL alone does not inactivate Ebola virus in a representative clinical sample type. J. Clin. Microbiol. 2015, 53, 3148–3154. [Google Scholar] [CrossRef] [PubMed]
- Blow, J.A.; Mores, C.N.; Dyer, J.; Dohm, D.J. Viral nucleic acid stabilization by RNA extraction reagent. J. Virol. Methods 2008, 150, 41–44. [Google Scholar] [CrossRef]
- Invitrogen. User Duide: TRIzol™ LS Reagent. 2016. Available online: https://tools.thermofisher.com/content/sfs/manuals/trizol_ls_reagent.pdf (accessed on 16 February 2025).
- Nguyen-Tran, H.; Reno, S.; Mwangi, E.; Mentel, M.; Hengartner, R.; Dominguez, S.R.; Messacar, K.; Jung, S.A. Qualitative detection of enterovirus D68 from PrimeStore® molecular transport medium: Implications for home- and self-collection. Diagn. Microbiol. Infect. Dis. 2023, 106, 115976. [Google Scholar] [CrossRef]
- Omar, S.V.; Peters, R.P.H.; Ismail, N.A.; Dreyer, A.W.; Said, H.M.; Gwala, T.; Ismail, N.; Fourie, P.B. Laboratory evaluation of a specimen transport medium for downstream molecular processing of sputum samples to detect Mycobacterium tuberculosis. J. Microbiol. Methods 2015, 117, 57–63. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chang, L.-Y.; Mohd Ali, A.R.; Hassan, S.S.; AbuBakar, S. Quantitative estimation of Nipah virus replication kinetics in vitro. Virol. J. 2006, 3, 47. [Google Scholar] [CrossRef]
- Prasad, A.N.; Woolsey, C.; Geisbert, J.B.; Agans, K.N.; Borisevich, V.; Deer, D.J.; Mire, C.E.; Cross, R.W.; Fenton, K.A.; Broder, C.C.; et al. Resistance of cynomolgus monkeys to Nipah and Hendra virus disease is associated with cell-mediated and humoral immunity. J. Infect. Dis. 2020, 221 (Suppl. 4), S436–S447. [Google Scholar] [CrossRef]
- Crameri, G.; Wang, L.-F.; Morrissy, C.; White, J.; Eaton, B.T. A rapid immune plaque assay for the detection of Hendra and Nipah viruses and anti-virus antibodies. J. Virol. Methods 2002, 99, 41–51. [Google Scholar] [CrossRef]
- Marsh, G.A.; Haining, J.; Hancock, T.J.; Robinson, R.; Foord, A.J.; Barr, J.A.; Riddell, S.; Heine, H.G.; White, J.R.; Crameri, G.; et al. Experimental infection of horses with Hendra virus/Australia/horse/2008/Redlands. Emerg. Infect. Dis. 2011, 17, 2232–2238. [Google Scholar] [CrossRef]
- Albe, J.R.; Ma, H.; Gilliland, T.H.; McMillen, C.M.; Gardner, C.L.; Boyles, D.A.; Cottle, E.L.; Dunn, M.D.; Lundy, J.D.; O’Malley, K.J.; et al. Physiological and immunological changes in the brain associated with lethal eastern equine encephalitis virus in macaques. PLoS Pathog. 2021, 17, e1009308. [Google Scholar] [CrossRef]
- Sikazwe, C.; Neave, M.J.; Michie, A.; Mileto, P.; Wang, J.; Cooper, N.; Levy, A.; Imrie, A.; Baird, R.W.; Currie, B.J.; et al. Molecular detection and characterisation of the first Japanese encephalitis virus belonging to genotype IV acquired in Australia. PLoS Negl. Trop. Dis. 2022, 16, e0010754. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Fu, S.; Ma, X.; Chen, X.; Wu, D.; Zhou, L.; Yin, Q.; Li, F.; He, Y.; Lei, W.; et al. An outbreak of Japanese encephalitis caused by genotype Ib Japanese encephalitis virus in China, 2018: A laboratory and field investigation. PLoS Negl. Trop. Dis. 2020, 14, e0008312. [Google Scholar] [CrossRef] [PubMed]
- Duh, D.; Saksida, A.; Petrovec, M.; Ahmeti, S.; Dedushaj, I.; Panning, M.; Drosten, C.; Avšič-Županc, T. Viral load as predictor of Crimean-Congo hemorrhagic fever outcome. Emerg. Infect. Dis. 2007, 13, 1769–1772. [Google Scholar] [CrossRef]
- Ergunay, K.; Kocak Tufan, Z.; Bulut, C.; Kinikli, S.; Demiroz, A.P.; Ozkul, A. Antibody responses and viral load in patients with Crimean-Congo hemorrhagic fever: A comprehensive analysis during the early stages of the infection. Diagn. Microbiol. Infect. Dis. 2014, 79, 31–36. [Google Scholar] [CrossRef]
- Bhat, T.; Cao, A.; Yin, J. Virus-like particles: Measures and biological functions. Viruses 2022, 14, 383. [Google Scholar] [CrossRef] [PubMed]
- Cutts, T.; Leung, A.; Banadyga, L.; Krishnan, J. Inactivation validation of Ebola, Marburg, and Lassa viruses in AVL and ethanol-treated viral cultures. Viruses 2024, 16, 1354. [Google Scholar] [CrossRef]
- Kumar, M.; Mazur, S.; Ork, B.L.; Postnikova, E.; Hensley, L.E.; Jahrling, P.B.; Johnson, R.; Holbrook, M.R. Inactivation and safety testing of Middle East respiratory syndrome coronavirus. J. Virol. Methods 2015, 223, 13–18. [Google Scholar] [CrossRef]
- Widerspick, L.; Vázquez, C.A.; Niemetz, L.; Heung, M.; Olal, C.; Bencsik, A.; Henkel, C.; Pfister, A.; Brunetti, J.E.; Kucinskaite-Kodze, I.; et al. Inactivation methods for experimental Nipah virus infection. Viruses 2022, 14, 1052. [Google Scholar] [CrossRef]
- Olschewski, S.; Thielebein, A.; Hoffmann, C.; Blake, O.; Müller, J.; Bockholt, S.; Pallasch, E.; Hinzmann, J.; Wurr, S.; Neddersen, N.; et al. Validation of inactivation methods for arenaviruses. Viruses 2021, 13, 968. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).