Investigation of Natural Resistance to Fostemsavir and Lenacapavir in Naïve Primary Infections by Ultra-Deep Sequencing of near Full-Length HIV-1 Genomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Virological Assessment
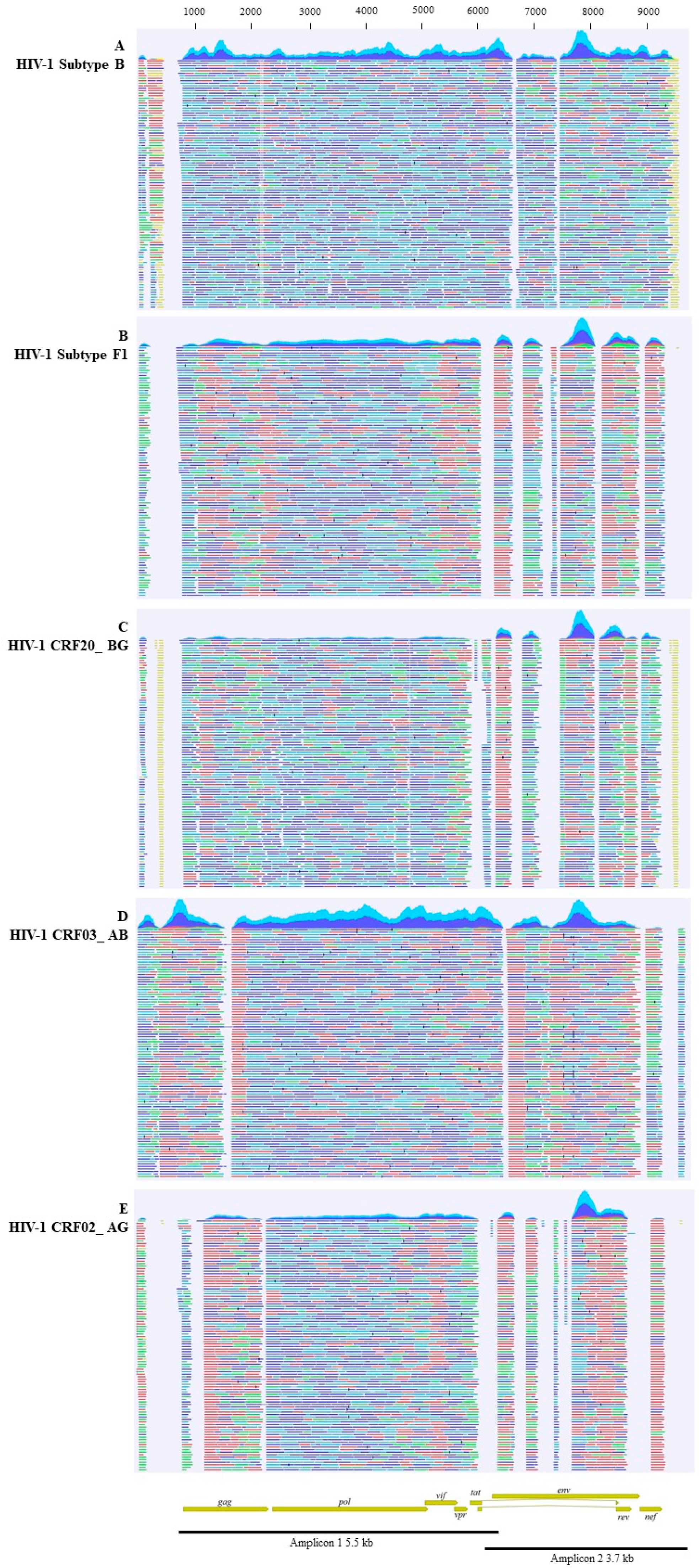
2.2. Near-Full-Length PCR and Bioinformatic Analysis
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Galli, L.; Parisi, M.R.; Poli, A.; Menozzi, M.; Fiscon, M.; Garlassi, E.; Francisci, D.; Di Biagio, A.; Sterrantino, G.; Fornabaio, C.; et al. Burden of Disease in PWH Harboring a Multidrug-Resistant Virus: Data from the PRESTIGIO Registry. Open Forum Infect. Dis. 2020, 7, ofaa456. [Google Scholar] [CrossRef] [PubMed]
- Hitchcock, A.M.; Kufel, W.D.; Dwyer, K.A.M.; Sidman, E.F. Lenacapavir: A novel injectable HIV-1 capsid inhibitor. Int. J. Antimicrob. Agents 2024, 63, 107009. [Google Scholar] [CrossRef] [PubMed]
- Heidary, M.; Shariati, S.; Nourigheimasi, S.; Khorami, M.; Moradi, M.; Motahar, M.; Bahrami, P.; Akrami, S.; Hassan Kaviar, V. Mechanism of action, resistance, interaction, pharmacokinetics, pharmacodynamics, and safety of fostemsavir. BMC Infect. Dis. 2024, 24, 250. [Google Scholar] [CrossRef] [PubMed]
- Dvory-Sobol, H.; Shaik, N.; Callebaut, C.; Rhee, M.S. Lenacapavir: A first-in-class HIV-1 capsid inhibitor. Curr. Opin. HIV AIDS 2022, 17, 15. [Google Scholar] [CrossRef] [PubMed]
- Bester, S.M.; Wei, G.; Zhao, H.; Adu-Ampratwum, D.; Iqbal, N.; Courouble, V.V.; Francis, A.C.; Annamalai, A.S.; Singh, P.K.; Shkriabai, N.; et al. Structural and mechanistic bases for a potent HIV-1 capsid inhibitor. Science 2020, 370, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Bouba, Y.; Berno, G.; Fabeni, L.; Carioti, L.; Salpini, R.; Aquaro, S.; Svicher, V.; Perno, C.F.; Ceccherini-Silberstein, F.; Santoro, M.M. Identification of gp120 polymorphisms in HIV-1 B subtype potentially associated with resistance to fostemsavir. J. Antimicrob. Chemother. 2020, 75, 1778–1786. [Google Scholar] [CrossRef] [PubMed]
- Gartland, M.; Arnoult, E.; Foley, B.T.; Lataillade, M.; Ackerman, P.; Llamoso, C.; Krystal, M. Prevalence of gp160 polymorphisms known to be related to decreased susceptibility to temsavir in different subtypes of HIV-1 in the Los Alamos National Laboratory HIV Sequence Database. J. Antimicrob. Chemother. 2021, 76, 2958–2964. [Google Scholar] [CrossRef] [PubMed]
- Zuze, B.J.L.; Radibe, B.T.; Choga, W.T.; Bareng, O.T.; Moraka, N.O.; Maruapula, D.; Seru, K.; Mokgethi, P.; Mokaleng, B.; Ndlovu, N.; et al. Fostemsavir resistance-associated polymorphisms in HIV-1 subtype C in a large cohort of treatment-naïve and treatment-experienced individuals in Botswana. Microbiol. Spectr. 2023, 11, e01251-23. [Google Scholar] [CrossRef] [PubMed]
- Fabeni, L.; Berno, G.; Fokam, J.; Bertoli, A.; Alteri, C.; Gori, C.; Forbici, F.; Takou, D.; Vergori, A.; Zaccarelli, M.; et al. Comparative Evaluation of Subtyping Tools for Surveillance of Newly Emerging HIV-1 Strains. J. Clin. Microbiol. 2017, 55, 2827–2837. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, S.; Nowak, P.; Neogi, U. Subtype-independent near full-length HIV-1 genome sequencing and assembly to be used in large molecular epidemiological studies and clinical management. J. Int. AIDS Soc. 2015, 18, 20035. [Google Scholar] [CrossRef] [PubMed]
- Kozal, M.; Aberg, J.; Pialoux, G.; Cahn, P.; Thompson, M.; Molina, J.-M.; Grinsztejn, B.; Diaz, R.; Castagna, A.; Kumar, P.; et al. Fostemsavir in Adults with Multidrug-Resistant HIV-1 Infection. N. Engl. J. Med. 2020, 382, 1232–1243. [Google Scholar] [CrossRef] [PubMed]
- Margot, N.; Pennetzdorfer, N.; Naik, V.; Rhee, M.; Callebaut, C. Cross-Resistance to Entry Inhibitors and Lenacapavir Resistance through Week 52 in Study CAPELLA. Antivir. Ther. 2023, 28, 13596535231220754. [Google Scholar] [CrossRef] [PubMed]
- Lepore, L.; Fabrizio, C.; Bavaro, D.F.; Milano, E.; Volpe, A.; Lagioia, A.; Angarano, A.; Saracino, A.; Monno, L. Gp120 substitutions at positions associated with resistance to fostemsavir in treatment-naive HIV-1-positive individuals. J. Antimicrob. Chemother. 2020, 75, 1580–1587. [Google Scholar] [CrossRef] [PubMed]
- Marcelin, A.G.; Charpentier, C.; Jary, A.; Perrier, M.; Margot, N.; Callebaut, C.; Calvez, V.; Descamps, D. Frequency of capsid substitutions associated with GS-6207 in vitro resistance in HIV-1 from antiretroviral-naive and -experienced patients. J. Antimicrob. Chemother. 2020, 75, 1588–1590. [Google Scholar] [CrossRef] [PubMed]
| Drug | Gene | Mutation | Type of Resistance-Associated Variants | References |
|---|---|---|---|---|
| FTR | env | L116P/Q | in vitro selected | [6,7] |
| FTR | env | A204D | in vitro selected | [7] |
| FTR | env | S375H/I/M/N/T | clinically relevant resistance | [3,6,7,8] |
| FTR | env | S375Y | in vitro selected | [7] |
| FTR | env | M426L | clinically relevant resistance | [3,6,7,8] |
| FTR | env | M434I | clinically relevant resistance | [3,6,7,8] |
| FTR | env | M434K | in vitro selected | [7] |
| FTR | env | M475I | clinically relevant resistance | [3,6,7,8] |
| FTR | env | V506M | in vitro selected | [7] |
| LEN | gag (p24) | L56I | in vitro selected | [4,10] |
| LEN | gag (p24) | M66I | in vitro selected | [4,5,10] |
| LEN | gag (p24) | Q67H | in vitro selected | [4,5,10] |
| LEN | gag (p24) | K70N | in vitro selected | [4,10] |
| LEN | gag (p24) | N74D/S | in vitro selected | [4,5,10] |
| LEN | gag (p24) | A105T | in vitro selected | [4,10] |
| LEN | gag (p24) | T107N | in vitro selected | [4,10] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lazzari, E.; Rozera, G.; Gagliardini, R.; Mazzotta, V.; Fabeni, L.; Forbici, F.; Berno, G.; Cosentino, C.; Girardi, E.; Antinori, A.; et al. Investigation of Natural Resistance to Fostemsavir and Lenacapavir in Naïve Primary Infections by Ultra-Deep Sequencing of near Full-Length HIV-1 Genomes. Viruses 2025, 17, 636. https://doi.org/10.3390/v17050636
Lazzari E, Rozera G, Gagliardini R, Mazzotta V, Fabeni L, Forbici F, Berno G, Cosentino C, Girardi E, Antinori A, et al. Investigation of Natural Resistance to Fostemsavir and Lenacapavir in Naïve Primary Infections by Ultra-Deep Sequencing of near Full-Length HIV-1 Genomes. Viruses. 2025; 17(5):636. https://doi.org/10.3390/v17050636
Chicago/Turabian StyleLazzari, Elisabetta, Gabriella Rozera, Roberta Gagliardini, Valentina Mazzotta, Lavinia Fabeni, Federica Forbici, Giulia Berno, Cristian Cosentino, Enrico Girardi, Andrea Antinori, and et al. 2025. "Investigation of Natural Resistance to Fostemsavir and Lenacapavir in Naïve Primary Infections by Ultra-Deep Sequencing of near Full-Length HIV-1 Genomes" Viruses 17, no. 5: 636. https://doi.org/10.3390/v17050636
APA StyleLazzari, E., Rozera, G., Gagliardini, R., Mazzotta, V., Fabeni, L., Forbici, F., Berno, G., Cosentino, C., Girardi, E., Antinori, A., Maggi, F., & Abbate, I. (2025). Investigation of Natural Resistance to Fostemsavir and Lenacapavir in Naïve Primary Infections by Ultra-Deep Sequencing of near Full-Length HIV-1 Genomes. Viruses, 17(5), 636. https://doi.org/10.3390/v17050636








