The Role of the Tyrosine-Based Sorting Signals of the ORF3a Protein of SARS-CoV-2 in Intracellular Trafficking and Pathogenesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells, Viruses, and Plasmids
2.2. Site-Directed Mutagenesis of ORF3a
2.3. Immunofluorescence Studies
2.4. Surface/Internal Immunofluorescence Assays
2.5. Isolation of Late Endosomes/Lysosomes from the Cells
2.6. Construction of the Infectious Clone of SARS-CoV-2 Delta
2.7. Construction of Mutant BACs
2.8. Rescue of Infectious SARS-CoV-2 Delta and Mutant ORF3a Viruses from BACs
2.9. Virus Growth Curves
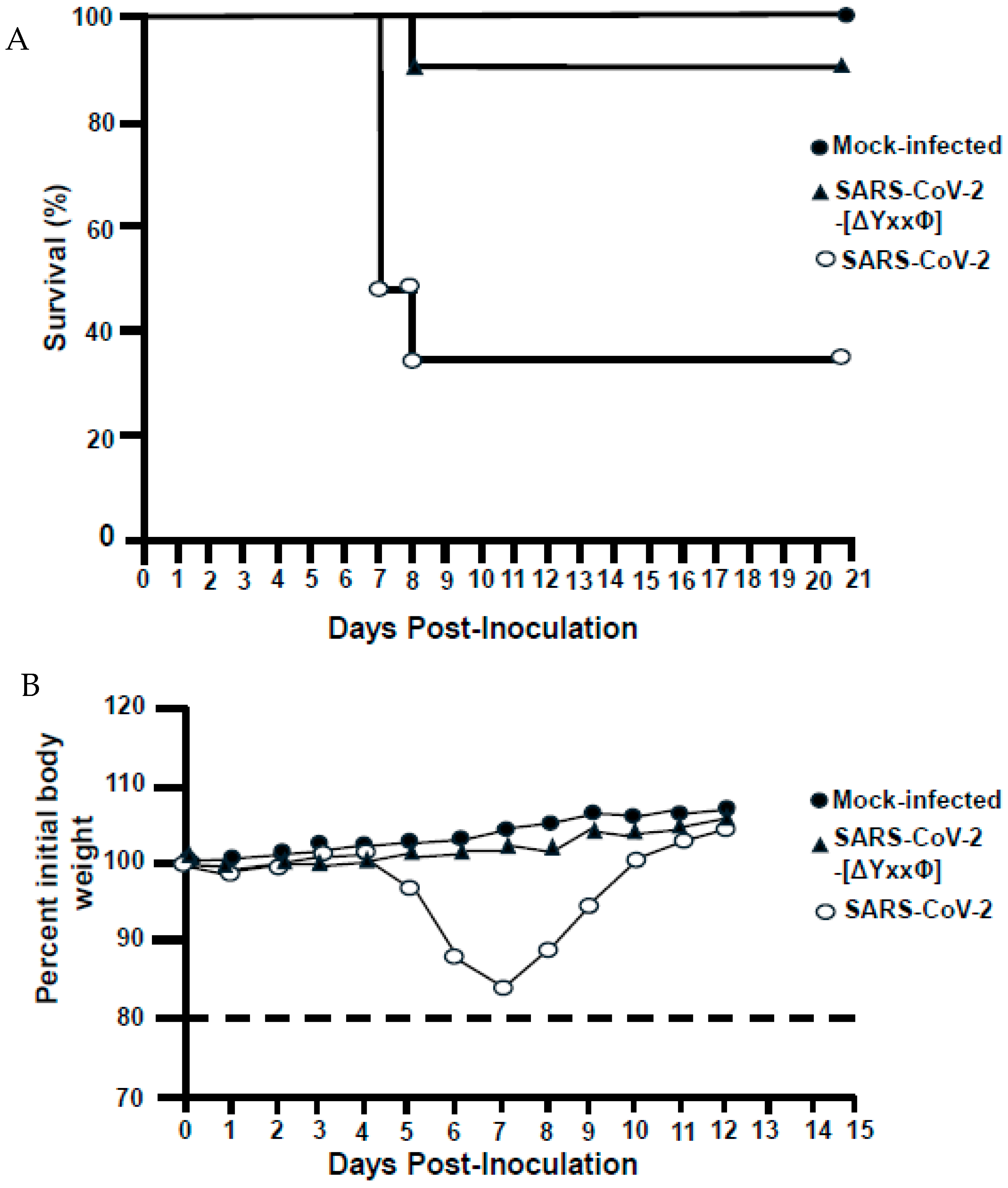
2.10. Pathogenicity with SARS-CoV-2 Viruses Expressing the Unmodified ORF3a or the ORF3a-[ΔYxxΦ] in K18hACE2 Mice
3. Results
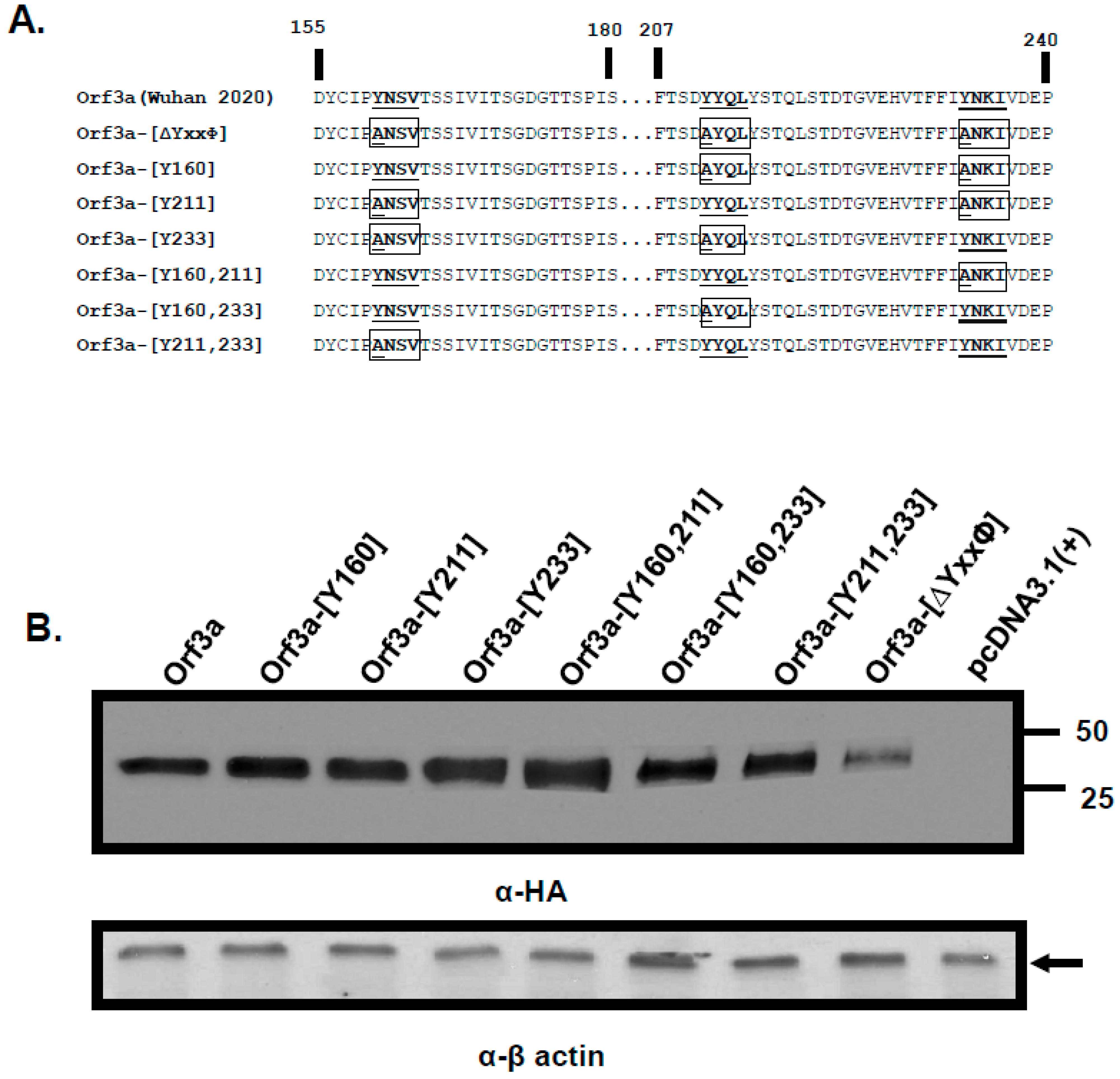
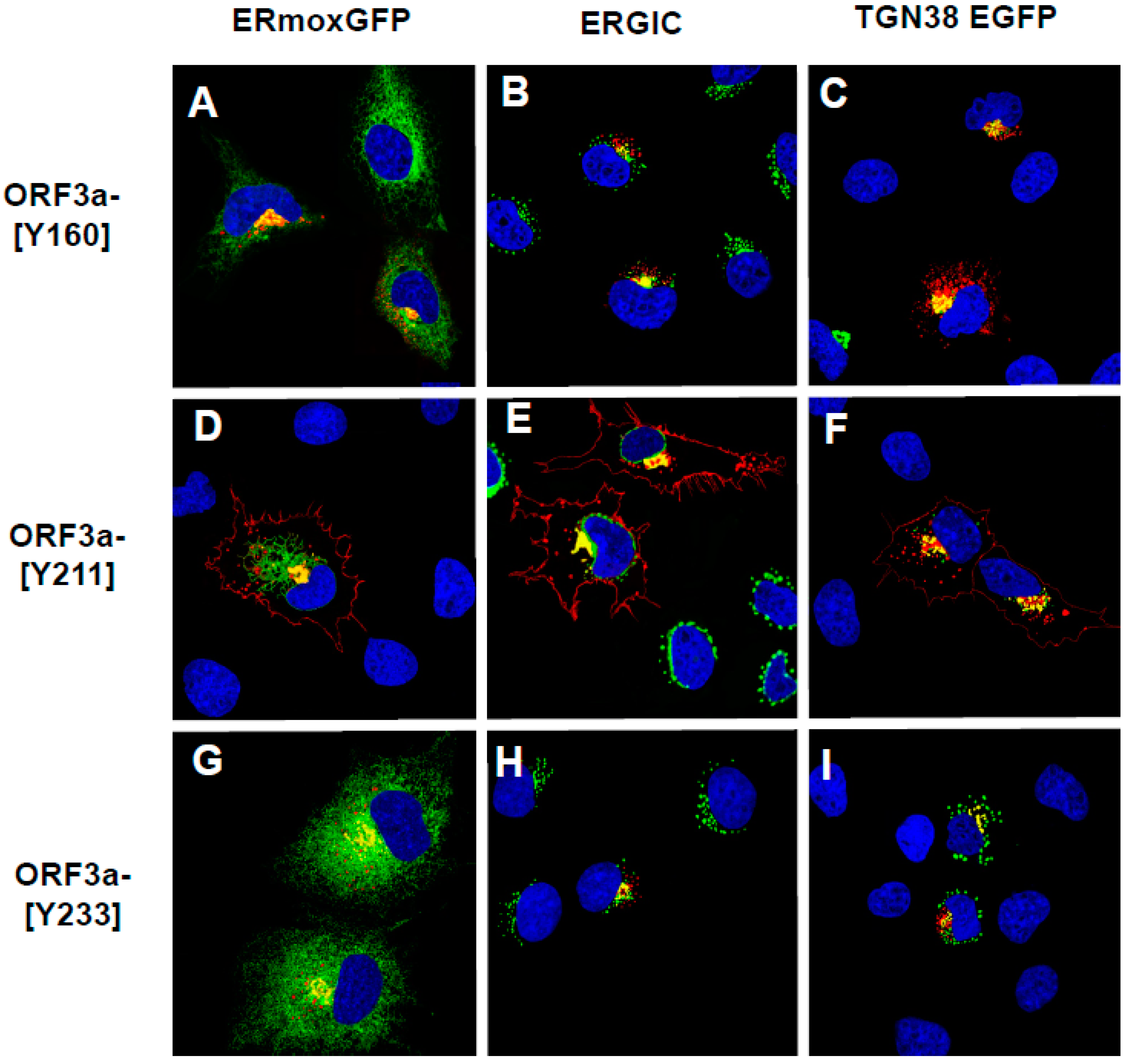
3.1. The ORF3a Has More than One Potential Tyrosine-Based Sorting Motif
3.2. Expression of SARS-CoV-2 ORF3a and Various Mutants
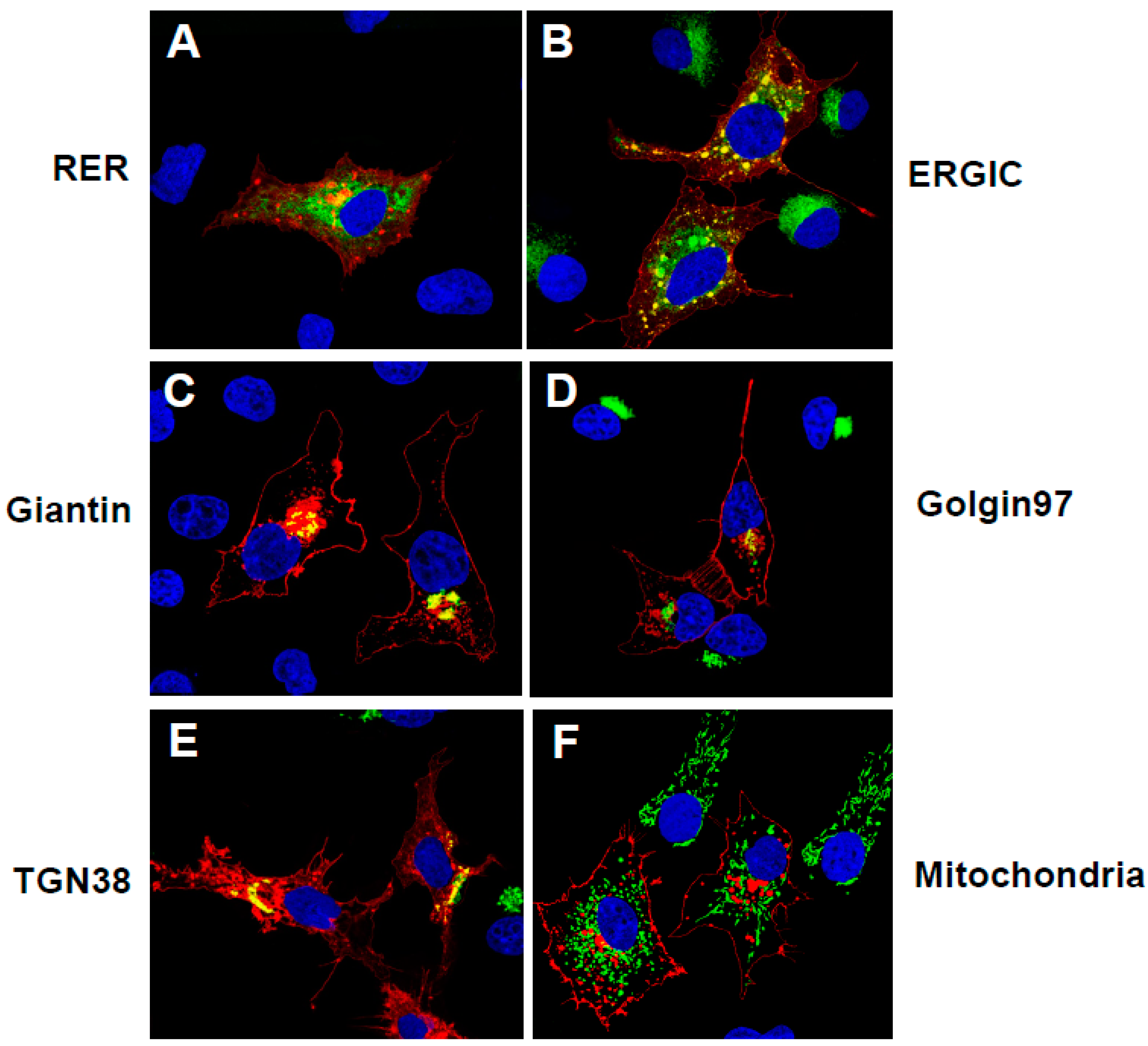
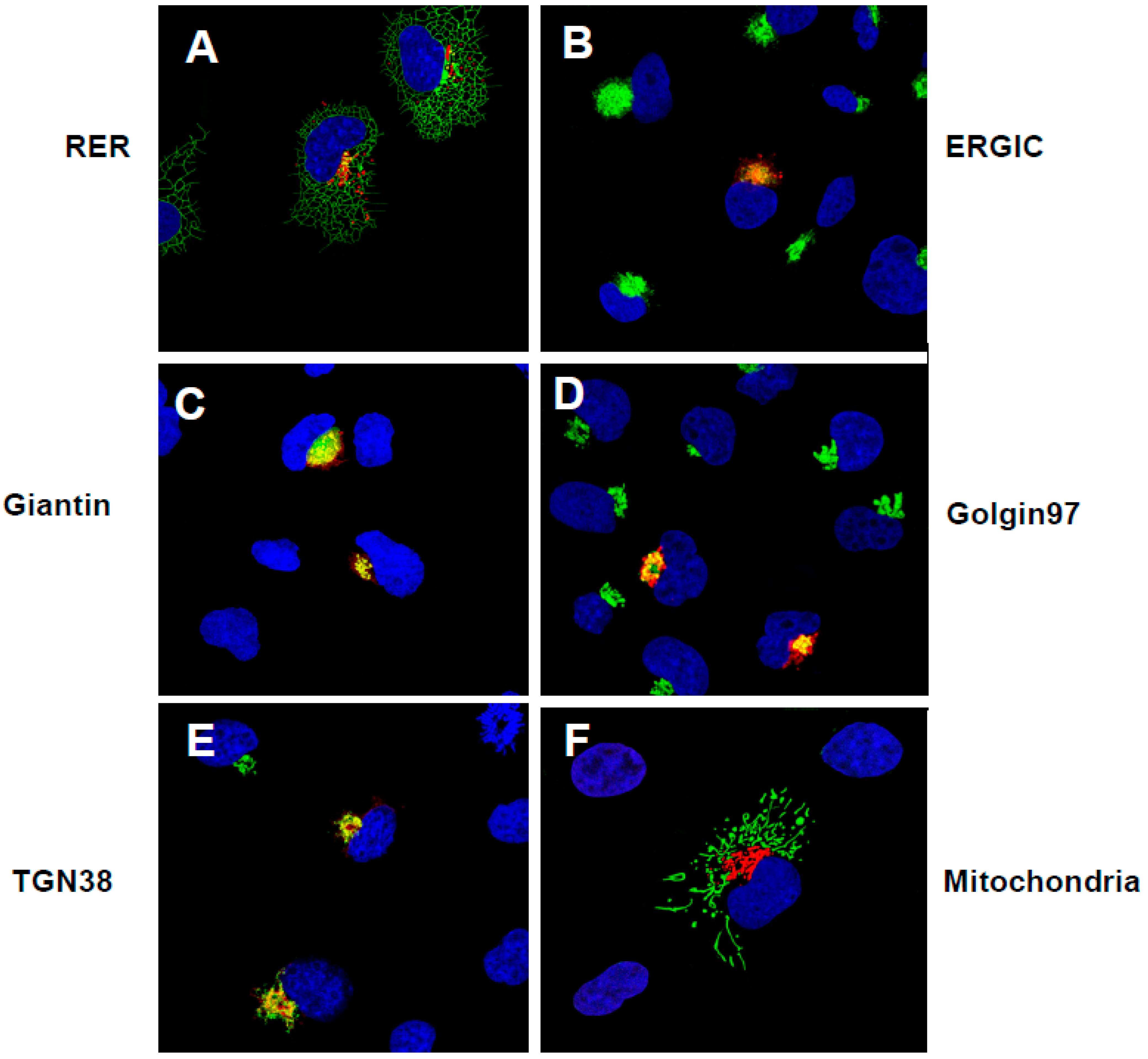
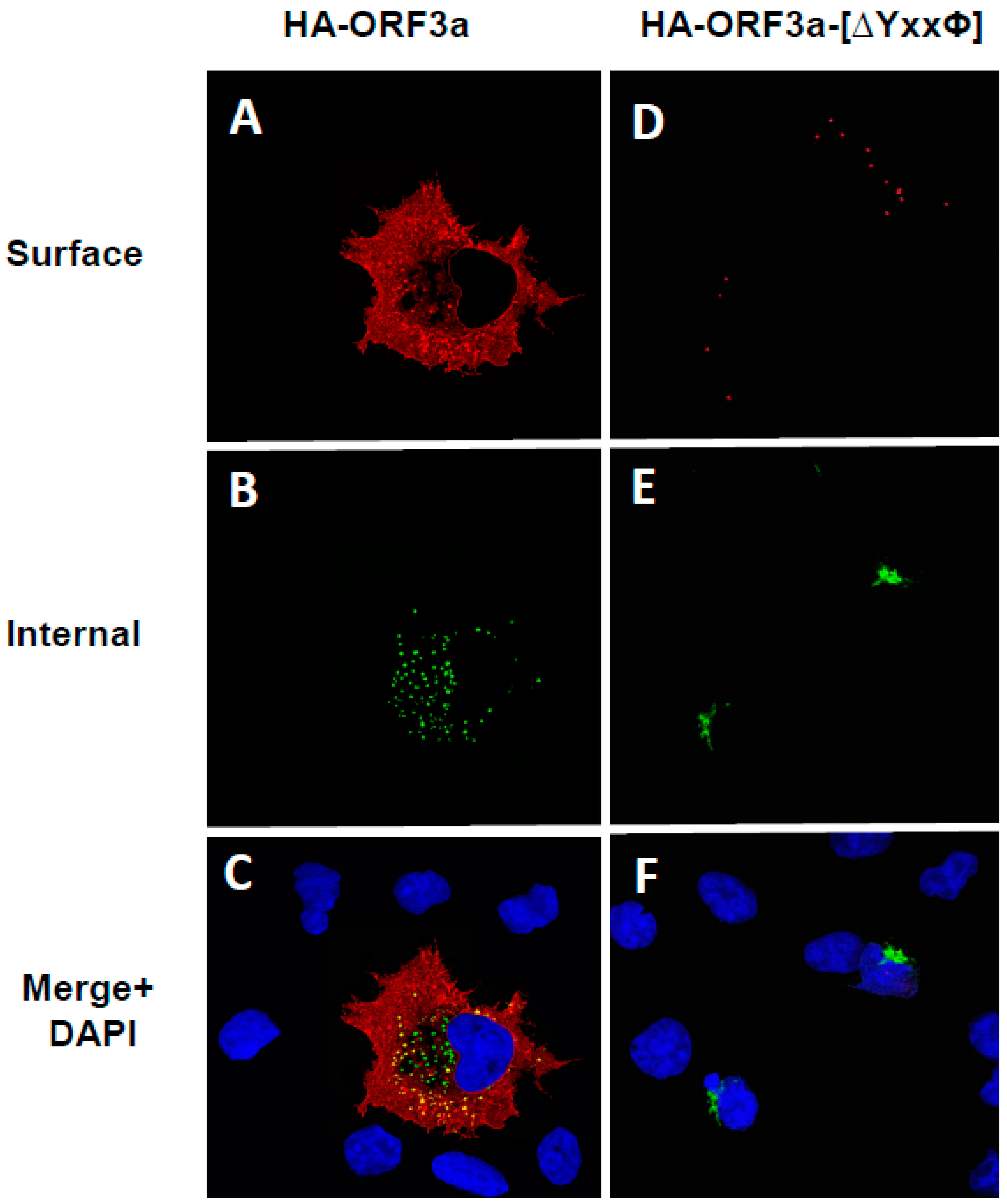
3.3. The Unmodified SARS-CoV-2 ORF3a Protein Is Transported Through the Secretory Pathway to the Cell-Plasma Membrane
3.4. The ORF3a-[ΔYxxΦ] Mutant Is Not Expressed at the Cell Surface
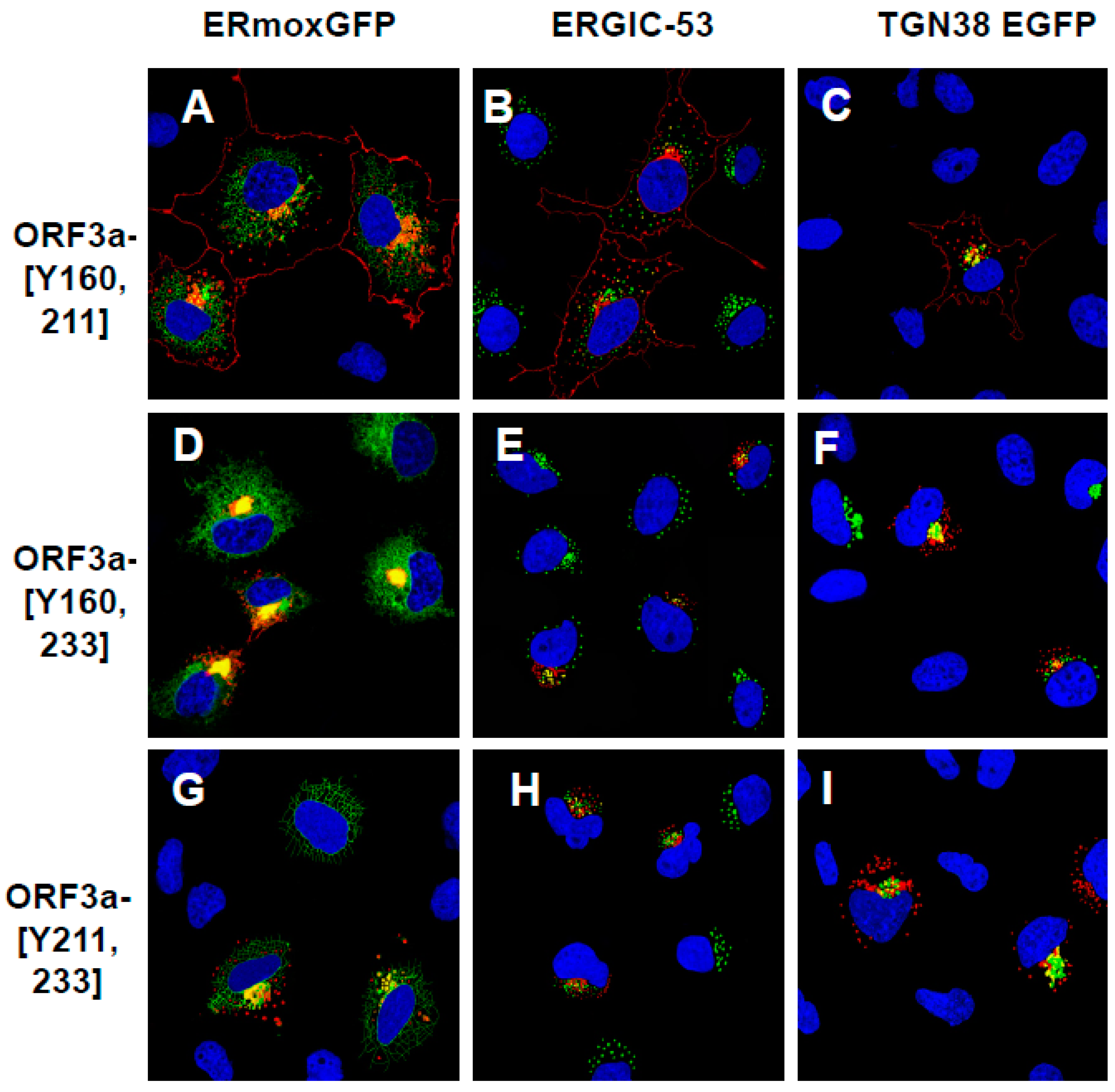
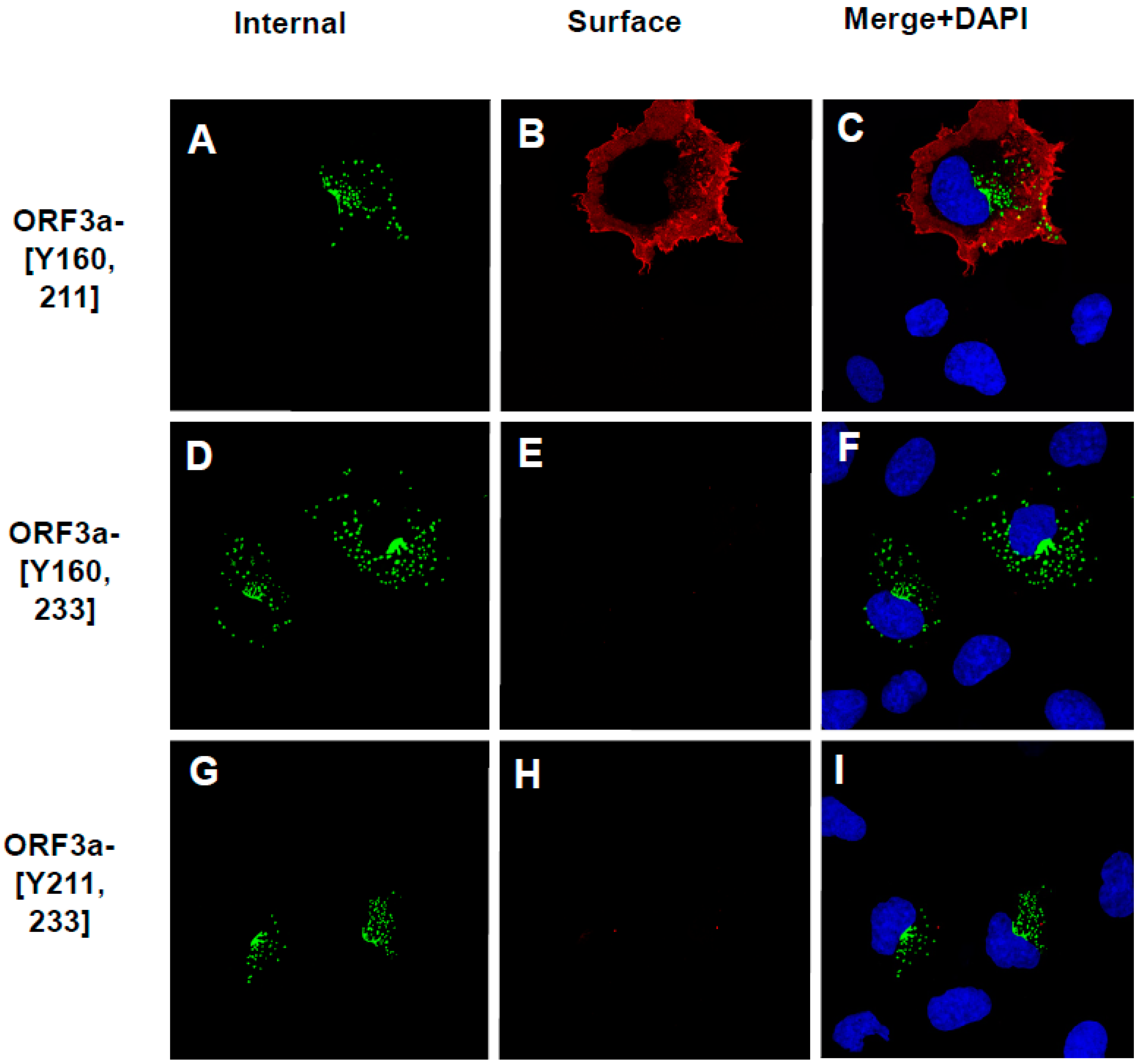
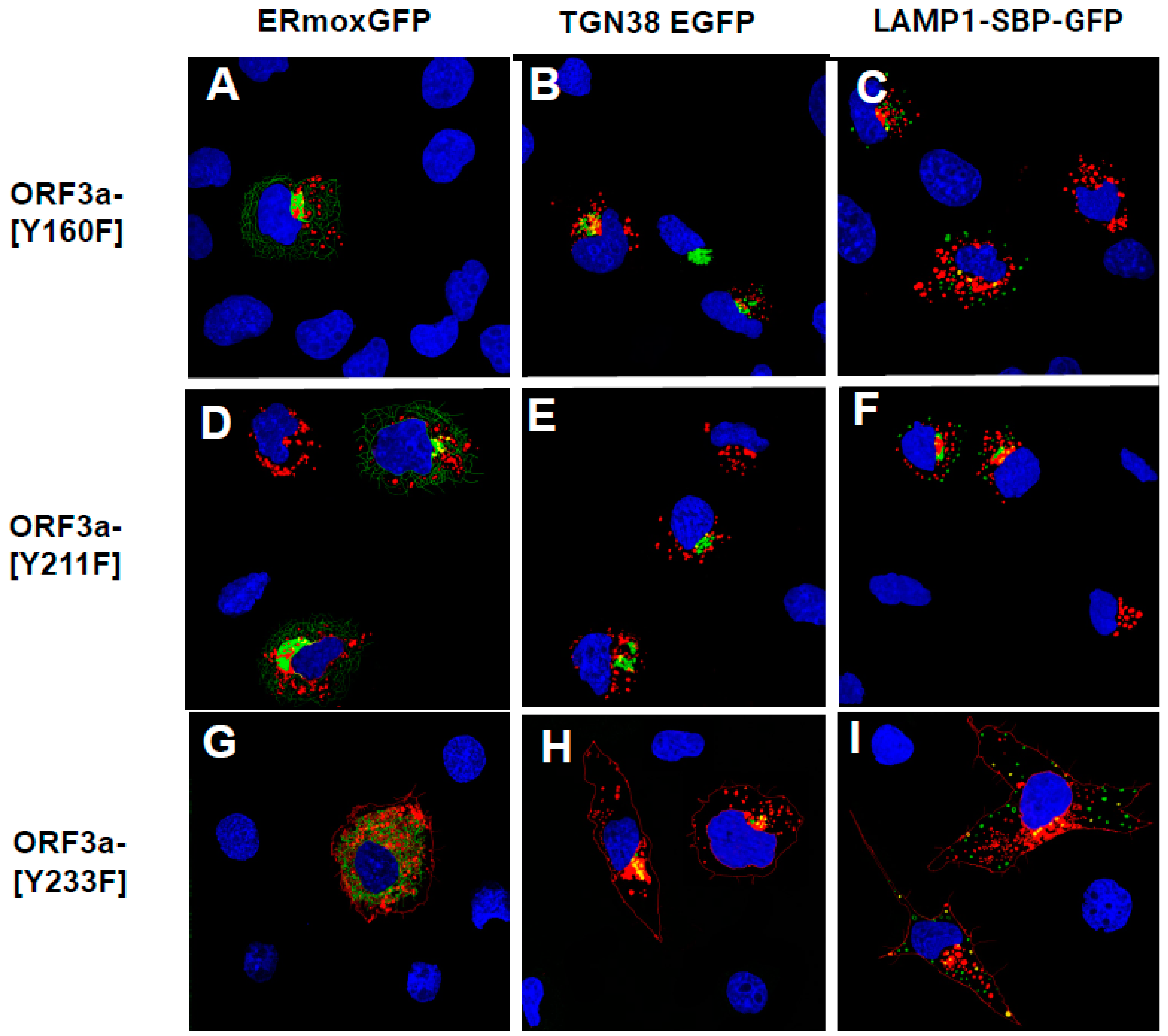
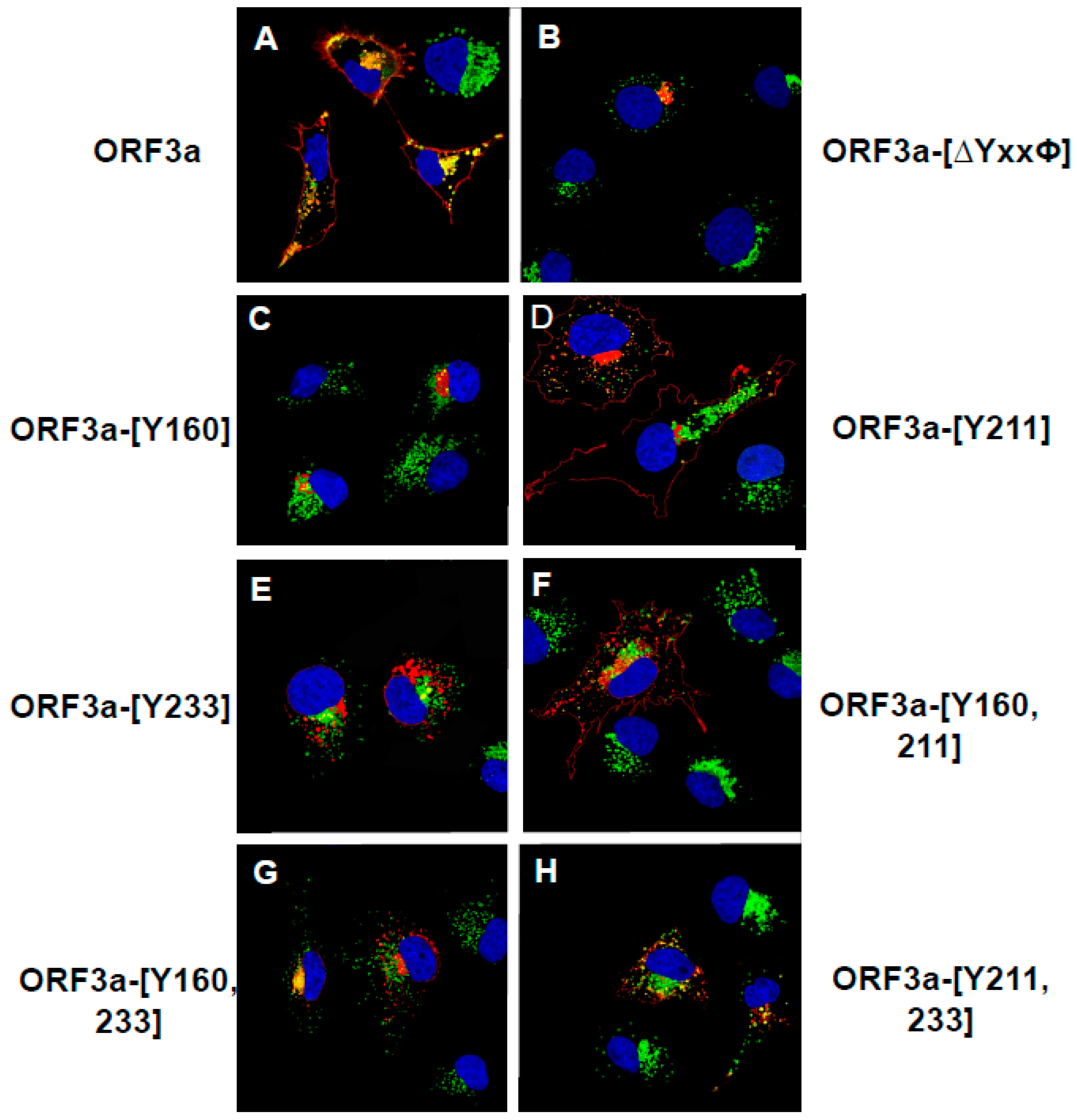
3.5. Expression of ORF3a Mutants with One or Two Potential Tyrosine-Based Sorting Motifs Intact
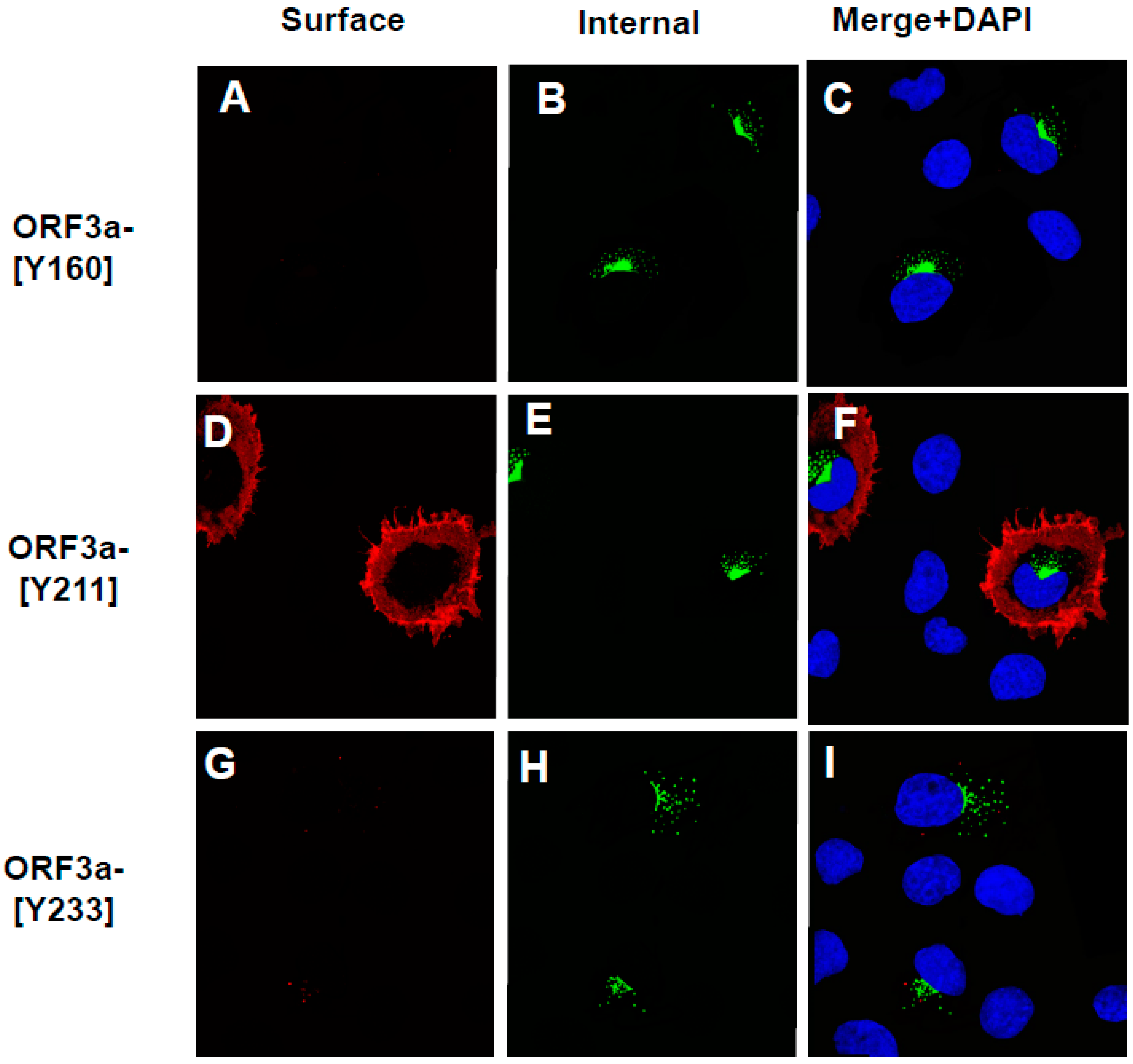
3.6. The Surface Immunostaining of Cells Transfected with Vectors Expressing the ORF3a and ORF3a Mutants Confirms the Cell Surface Expression Patterns
3.7. Substitution of the Tyrosine Residues in the Three Motifs with Phenylalanine Residues
3.8. Lysosome Localization of the ORF3a Mutants
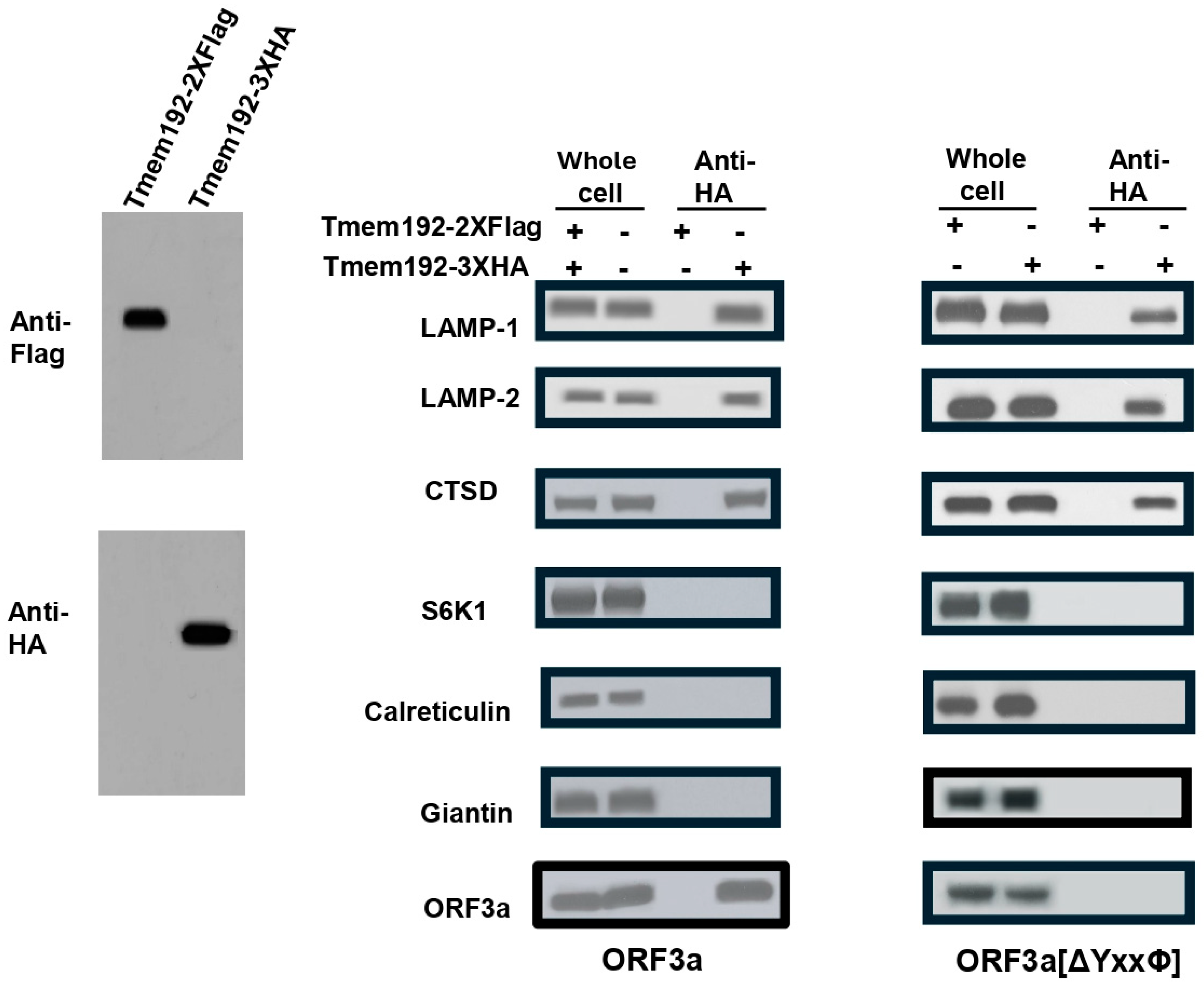
3.9. Purification of Late Endosomes/Lysosomes Reveals That ORF3a-[ΔYxxФ] Does Not Traffic to These Compartments of the Cell
3.10. Replication of SARS-CoV-2 and SARS-CoV-2 Mutants in Vero E6 Cells
3.11. A SARS-CoV-2 Virus with All Three Tyrosine Motifs Removed (SARS-CoV-2-[ORF3a-[ΔYxxΦ]) Was Less Pathogenic in K18-hACE2 Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tan, Y.J.; Teng, E.; Shen, S.; Tan, T.H.; Goh, P.Y.; Fielding, B.C.; Ooi, E.E.; Tan, H.C.; Lim, S.G.; Hong, W. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 2004, 78, 6723–6734. [Google Scholar] [CrossRef] [PubMed]
- Chien, T.H.; Chiang, Y.L.; Chen, C.P.; Henklein, P.; Hänel, K.; Hwang, I.S.; Willbold, D.; Fischer, W.B. Assembling an ion channel: ORF 3a from SARS-CoV. Biopolymers 2013, 99, 628–635. [Google Scholar] [CrossRef] [PubMed]
- Kern, D.M.; Sorum, B.; Mali, S.S.; Hoel, C.M.; Sridharan, S.; Remis, J.P.; Toso, D.B.; Kotecha, A.; Bautista, D.M.; Brohawn, S.G. Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs. Nat. Struct. Mol. Biol. 2021, 28, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Zheng, B.J.; Xu, K.; Du, L.; Wong, C.K.; Chen, J.; Duan, S.; Deubel, V.; Sun, B. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. USA 2006, 103, 12540–12545. [Google Scholar]
- Miller, A.N.; Houlihan, P.R.; Matamala, E.; Cabezas-Bratesco, D.; Lee, G.Y.; Cristofori-Armstrong, B.; Dilan, T.L.; Sanchez-Martinez, S.; Matthies, D.; Yan, R.; et al. The SARS-CoV-2 accessory protein Orf3a is not an ion channel but does interact with trafficking proteins. eLife 2023, 12, e84477. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Akinyemi, I.A.; Chitre, S.A.; Loeb, J.C.; Lednicky, J.A.; McIntosh, M.T.; Bhaduri-McIntosh, S. SARS-CoV-2 viroporin encoded by ORF3a triggers the NLRP3 inflammatory pathway. Virology 2022, 568, 13–22. [Google Scholar] [CrossRef] [PubMed]
- McClenaghan, C.; Hanson, A.; Lee, S.J.; Nichols, C.G. Coronavirus proteins as ion channels: Current and potential research. Front. Immunol. 2020, 11, 573339. [Google Scholar] [CrossRef]
- Oliveira-Mendes, B.B.R.; Alameh, M.; Ollivier, B.; Montnach, J.; Bidère, N.; Souazé, F.; Escriou, N.; Charpentier, F.; Baró, I.; De Waard, M.; et al. SARS-CoV-2 E and 3a proteins are inducers of pannexin currents. Cells 2023, 12, 1474. [Google Scholar] [CrossRef] [PubMed]
- Anitei, M.; Hoflack, B. Exit from the trans-Golgi network: From molecules to mechanisms. Curr. Opin. Cell Biol. 2011, 23, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Bonifacino, J.S.; Traub, L.M. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 2003, 72, 395–447. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.J.; Goldstein, J.L.; Brown, M.S. NPXY, a sequence often found in cytoplasmic tails, is required for coated pit-mediated internalization of the low density lipoprotein receptor. J. Biol. Chem. 1990, 265, 3116–3123. [Google Scholar]
- Traub, L.M. Sorting it out: AP-2 and alternate clathrin adaptors in endocytic cargo selection. J. Cell Biol. 2003, 163, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Kirchhausen, T.; Pines, J.; Toldo, L.; Lafont, F. Membranes and sorting. membrane permeability. Curr. Opin. Cell Biol. 1997, 9, 473. [Google Scholar] [CrossRef] [PubMed]
- Kirchhausen, T. Adaptors for clathrin-mediated traffic. Annu. Rev. Cell Dev. Biol. 1999, 15, 705–732. [Google Scholar]
- Marks, M.S.; Roche, P.A.; van Donselaar, E.; Woodruff, L.; Peters, P.J.; Bonifacino, J.S. A lysosomal targeting signal in the cytoplasmic tail of the beta chain directs HLA-DM to MHC class II compartments. J. Cell Biol. 1995, 131, 351–369. [Google Scholar] [PubMed]
- Marks, M.S.; Ohno, H.; Kirchausen, T.; Bonifacino, J.S. Protein sorting by tyrosine-based signals: Adapting to the Ys and wherefores. Trends Cell Biol. 1997, 7, 124–128. [Google Scholar]
- Rous, B.A.; Reaves, B.J.; Ihrke, G.; Briggs, J.A.; Gray, S.R.; Stephens, D.J.; Banting, G.J.; Luzio, J.P. Role of adaptor complex AP-3 in targeting wild-type and mutated CD63 to lysosomes. Mol. Biol. Cell 2002, 13, 1071–1082. [Google Scholar] [PubMed]
- Stolt, P.C.; Bock, H.H. Modulation of lipoprotein receptor functions by intracellular adaptor proteins. Cell. Signal. 2006, 18, 1560–1571. [Google Scholar] [PubMed]
- Traub, L.M.; Bonificano, J.S. Cargo recognition in clathrin-mediated endocytosis. Cold Spring Harb. Perspect. Biol. 2013, 5, a016790. [Google Scholar]
- Williams, M.A.; Fukuda, M. Accumulation of membrane glycoproteins in lysosomes requires a tyrosine residue at a particular position in the cytoplasmic tail. J. Cell Biol. 1990, 111, 955–966. [Google Scholar] [PubMed]
- Braulke, T.; Bonifacino, J.S. Sorting of lysosomal proteins. Biochim. Biophys. Acta 2009, 1793, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Höning, S.; Griffith, J.; Geuze, H.; Hunziker, W. The tyrosine-based lysosomal targeting signal in LAMP-1 mediates sorting into Golgi-derived clathrin-coated vesicles. EMBO J. 1996, 15, 5230–5239. [Google Scholar] [CrossRef] [PubMed]
- Rohrer, J.; Schweizer, A.; Russell, D.; Kornfeld, S. The targeting of Lamp1 to lysosomes is dependent on the spacing of its cytoplasmic tail tyrosine sorting motif relative to the membrane. J. Cell Biol. 1996, 132, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Blot, V.; Lopez-Vergès, S.; Breton, M.; Pique, C.; Berlioz-Torrent, C.; Grange, M.P. The conserved dileucine- and tyrosine-based motifs in MLV and MPMV envelope glycoproteins are both important to regulate a common Env intracellular trafficking. Retrovirology 2006, 3, 62. [Google Scholar] [CrossRef] [PubMed]
- Boll, W.; Ohno, H.; Songyang, Z.; Rapoport, I.; Cantley, L.C.; Bonifacino, J.S.; Kirchhausen, T. Sequence requirements for the recognition of tyrosine-based endocytic signals by clathrin AP-2 complexes. EMBO J. 1996, 15, 5789–5795. [Google Scholar] [CrossRef]
- Brewer, C.B.; Roth, M.G. A single amino acid change in the cytoplasmic domain alters the polarized delivery of influenza virus hemagglutinin. J Cell Biol. 1991, 114, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Day, J.R.; Münk, C.; Guatelli, J.C. The membrane-proximal tyrosine-based sorting signal of human immunodeficiency virus type 1 gp41 is required for optimal viral infectivity. J. Virol. 2004, 78, 1069–1079. [Google Scholar] [CrossRef]
- Favoreel, H.W. The why’s of Y-based motifs in alphaherpesvirus envelope proteins. Virus Res. 2006, 117, 202–208. [Google Scholar] [CrossRef]
- Hou, Y.; Meulia, T.; Gao, X.; Saif, L.J.; Wang, Q.J. Deletion of both the tyrosine-based endocytosis signal and the endoplasmic reticulum retrieval signal in the cytoplasmic tail of spike protein attenuates porcine epidemic diarrhea virus in pigs. J. Virol. 2019, 93, e01758-18. [Google Scholar] [CrossRef] [PubMed]
- Roush, D.L.; Gottardi, C.J.; Naim, Y.; Roth, M.G.; Caplan, M.J. Tyrosine-based membrane protein sorting signals are differentially interpreted by polarized Madin-Darby canine kidney and LLC-PK1 epithelial cells. J. Biol. Chem. 1998, 273, 26862–26869. [Google Scholar] [CrossRef]
- Ren, Y.; Shu, T.; Wu, D.; Mu, J.; Wang, C.; Huang, M.; Han, Y.; Zhang, X.Y.; Zhou, W.; Qiu, Y.; et al. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell. Mol. Immunol. 2020, 17, 881–883. [Google Scholar] [CrossRef] [PubMed]
- Minakshi, R.; Padhan, K. The YXXΦ motif within the severe acute respiratory syndrome coronavirus (SARS-CoV) 3a protein is crucial for its intracellular transport. Virol. J. 2014, 11, 75. [Google Scholar] [CrossRef] [PubMed]
- Abu-Remaileh, M.; Wyant, G.A.; Kim, C.; Laqtom, N.N.; Abbasi, M.; Chan, S.H.; Freinkman, E.; Sabatini, D.M. Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science 2017, 358, 807–813. [Google Scholar] [CrossRef]
- Noskov, V.; Kouprina, N.; Leem, S.H.; Koriabine, M.; Barrett, J.C.; Larionov, V. A genetic system for direct selection of gene-positive clones during recombinational cloning in yeast. Nucleic Acids Res. 2002, 30, E8. [Google Scholar] [PubMed]
- Thao, T.T.N.; Labroussaa, F.; Ebert, N.; Jores, J.; Thiel, V. In-yeast assembly of coronavirus infectious cDNA clones using a synthetic genomics pipeline. Methods Mol. Biol. 2020, 2203, 167–184. [Google Scholar]
- Tischer, B.K.; von Einem, J.; Kaufer, B.; Osterrieder, N. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. BioTechniques 2006, 40, 191–197. [Google Scholar] [PubMed]
- Trimpert, J.; Dietert, K.; Firsching, T.C.; Ebert, N.; Thi Nhu Thao, T.; Vladimirova, D.; Kaufer, S.; Labroussaa, F.; Abdelgawad, A.; Conradie, A.; et al. Development of safe and highly protective live-attenuated SARS-CoV-2 vaccine candidates by genome recoding. Cell Rep. 2021, 36, 109493. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty-percent endpoints. Am. J. Hyg. 1938, 27, 493–497. [Google Scholar]
- Silvas, J.A.; Vasquez, D.M.; Park, J.G.; Chiem, K.; Allué-Guardia, A.; Garcia-Vilanova, A.; Platt, R.N.; Miorin, L.; Kehrer, T.; Cupic, A.; et al. Contribution of SARS-CoV-2 accessory proteins to viral pathogenicity in K18 human ACE2 transgenic mice. J. Virol. 2021, 95, e0040221. [Google Scholar] [CrossRef] [PubMed]
- Law, P.T.; Wong, C.H.; Au, T.C.; Chuck, C.P.; Kong, S.K.; Chan, P.K.S.; To, K.F.; Lo, A.W.I.; Chan, J.Y.W.; Suen, Y.K.; et al. The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J. Gen. Virol. 2005, 86, 1921–1930. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.M.; Tsoi, H.; Chan, W.M.; Zhai, S.; Wong, C.O.; Yao, X.; Chan, W.V.; Tsui, S.K.W.; Chan, H.Y.E. The ion channel activity of the SARS-coronavirus 3a protein is linked to its pro-apoptotic function. Int. J. Biochem. Cell Biol. 2019, 41, 2232–2239. [Google Scholar] [CrossRef]
- Miao, G.; Zhao, H.; Li, Y.; Ji, M.; Chen, Y.; Shi, Y.; Bi, Y.; Wang, P.; Zhang, H. ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation. Dev. Cell 2021, 56, 427–442. [Google Scholar]
- Qu, Y.; Wang, X.; Zhu, Y.; Wang, W.; Wang, Y. ORF3a-mediated incomplete autophagy facilitates severe acute respiratory syndrome coronavirus-2 replication. Front. Cell Dev. Biol. 2021, 9, 716208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, H.; Pei, R.; Mao, B.; Zhao, Z.; Li, H.; Lin, Y.; Lu, K. The SARS-CoV-2 protein ORF3a inhibits fusion of autophagosomes with lysosomes. Cell Discov. 2021, 7, 31. [Google Scholar] [CrossRef] [PubMed]
- Freundt, E.C.; Yu, L.; Goldsmith, C.S.; Welsh, S.; Cheng, A.; Yount, B.; Liu, W.; Frieman, M.B.; Buchholz, U.J.; Screaton, G.R.; et al. The open reading frame 3a protein of severe acute respiratory syndrome-associated coronavirus promotes membrane rearrangement and cell death. J. Virol. 2010, 84, 1097–1109. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Dellibovi-Ragheb, T.A.; Kerviel, A.; Pak, E.; Qiu, Q.; Fisher, M.; Takvorian, P.M.; Bleck, C.; Hsu, V.W.; Fehr, A.R.; et al. β-coronaviruses use lysosomes for egress instead of the biosynthetic secretory pathway. Cell 2020, 183, 1520–1535. [Google Scholar] [CrossRef]
- Guarnieri, J.W.; Angelin, A.; Murdock, D.G.; Schaefer, P.; Portluri, P.; Lie, T.; Huang, J.; Wallace, D.C. SARS-CoV-2 viroporins activate the NLRP3-inflammasome by the mitochondrial permeability transition pore. Front. Immunol. 2023, 14, 1064293. [Google Scholar]
- Siu, K.L.; Yuen, K.S.; Castaño-Rodriguez, C.; Ye, Z.W.; Yeung, M.L.; Fung, S.Y.; Yuan, S.; Chan, C.P.; Yuen, K.Y.; Enjuanes, L.; et al. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome bypromoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019, 33, 8865–8877. [Google Scholar] [CrossRef]
- Solinger, J.A.; Spang, A. Tethering complexes in the endocytic pathway: CORVET and HOPS. FEBS J. 2013, 280, 2743–2757. [Google Scholar] [CrossRef] [PubMed]
- Walia, K.; Sharma, A.; Paul, S.; Chouhan, P.; Kumar, G.; Ringe, R.; Sharma, M.; Tuli, A. SARS-CoV-2 virulence factor ORF3a blocks lysosome function by modulating TBC1D5-dependent Rab7 GTPase cycle. Nat. Commun. 2024, 15, 2053. [Google Scholar] [CrossRef] [PubMed]
- Yinda, C.K.; Port, J.R.; Bushmaker, T.; Offei Owusu, I.; Purushotham, J.N.; Avanzato, V.A.; Fischer, R.J.; Schulz, J.; Holbrook, M.G.; Hebner, M.J.; et al. K18-hACE2 mice develop respiratory disease resembling severe COVID-19. PLoS Pathog. 2021, 17, e1009195. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.A.; Xie, X.; Bailey, A.L.; Kalveram, B.; Lokugamage, K.G.; Muruato, A.; Zou, J.; Zhang, X.; Juelich, T.; Smith, J.K.; et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 2021, 591, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Valleriani, F.; Di Pancrazio, C.; Spedicato, M.; Di Teodoro, G.; Malatesta, D.; Petrova, T.; Profeta, F.; Colaianni, M.L.; Berjaoui, S.; Puglia, I.; et al. A cell-adapted SARS-CoV-2 mutant, showing a deletion in the spike protein spanning the furin cleavage site, has reduced virulence at the lung level in K18-hACE2 mice. Virology 2024, 592, 109997. [Google Scholar] [PubMed]

| Fragment | Primer | Sequence (5′–3′) | Size (bp) | Overlap with Next Fragment (bp) |
| 1 | Delta-F1-F | ATATTAGGTTTATACCTTCCCAGG | 3317 | 276 |
| Delta-F1-R | CTGGTGTAAGTTCCATCTCTAATTG | |||
| 2 | Delta-F2-F | CAGATGAGGATGAAGAAGAAGG | 3271 | 313 |
| Delta-F2-R | TTTGTGCTCCAAAGACAACGTATAC | |||
| 3 | Delta-F3-F | ATCTTGTACCAAACCAACCATATCC | 3016 | 301 |
| Delta-F3-R | TCAGCAGCCAAAACACAAGC | |||
| 4 | Delta-F4-F | GTGACATAGCATCTACAGATACTTG | 3249 | 303 |
| Delta-F4-R | CTAAGAGAATGTCATTGTGTAACTGG | |||
| 5 | Delta-F5-F | TCAACCGCTACTTTAGACTGAC | 2940 | 266 |
| Delta-F5-R | AATAGATTACCAGAAGCAGCGTG | |||
| 6 | Delta-F6-F | AACTGTTTGGATGACAGATGC | 3267 | 206 |
| Delta-F6-R | AACCAAAGCACTCACAGTG | |||
| 7 | Delta-F7-F | ATGCCAGATTACGTGCTAAGCAC | 2960 | 249 |
| Delta-F7-R | ACCTAACTGACTATGACTAAAATCTC | |||
| 8 | Delta-F8-F | GGAGTCACATTAATTGGAGAAGC | 3171 | 291 |
| Delta-F8-R | GCATCAGTAGTGTCAGCAATGTC | |||
| 9 | Delta-F9-F | ACCTTGTAATGGTGTTGAAGG | 2945 | 346 |
| Delta-F9-R | TCATGTTCAGAAATAGGACTTGTTG | |||
| 10 | Delta-F10-F | GATGGCAACTAGCACTCTCC | 3178 | 204 |
| Delta-F10-R | TTTGGCAATGTTGTTCCTTGAGG | |||
| 11 | Delta-F11-F | GAAAGATCTCAGTCCAAGATGG | 1301 | 123 |
| Delta-F11-R | TTTTTTGTCATTCTCCTAAGAAGCT | |||
| BAC-YAC | BAC-YAC-F | CGAGTGTACAGTGAACAATGC | 9016 | 64 |
| BAC-YAC-R | GAACAGATCTACAAGAGATCG |
| Primer | Sequence (5’–3’) |
|---|---|
| Y160A-F | CTTTGCTGGCATACTAATTGTTACGACTATTGTATACCTGCAAATAGTGTAACTTCTTCATAGGGATAACAGGGTAATCG |
| Y160A-R | ACCTGAAGTAATGACAATTGAAGAAGTTACACTATTTGCAGGTATACAATAGTCGTAAGCCAGTGTTACAACCAATTAAC |
| Y211A-F | AAAGACTGTGTTGTATTACACAGTTACTTCACTTCAGACGCATACCAGCTGTACTCAACTTAGGGATAACAGGGTAATCG |
| Y211A-R | AGTGTCTGTACTCAATTGAGTTGAGTACAGCTGGTATGCGTCTGAAGTGAAGTAACTGGCCAGTGTTACAACCAATTAAC |
| Y233A-F | AGTACAGACACTGGTGTTGAACATGTTACCTTCTTCATCGCAAATAAAATTGTTGATGAGTAGGGATAACAGGGTAATCG |
| Y233A-R | TTGGACATGTTCTTCAGGCTCATCAACAATTTTATTTGCGATGAAGAAGGTAACATGTGCCAGTGTTACAACCAATTAAC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stephens, E.B.; Kunec, D.; Henke, W.; Vidal, R.M.; Greishaber, B.; Saud, R.; Kalamvoki, M.; Singh, G.; Kafle, S.; Trujillo, J.D.; et al. The Role of the Tyrosine-Based Sorting Signals of the ORF3a Protein of SARS-CoV-2 in Intracellular Trafficking and Pathogenesis. Viruses 2025, 17, 522. https://doi.org/10.3390/v17040522
Stephens EB, Kunec D, Henke W, Vidal RM, Greishaber B, Saud R, Kalamvoki M, Singh G, Kafle S, Trujillo JD, et al. The Role of the Tyrosine-Based Sorting Signals of the ORF3a Protein of SARS-CoV-2 in Intracellular Trafficking and Pathogenesis. Viruses. 2025; 17(4):522. https://doi.org/10.3390/v17040522
Chicago/Turabian StyleStephens, Edward B., Dusan Kunec, Wyatt Henke, Ricardo Martin Vidal, Brandon Greishaber, Rabina Saud, Maria Kalamvoki, Gagandeep Singh, Sujan Kafle, Jessie D. Trujillo, and et al. 2025. "The Role of the Tyrosine-Based Sorting Signals of the ORF3a Protein of SARS-CoV-2 in Intracellular Trafficking and Pathogenesis" Viruses 17, no. 4: 522. https://doi.org/10.3390/v17040522
APA StyleStephens, E. B., Kunec, D., Henke, W., Vidal, R. M., Greishaber, B., Saud, R., Kalamvoki, M., Singh, G., Kafle, S., Trujillo, J. D., Ferreyra, F. M., Morozov, I., & Richt, J. A. (2025). The Role of the Tyrosine-Based Sorting Signals of the ORF3a Protein of SARS-CoV-2 in Intracellular Trafficking and Pathogenesis. Viruses, 17(4), 522. https://doi.org/10.3390/v17040522






