1. Introduction
Viral infection drives host diversity, densities and growth, and global biogeochemical cycles [
1,
2,
3]. This can occur through lytic infections where the viruses infect cells and then immediately replicate and lyse them, killing off infection-sensitive lineages [
4,
5]. Or it can proceed through temperate infection where viruses infect and then persist within cells for extended periods [
6,
7], allowing cells prolonged access to genes shared between hosts by the viruses. Altogether, this means that lytic dynamics can suppress bacterial population growth and abundances while temperate infection can enhance it [
8].
In lytic dynamics, infection and lysis can be thought of and modeled as synonymous [
5,
9,
10], similar to how predation is modeled in microorganismal predator–prey systems [
11,
12]. In this scenario, virus–host encounters lead to infection at a given efficiency that then leads to an equal amount of lysis because lytic viruses rapidly kill any cells they infect. This means that lytic dynamics are directly driven by encounter rates that are the product of virus and host densities. In contrast to density-dependent lytic dynamics, temperate infection appears to be modulated by host physiology, where lysis is suspended post-infection so long as hosts remain healthy [
8,
13,
14]. This effectively decouples infection and lysis in temperate dynamics.
Given that bacterial over-proliferation is a pervasive threat to ecosystems [
15], knowing which infection dynamic dominates a given ecosystem and predicting what the ecosystem impacts might be are central questions in viral ecology. However, the fields that describe and that test the rules of viral infection—theoretical ecology and environmental microbiology, respectively—remain distant. This is largely driven by a lack of overlap in expertise between empiricists and modelers that can result in empiricists avoiding the mathematics of models and in theoreticians making suggestions that are not empirically tractable [
16]. Further, difficulties in communication between empirical and theoretical researchers may hint at even deeper challenges, and the possibility that empirical and theoretical fields may currently be somewhat incommensurate [
17,
18,
19,
20]. Altogether, the decoupling of theory and data prohibits the improvement and refinement of models through empirical challenges, which in turn precludes the development of an empirically sound predictive understanding of viral infections and effects.
To remedy this, we examined three theoretical models mechanistically representing lytic, super-lytic, and sub-lytic/temperate infection dynamics. These models either had lysis rates that were unaffected, amplified, or suppressed by rapid host proliferation, respectively. In contrast to theoretical analyses used to assess models, we subjected model outputs to routine experimental data analyses and presentation to learn how to distinguish the models with empirical metrics. By doing so, we found that the models were remarkably similar. Virus and host parameters were similarly related to viral and host standing stocks. The relationship between lysis rates, host growth rates, viral and host abundances, and the virus: microbe ratio was conserved across all models. As a result, distinguishing between models appears to require experimental manipulation. Focusing on nutrient addition as a common experimental approach that also targets the mechanistic and mathematic differences between models, we found that simulating nutrient addition allowed us to bisect lytic and temperate models. In particular, lytic systems would experience no increase in host densities with nutrient addition but would show increasing instability of viral and host abundances. In contrast, temperate systems would remain consistently stable under nutrient addition but have increasing host densities. Building on this, we lay out a dichotomous key to allow researchers to determine whether lytic or temperate infection dynamics dominate their samples or ecosystem of interest using the commonly used approaches of counting viruses and hosts and measuring their responses to nutrient addition.
2. Methods
2.1. Models and Their Mechanistic Differences
Three mechanistically divergent theoretical models were compared (model parameters listed and summarized in
Table 1). The models included a lytic Lotka–Volterra model with a logistic growth term (“Logistic Lotka-Volterra” [
21]), a super-lytic model that enhances the viral killing of hosts when hosts are proliferating rapidly (“Weitz & Dushoff” [
22]), and a sub-lytic/temperate model that suppresses the viral killing of hosts when hosts are proliferating rapidly (“Piggyback-the-Winner” [
8]).
In all cases, the models comprise the following terms:
host population growth—viral lysis—host viral-independent mortality
viral production—viral decay
2.2. Logistic Lotka–Volterra Model
The Logistic Lotka–Volterra model is a basic predator–prey model [
21]. This model has the following equations (see
Table 1 for parameters):
These equations, like all the models, can be used to calculate steady-state solutions where the host and viral densities are unchanging (N* and V*, respectively; i.e.,
and
= 0, assuming that N and V ≠ 0). These values are also known as equilibrium values because the host and viral densities are at equilibrium with each other and not generating oscillations. These densities presumably approximate the densities that would be sampled at any given time in the environment by direct counts of viruses and hosts, or serve as values that the host and viral densities might return to after perturbation [
8,
23,
24,
25]. When solving for steady-state solutions, these equations become the following:
Therefore, in this model, host steady-state densities are driven by viral decay, burst size, and adsorption (i.e., steady-state host densities are an outcome of viral, not host parameters). Note that steady-state viral solutions (V*) in this and the following models are dependent on steady-state host densities (N*) and thus determined by these parameters as well. In addition, viral steady-state densities result from the host intrinsic growth rate, viral-independent mortality, and viral adsorption efficiency.
2.3. Weitz & Dushoff Model
In contrast with Piggyback-the-Winner, the “Weitz & Dushoff” model [
22] enhances lysis and viral production when host densities are low and increasing. In this model, lysis and viral production are functions of host physiology. The Weitz & Dushoff model has the following equations (see
Table 1 for parameters):
Note that this model introduces elements to the lysis and viral production term, compared to Equation (2). The part of this element suppresses the lysis and production terms as . The a term determines how much lysis is suppressed as N approaches K and host growth slows.
When solved for steady-state solutions (i.e.,
= 0), Equations (9) and (10) become the following:
As a result of introducing the element, there are many more parameters predicted to drive host and viral steady-state densities in this model, and the relationship between the parameters is much more complicated than in the other models. Further, the solution for N* involves a quadratic that therefore has two sets of solutions depending on whether the ± in Equation (7) is implemented as a plus or as a minus. In particular, implementing Equation (8) using a plus leads to the generation of pervasive negative steady-state values for viruses and hosts. Altogether, compared to other models, it is much less clear what drives viral and host steady-state densities and what those densities should be in this model.
2.4. Piggyback-the-Winner Model
The “Piggyback-the-Winner” model [
8] simulates a temperate infection dynamic where viruses choose to not kill their rapidly growing hosts far below crowded and nutrient-limited densities. It does so by suppressing lysis at low host densities. The Piggyback-the-Winner model has the following equations (see
Table 1 for parameters):
Note that this model introduces to the viral production term, compared to Equation (2). This has the effect of suppressing viral production, and therefore infection, in low-density host conditions (i.e., ). This delays the onset of lytic dynamics until hosts are dense (i.e., ), presenting a de facto temperate to lytic transition as systems go from having low to high host densities.
When solved for steady-state solutions (i.e.,
= 0), Equations (5) and (6) become the following:
Compared to the Logistic Lotka–Volterra model (Equations (3) and (4)), host steady-state densities in the Piggyback-the-Winner model are also driven by the ecosystem host carrying capacity (K), especially with N* rising with K. Viral steady-state solutions, V*, are the same as for Equation (4), but are driven differently by their dependence on N*, which varies between Equations (3) and (11), especially through the inclusion of K in the model.
2.5. The Effect of Varying All Parameters at Once
To investigate how N* and V* values would look across different conditions (e.g., samples with different growth rates, burst sizes, etc.), we made arrays of random values for each parameter (
) with 10-fold ranges, and solved for steady-state host (N*) and viral (V*) densities (see
Table 1), building off Weitz et al. (2017) [
20]. In total, we solved for 10,000 N* and V* values per model. We first quantified the effect of each parameter and term on N* and V* using Principal Component Analysis on the 10,000-solution dataset (PCA; prcomp() function with scale = TRUE;
Figure 1). We then used Spearman’s Rho (the corr.test() function in R; with data log
10-transformed and exact = FALSE) to summarize the relationship between N* and the host proliferation rate and lysis rates for each model across all 10,000 solutions (
Figure 2), as well as for the relationship between N* and V* and the virus-to-microbe ratio (VMR; V*/N*;
Figure 3). Note that the Spearman’s Rho coefficient quantifies how monotonic or consistent the correlation between two variables is and varies between −1 and 1 for strong negative and positive correlations, with 0 showing a lack of monotonic correlation. We considered correlations with different signs to be diagnostic between models (e.g., if the lytic models showed positive correlations but the temperate model had a negative correlation).
2.6. Effect of Altered Carrying Capacity (K) on Steady-State N* and V* and Stability
To simulate the impact of nutrient enrichment, a widely used experimental approach that targets the mechanistic differences in the models, we modified the range of carrying capacity (K) for all models (
Figure 4). Following the same methodology as in
Section 2.5, we systematically increased K across multiple scenarios, allowing K to range between 1 × 10
5–1 × 10
6, 1 × 10
6–1 × 10
7, 1 × 10
7–1 × 10
8, 1 × 10
8–1 × 10
9, and 1 × 10
9–1 × 10
10 in each simulation. The ranges of all other parameters remained the same as in
Section 2.5. We then computed the resulting steady-state host (N*) and viral (V*) densities to evaluate how increasing resource availability influences microbial and viral population dynamics under the same model framework. We performed stability analysis on these steady states to assess their robustness under different nutrient conditions. For each model, we computed the Jacobian matrix at the steady-state points and determined its eigenvalues. Calculations were conducted in Python using the sympy, numpy, and pandas package (Code is available at
https://tinyurl.com/EmpiricistGuide). Each stability analysis resulted in two eigenvalues due to models being two dimensional systems. The stability of each steady-state point was classified as either stable or unstable based on the signs of the real parts of the eigenvalues (
Figure 4;
Supplementary Table S1). We categorized stable nodes and stable spirals as stable, and saddle points, unstable nodes, and unstable spirals as unstable. We did not observe any centers in our analysis, as no real parts of the eigenvalues were equal to 0.
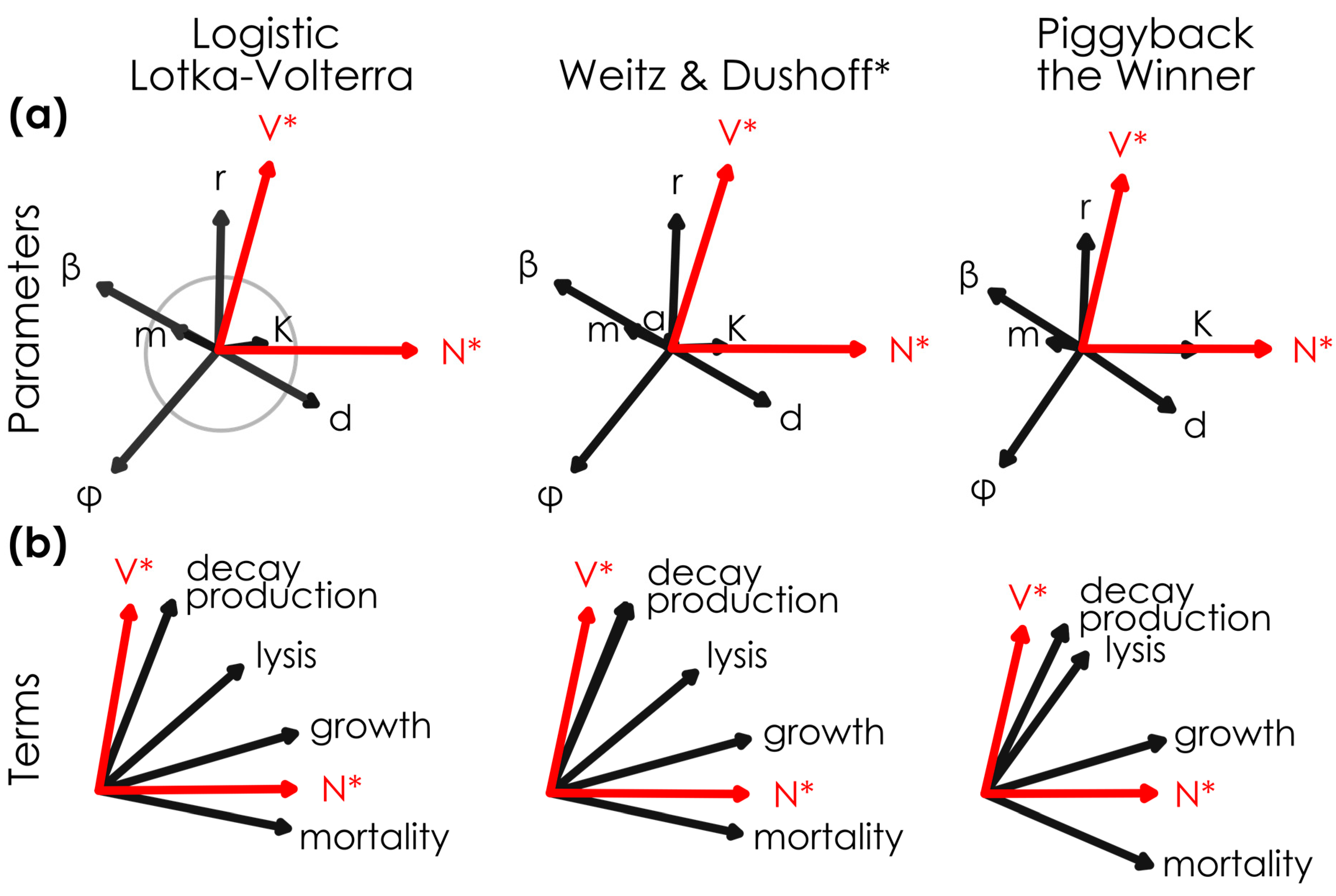
Figure 1.
Mechanistically divergent lytic and sub-lytic models had similar relationships between viral density, host density, and permuted parameters and terms. The relationship between model (
a) parameters, (
b) terms, and steady-state viral and host abundances (V* and N*) in mechanistically-divergent models when parameters are randomly varied across 10,000 permutations (see
Table 1 for the range of parameter values), where
n = 7304,
n = 7171, and
n = 6275 solutions had positive virus and host densities in the Logistic Lotka–Volterra, Weitz & Dushoff models, and Piggyback-the-Winner models, respectively. Steady-state viral and host densities (red arrows) and parameters and terms (black arrows) are shown for lytic Logistic Lotka–Volterra, super-lytic Weitz & Dushoff, and sub-lytic Piggyback-the-Winner models. Vectors (i.e., arrows) were normalized across models; the gray circle in the top left panel has a loading value = 25 a.u. diameter. PCA outputs were rotated so that N* vectors had similar directions across models. * Note that the Weitz & Dushoff model results are for the ‘negative sign’ quadratic solution, as the ‘positive sign’ solution showed pervasive negative host and viral densities.
Figure 1.
Mechanistically divergent lytic and sub-lytic models had similar relationships between viral density, host density, and permuted parameters and terms. The relationship between model (
a) parameters, (
b) terms, and steady-state viral and host abundances (V* and N*) in mechanistically-divergent models when parameters are randomly varied across 10,000 permutations (see
Table 1 for the range of parameter values), where
n = 7304,
n = 7171, and
n = 6275 solutions had positive virus and host densities in the Logistic Lotka–Volterra, Weitz & Dushoff models, and Piggyback-the-Winner models, respectively. Steady-state viral and host densities (red arrows) and parameters and terms (black arrows) are shown for lytic Logistic Lotka–Volterra, super-lytic Weitz & Dushoff, and sub-lytic Piggyback-the-Winner models. Vectors (i.e., arrows) were normalized across models; the gray circle in the top left panel has a loading value = 25 a.u. diameter. PCA outputs were rotated so that N* vectors had similar directions across models. * Note that the Weitz & Dushoff model results are for the ‘negative sign’ quadratic solution, as the ‘positive sign’ solution showed pervasive negative host and viral densities.
![Viruses 17 00513 g001]()
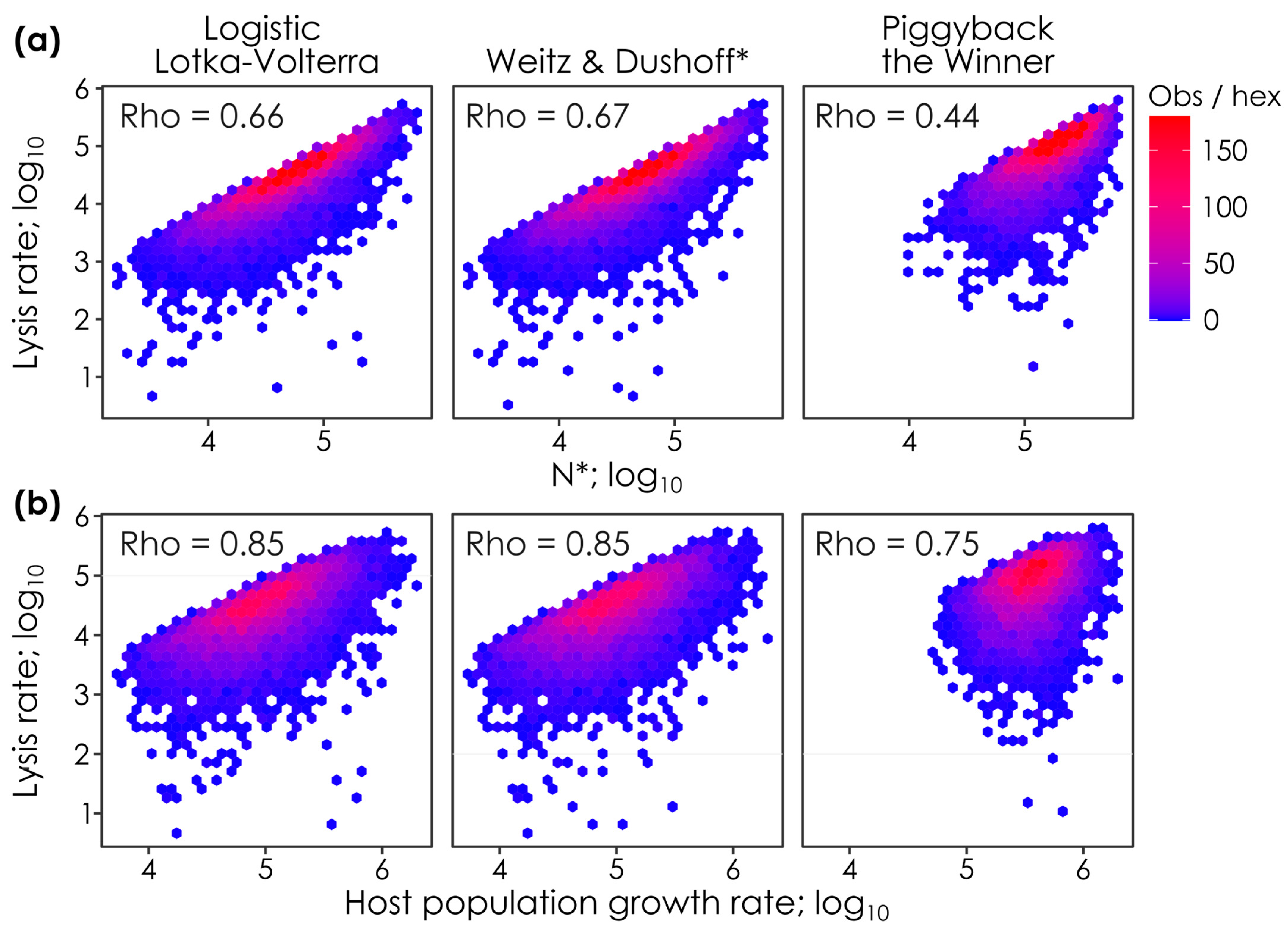
Figure 2.
Lysis rates covary with steady-state host densities (N*) and host population growth rates in all models. The relationship between lysis rates and (
a) steady-state host abundances (N*) and (
b) host population growth rates when parameter values are permuted in lytic Logistic Lotka–Volterra, super-lytic Weitz & Dushoff, and sub-lytic Piggyback-the-Winner models. Each panel shows the outcomes of 10,000 parameter permutations (see
Table 1), where
n = 7304,
n = 7171, and
n = 6275 solutions had positive virus and host densities in the Logistic Lotka–Volterra, Weitz & Dushoff, and Piggyback-the-Winner models, respectively. Colors show the number of observations in each hex (i.e., observation density). Spearman’s Rho values, the monotonicity of the relationship between lysis rates and N* or host population growth rates, are shown on each panel. * Note that the Weitz & Dushoff model results are for the ‘negative sign’ quadratic solution, as the ‘positive sign’ solution showed pervasive negative host and viral densities. Note that correlations with Spearman’s Rho values < 0.2 are considered weak, 0.2. to 0.5 are moderate, and >0.5 strong [
26].
Figure 2.
Lysis rates covary with steady-state host densities (N*) and host population growth rates in all models. The relationship between lysis rates and (
a) steady-state host abundances (N*) and (
b) host population growth rates when parameter values are permuted in lytic Logistic Lotka–Volterra, super-lytic Weitz & Dushoff, and sub-lytic Piggyback-the-Winner models. Each panel shows the outcomes of 10,000 parameter permutations (see
Table 1), where
n = 7304,
n = 7171, and
n = 6275 solutions had positive virus and host densities in the Logistic Lotka–Volterra, Weitz & Dushoff, and Piggyback-the-Winner models, respectively. Colors show the number of observations in each hex (i.e., observation density). Spearman’s Rho values, the monotonicity of the relationship between lysis rates and N* or host population growth rates, are shown on each panel. * Note that the Weitz & Dushoff model results are for the ‘negative sign’ quadratic solution, as the ‘positive sign’ solution showed pervasive negative host and viral densities. Note that correlations with Spearman’s Rho values < 0.2 are considered weak, 0.2. to 0.5 are moderate, and >0.5 strong [
26].
![Viruses 17 00513 g002]()
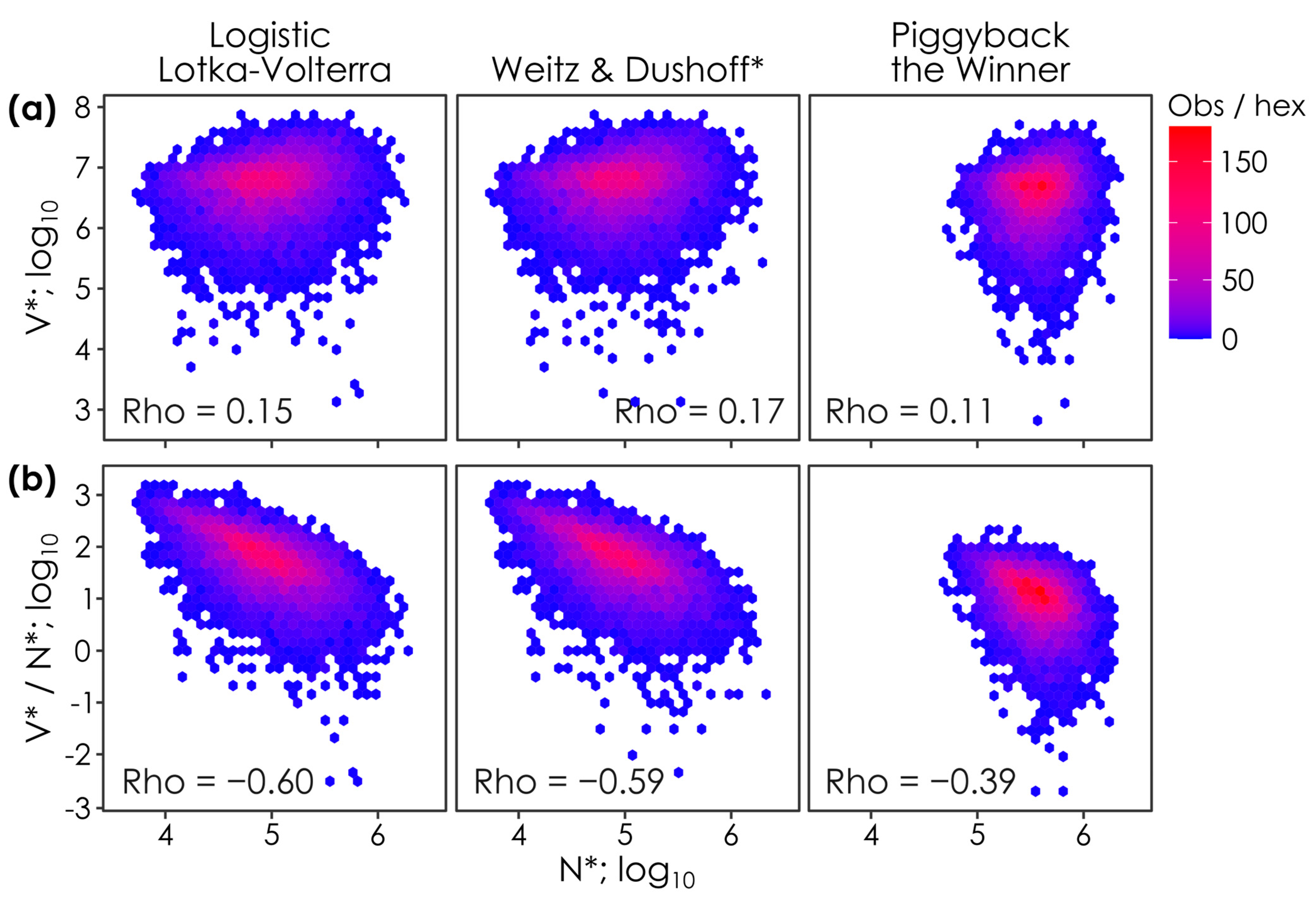
Figure 3.
Host densities (N*) are positively correlated with viral densities (V*) and negatively correlated with virus-to-microbe ratios (V*/N*) in all models. The relationship between steady-state host (N*) and (
a) viral densities (V*) and (
b) virus-to-microbe ratios (V*/N*) when parameter values are permuted in lytic Logistic Lotka–Volterra, super-lytic Weitz & Dushoff, and sub-lytic/temperate Piggyback-the-Winner models. Each panel shows the outcomes of 10,000 parameter permutations (see
Table 1), where
n = 7304,
n = 7171, and
n = 6275 solutions had positive virus and host densities in the Logistic Lotka–Volterra, Weitz & Dushoff, and Piggyback-the-Winner models, respectively. Colors show the number of observations in each hex (i.e., observation density). Spearman’s Rho values, the monotonicity of the relationship between N* and V* or V*/N*, are shown on each panel. * Note that the Weitz & Dushoff model results are for the ‘negative sign’ quadratic solution, as the ‘positive sign’ solution showed pervasive negative host and viral densities. Note that correlations with Spearman’s Rho values < 0.2 are considered weak, 0.2. to 0.5 are moderate, and >0.5 strong [
26].
Figure 3.
Host densities (N*) are positively correlated with viral densities (V*) and negatively correlated with virus-to-microbe ratios (V*/N*) in all models. The relationship between steady-state host (N*) and (
a) viral densities (V*) and (
b) virus-to-microbe ratios (V*/N*) when parameter values are permuted in lytic Logistic Lotka–Volterra, super-lytic Weitz & Dushoff, and sub-lytic/temperate Piggyback-the-Winner models. Each panel shows the outcomes of 10,000 parameter permutations (see
Table 1), where
n = 7304,
n = 7171, and
n = 6275 solutions had positive virus and host densities in the Logistic Lotka–Volterra, Weitz & Dushoff, and Piggyback-the-Winner models, respectively. Colors show the number of observations in each hex (i.e., observation density). Spearman’s Rho values, the monotonicity of the relationship between N* and V* or V*/N*, are shown on each panel. * Note that the Weitz & Dushoff model results are for the ‘negative sign’ quadratic solution, as the ‘positive sign’ solution showed pervasive negative host and viral densities. Note that correlations with Spearman’s Rho values < 0.2 are considered weak, 0.2. to 0.5 are moderate, and >0.5 strong [
26].
![Viruses 17 00513 g003]()
Figure 4.
Increased nutrient availability (K) drives increasing instability in lytic models and increasing host densities in the temperate model. Increasing carrying capacity (equivalent to nutrient availability; K) from 1 × 10
5 to 1 × 10
10 cells per ml (
a–
e) leads to elevated host densities (N*) in the temperate Piggyback-the-Winner model, but not in the lytic Logistic Lotka–Volterra and Weitz & Dushoff models. In contrast, the simulated nutrient addition leads to increased instability in the lytic models (pink dots and text for unstable steady-states vs. black dots and text for stable steady states), with no change in stability in the temperate model. See each panel for the number of stable and unstable steady states (
n = ; pink for unstable and black for stable solutions). Each panel shows the outcomes of 10,000 parameter permutations (see
Table 1), where solutions had positive virus and host densities in each model. Note that K in this simulation assumes that the carrying capacity (i.e., nutrient availability) is constant or constantly added, in contrast with batch or pulse experiments. * Note that the Weitz & Dushoff model results are for the ‘negative sign’ quadratic solution, as the ‘positive sign’ solution showed pervasive negative host and viral densities.
Figure 4.
Increased nutrient availability (K) drives increasing instability in lytic models and increasing host densities in the temperate model. Increasing carrying capacity (equivalent to nutrient availability; K) from 1 × 10
5 to 1 × 10
10 cells per ml (
a–
e) leads to elevated host densities (N*) in the temperate Piggyback-the-Winner model, but not in the lytic Logistic Lotka–Volterra and Weitz & Dushoff models. In contrast, the simulated nutrient addition leads to increased instability in the lytic models (pink dots and text for unstable steady-states vs. black dots and text for stable steady states), with no change in stability in the temperate model. See each panel for the number of stable and unstable steady states (
n = ; pink for unstable and black for stable solutions). Each panel shows the outcomes of 10,000 parameter permutations (see
Table 1), where solutions had positive virus and host densities in each model. Note that K in this simulation assumes that the carrying capacity (i.e., nutrient availability) is constant or constantly added, in contrast with batch or pulse experiments. * Note that the Weitz & Dushoff model results are for the ‘negative sign’ quadratic solution, as the ‘positive sign’ solution showed pervasive negative host and viral densities.
![Viruses 17 00513 g004]()
2.7. Code and Implementation
All models were implemented in Python 3.6. Code is available at
https://tinyurl.com/EmpiricistGuide. Steady-state predictions were generated for 10,000 iterations using Equations (3), (4), (7), (8), (11), and (12) with randomly generated parameter values within the ranges shown in
Table 1 using the random.random() command for values < 1 and >0 and random.uniform() command for values > 1. Parameter values as well as predicted steady-state host and viral densities (N* and V*, respectively) and solutions for terms in Equations (1), (2), (5), (6), (9), and (10) were recorded. Terms (e.g., proliferation rate) were solved using the randomly generated parameters in each iteration. Stability analysis was conducted for each model by calculating Jacobians and eigenvalues using Python packages sympy, numpy, and pandas.
4. Discussion
Viral infection of bacteria is one of the most common interactions on the planet. It is a major form of death in nature, with an estimated 10
23 cells infected and lysed per second in the oceans [
1]. Lytic infection can drive bacterial community composition by killing off lineages sensitive to infection and can drive bacterial evolution by selecting for resistant individuals [
5,
27,
28]. This specific lysis of cells can also move large pools of resources between guilds. For example, the viral shunt describes the liberation of photosynthates and metabolites from algae to heterotrophic bacteria [
29].
However, in addition to lytic infection, temperate infection can also take place. In temperate infection, the viruses decide whether to proceed with lysis after infecting the cell [
30]. In this scenario, the viruses respond to host physiology in their decision-making and can reside for extended periods within the host genome. The lytic program can remain suspended until a physiological stress cue is received, inducing the restoration of the lytic program with intracellular viral production and subsequent lysis of the host. This suspended death can allow host densities to rise, in contrast to lytic infection which suppresses host densities [
8].
Bacterial overgrowth has been associated with ecosystem degradation from coral reefs to the human lung and gut [
15,
31,
32,
33]. Given the divergent effects of lytic and temperate infection and how each lifestyle might be affecting ecosystem dysbiosis, understanding the role and prevalence of lytic and temperate infection has emerged as a central question in viral ecology. However, the fields of theoretical and empirical viral ecology—that lay out how these infection dynamics should look and that gather data on whether the dynamics are observed in nature, respectively—are disconnected and perhaps even incommensurate. This hinders theoreticians from refining improved models as well as empirical attempts to test the models. To address the gulf between theory and empirical approaches, we implemented three mechanistically divergent models—super-lytic, lytic, and sub-lytic/temperate—and explored their predictions as if they were empirical data to integrate theoretical and empirical approaches, hopefully accelerating progress in the field.
The models are surprisingly similar, especially given how mechanistically divergent they are. The similarity was observed across several analyses. First, we found that viral and host densities were linked similarly to model parameters like viral burst size, intrinsic growth rates, etc., and to model terms like the host population growth rate, viral production, etc. We also found that the relationship between host densities and viral densities and between host densities and the virus-to-microbe ratio were conserved across all models. Several of these, especially intrinsic and population growth rates and the virus-to-microbe ratio, have been used to differentiate between ecological dynamics like lytic and temperate infection and host infection/growth rate tradeoffs [
20,
34,
35]. Our modeling suggests, however, that these metrics cannot effectively be used to differentiate even mechanistically divergent models.
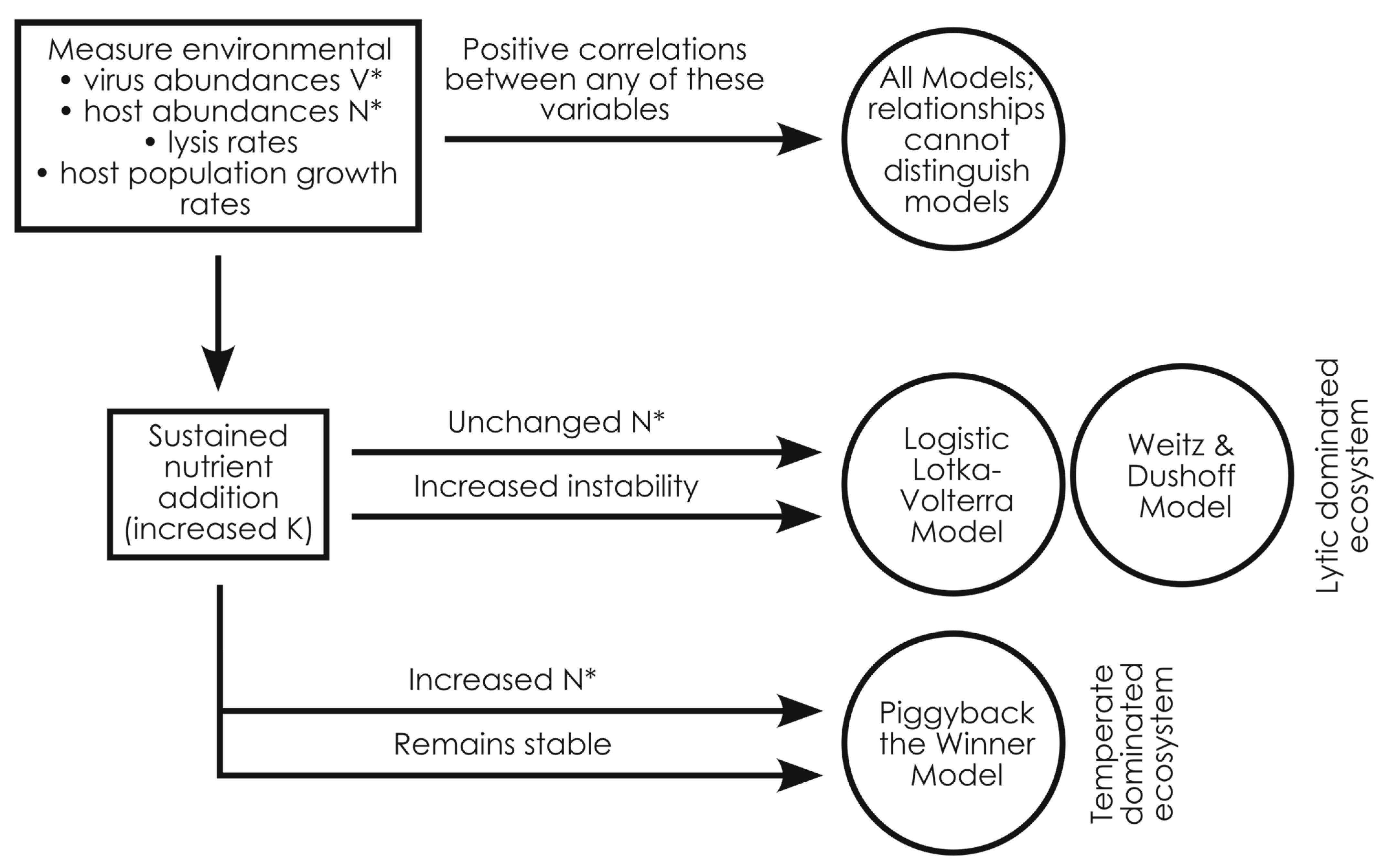
However, the main mechanistic differences between the models hinge on how they drive lysis and viral production when cell densities are far from or close to carrying capacity (K). Nutrient addition—experimentally altering K to change the distance between the current cell abundance and carrying capacity—is also a commonly used empirical approach. We therefore focused on nutrient addition to try to distinguish the models. When we simulated nutrient addition, we were readily able to bisect the lytic and temperate models. The lytic models had no notable increase in host densities with nutrient addition, but became markedly less stable (
Figure 5). In contrast, the temperate model showed elevated host densities and sustained stability with nutrient addition (
Figure 5). It appears that, with all else being equal, temperate infection is required to allow ecosystems to have high host densities with nutrient addition (i.e., high carrying capacity).
Altogether our analyses sift through the remarkably similar predictions of the super-lytic Weitz & Dushoff, lytic Logistic Lotka–Volterra, or sub-lytic/temperate Piggyback-the-Winner models. Although it is likely that lytic and temperate infection takes place simultaneously in ecosystems, by highlighting the similarities and differences between the models, our work provides a guide for empiricists to assess whether their ecosystem or datasets are dominated by super-lytic, lytic, or sub-lytic infection dynamics through commonly used empirical metrics and experimental approaches (
Figure 5). Ultimately, the ability to differentiate between these models using the tractable method of nutrient addition allows researchers to better describe the conditions favorable to lytic and temperate infections and to better predict the effects of viral infection in nature.