The ACE2 Receptor from Common Vampire Bat (Desmodus rotundus) and Pallid Bat (Antrozous pallidus) Support Attachment and Limited Infection of SARS-CoV-2 Viruses in Cell Culture
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses Used and Growth
2.2. Cell Culture
2.3. Bat ACE2 Plasmid Construction Using the PiggyBac Transposon Vector
2.4. Transfection of Bat ACE2 Plasmids in E. coli
2.5. Transgenic Cell Line Development Using the PiggyBac Plasmid Transposon System
2.6. Fluorescence-Activated Cell Sorting (FACS)
2.7. RNA Extraction and qRT-PCR Conditions for Confirmation of Bat ACE2:hTMPRSS2 Cell Lines
2.8. ACE2 and TMPRSS2 Protein Analysis by Western Blotting
2.9. Immunofluorescence Analysis of Viral Infection
2.10. Infection and Replication Dynamics of SC2 and Variants in CVB-ACE2, PB-ACE2 Cell Lines
2.11. ACE2 Sequence Analysis
2.12. Statistics
3. Results
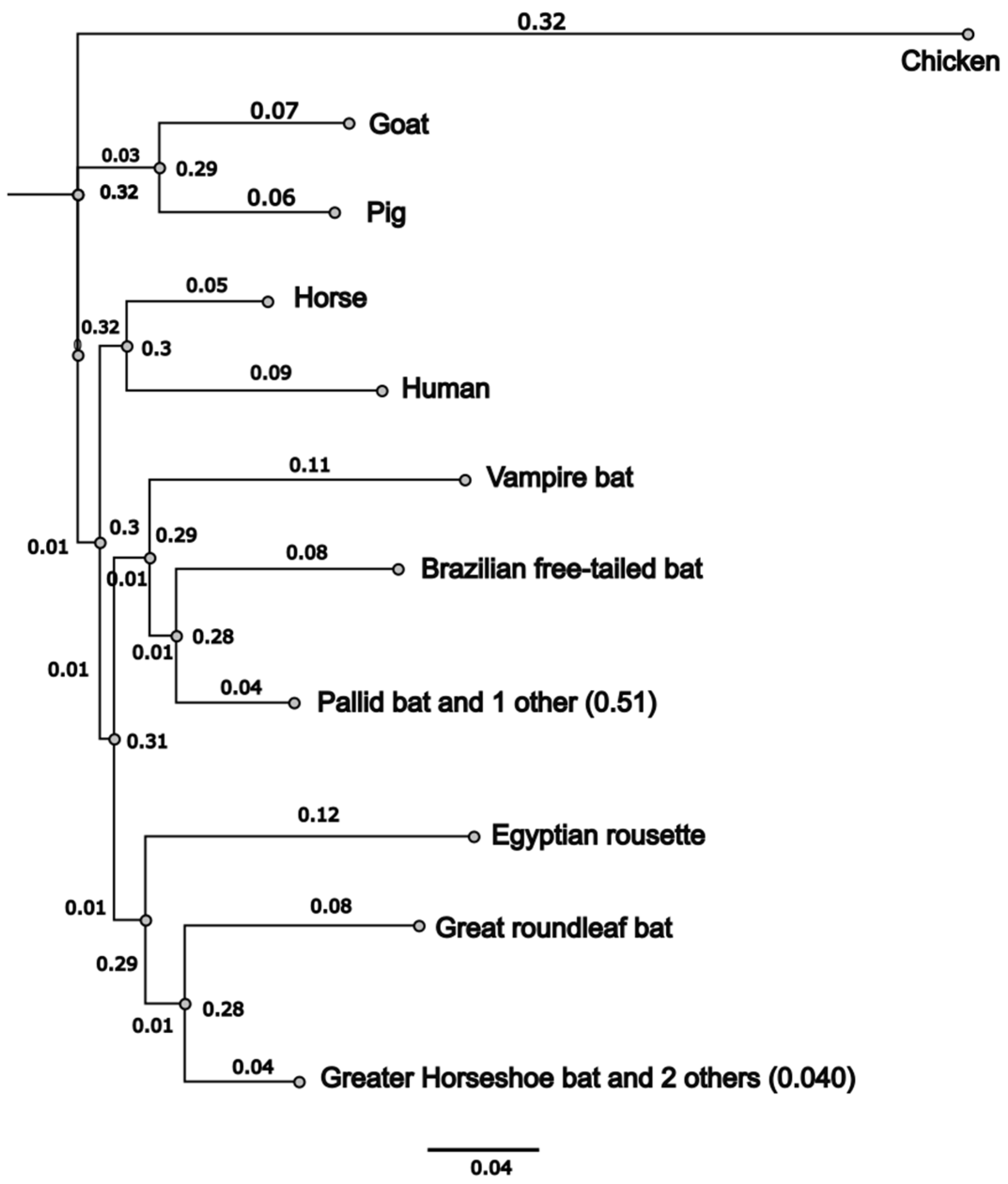
3.1. Common Vampire Bat and Pallid Bat ACE2 Sequences Cluster Independently
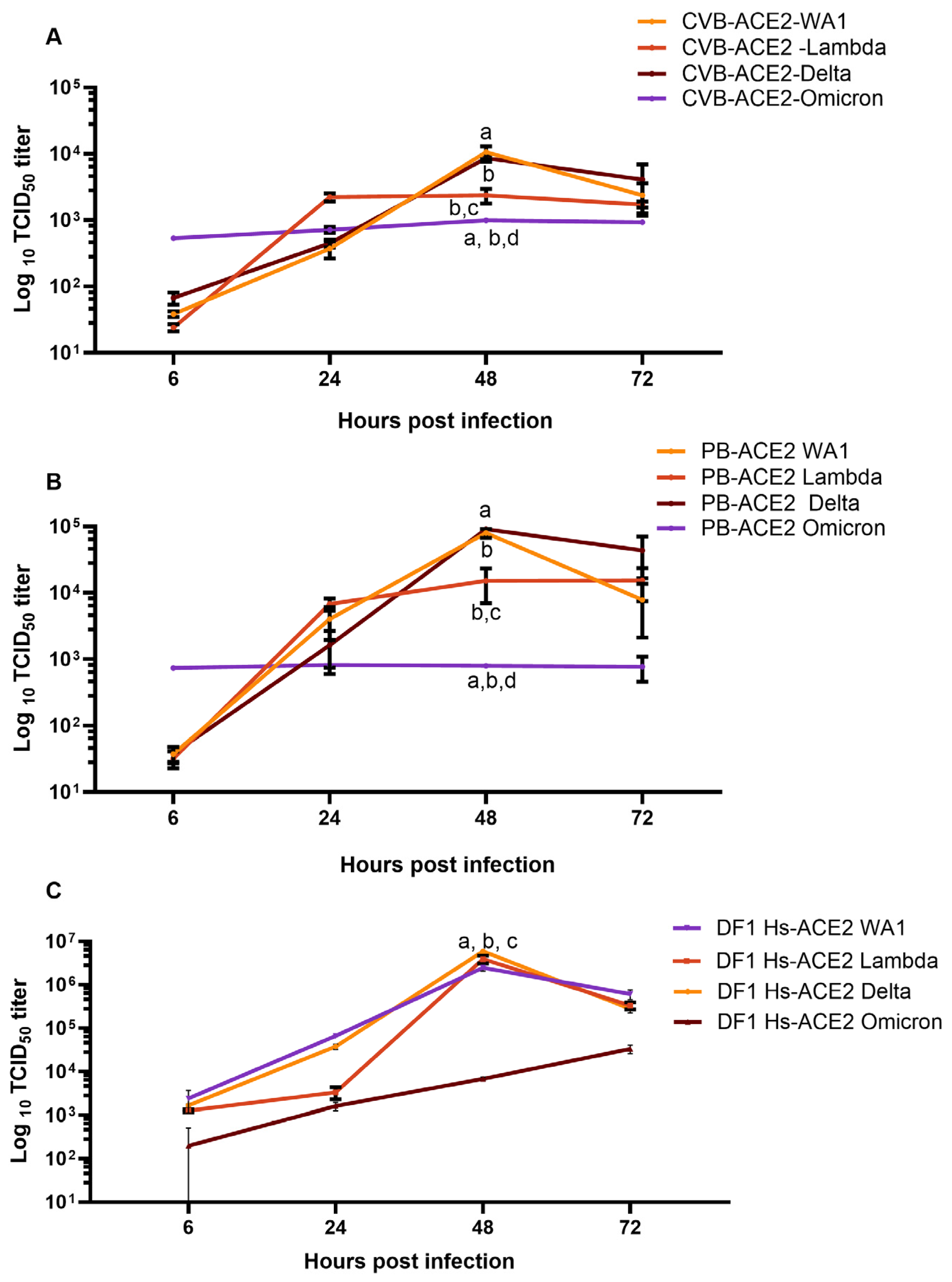
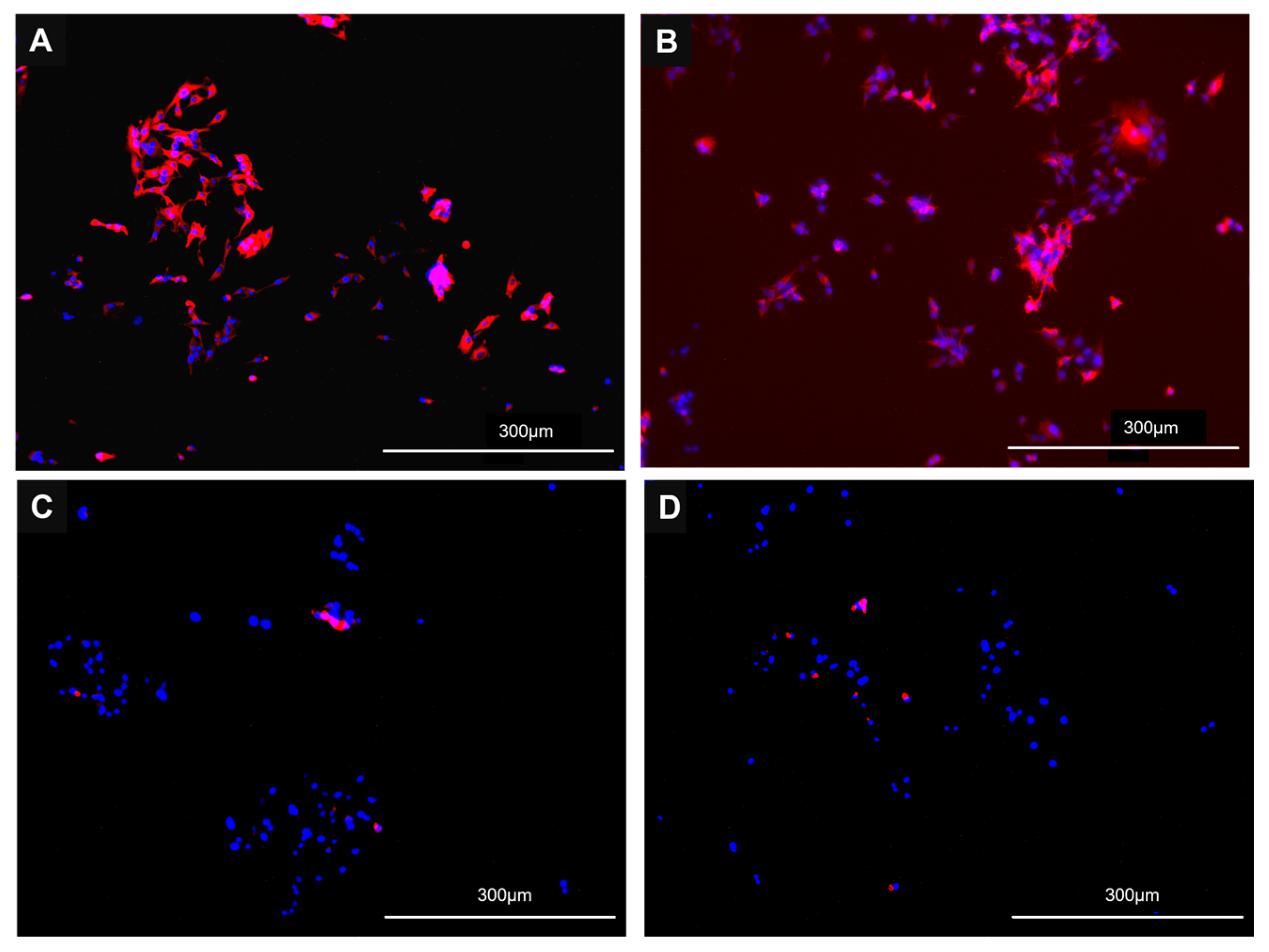
3.2. Cells Lines Expressing CVB or PB ACE2 Support Replication of Multiple SC2 Variants
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Holmes, E.C. The Emergence and Evolution of SARS-CoV-2. Annu. Rev. Virol. 2024, 11, 21–42. [Google Scholar] [CrossRef] [PubMed]
- Machkovech, H.M.; Hahn, A.M.; Garonzik Wang, J.; Grubaugh, N.D.; Halfmann, P.J.; Johnson, M.C.; Lemieux, J.E.; O’Connor, D.H.; Piantadosi, A.; Wei, W.; et al. Persistent SARS-CoV-2 infection: Significance and implications. Lancet Infect. Dis. 2024, 24, e453–e462. [Google Scholar] [CrossRef] [PubMed]
- Steiner, S.; Kratzel, A.; Barut, G.T.; Lang, R.M.; Aguiar Moreira, E.; Thomann, L.; Kelly, J.N.; Thiel, V. SARS-CoV-2 biology and host interactions. Nat. Rev. Microbiol. 2024, 22, 206–225. [Google Scholar] [CrossRef] [PubMed]
- Multiple. A Global Open Access Dataset of Reported SARS-CoV-2 Events in Animals. Available online: https://vis.csh.ac.at/sars-ani/#infections (accessed on 27 March 2025).
- Dhama, K.; Patel, S.K.; Sharun, K.; Pathak, M.; Tiwari, R.; Yatoo, M.I.; Malik, Y.S.; Sah, R.; Rabaan, A.A.; Panwar, P.K.; et al. SARS-CoV-2 jumping the species barrier: Zoonotic lessons from SARS, MERS and recent advances to combat this pandemic virus. Travel Med. Infect. Dis. 2020, 37, 101830. [Google Scholar] [CrossRef]
- do Vale, B.; Lopes, A.P.; Fontes, M.D.C.; Silvestre, M.; Cardoso, L.; Coelho, A.C. Bats, pangolins, minks and other animals—Villains or victims of SARS-CoV-2? Vet. Res. Commun. 2021, 45, 1–19. [Google Scholar] [CrossRef]
- Eslami, N.; Aghbash, P.S.; Shamekh, A.; Entezari-Maleki, T.; Nahand, J.S.; Sales, A.J.; Baghi, H.B. SARS-CoV-2: Receptor and Co-receptor Tropism Probability. Curr. Microbiol. 2022, 79, 133. [Google Scholar] [CrossRef]
- Gao, S.; Zhang, L. ACE2 partially dictates the host range and tropism of SARS-CoV-2. Comput. Struct. Biotechnol. J. 2020, 18, 4040–4047. [Google Scholar] [CrossRef]
- Hossain, M.G.; Javed, A.; Akter, S.; Saha, S. SARS-CoV-2 host diversity: An update of natural infections and experimental evidence. J. Microbiol. Immunol. Infect. 2021, 54, 175–181. [Google Scholar] [CrossRef]
- Jo, W.K.; de Oliveira-Filho, E.F.; Rasche, A.; Greenwood, A.D.; Osterrieder, K.; Drexler, J.F. Potential zoonotic sources of SARS-CoV-2 infections. Transbound. Emerg. Dis. 2021, 68, 1824–1834. [Google Scholar] [CrossRef]
- Leist, S.R.; Schäfer, A.; Martinez, D.R. Cell and animal models of SARS-CoV-2 pathogenesis and immunity. Dis. Model Mech. 2020, 13, dmm046581. [Google Scholar] [CrossRef]
- Nooruzzaman, M.; Diel, D.G. Infection Dynamics, Pathogenesis, and Immunity to SARS-CoV-2 in Naturally Susceptible Animal Species. J. Immunol. 2023, 211, 1195–1201. [Google Scholar] [CrossRef] [PubMed]
- Sharun, K.; Dhama, K.; Pawde, A.M.; Gortázar, C.; Tiwari, R.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J.; de la Fuente, J.; Michalak, I.; Attia, Y.A. SARS-CoV-2 in animals: Potential for unknown reservoir hosts and public health implications. Vet. Q. 2021, 41, 181–201. [Google Scholar] [CrossRef] [PubMed]
- Oude Munnink, B.B.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; van der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M.; et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 2021, 371, 172–177. [Google Scholar] [CrossRef]
- Lu, L.; Sikkema, R.S.; Velkers, F.C.; Nieuwenhuijse, D.F.; Fischer, E.A.J.; Meijer, P.A.; Bouwmeester-Vincken, N.; Rietveld, A.; Wegdam-Blans, M.C.A.; Tolsma, P.; et al. Adaptation, spread and transmission of SARS-CoV-2 in farmed minks and associated humans in the Netherlands. Nat. Commun. 2021, 12, 6802. [Google Scholar] [CrossRef]
- Kok, K.H.; Wong, S.C.; Chan, W.M.; Wen, L.; Chu, A.W.; Ip, J.D.; Lee, L.K.; Wong, I.T.; Lo, H.W.; Cheng, V.C.; et al. Co-circulation of two SARS-CoV-2 variant strains within imported pet hamsters in Hong Kong. Emerg. Microbes Infect. 2022, 11, 689–698. [Google Scholar] [CrossRef]
- Moreno, A.; Lelli, D.; Trogu, T.; Lavazza, A.; Barbieri, I.; Boniotti, M.; Pezzoni, G.; Salogni, C.; Giovannini, S.; Alborali, G.; et al. SARS-CoV-2 in a Mink Farm in Italy: Case Description, Molecular and Serological Diagnosis by Comparing Different Tests. Viruses 2022, 14, 1738. [Google Scholar] [CrossRef]
- Pappas, G.; Vokou, D.; Sainis, I.; Halley, J.M. SARS-CoV-2 as a Zooanthroponotic Infection: Spillbacks, Secondary Spillovers, and Their Importance. Microorganisms 2022, 10, 2166. [Google Scholar] [CrossRef]
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef]
- Dong, W.Q.; Bai, B.; Lin, Y.P.; Gao, J.; Yu, N.S. [Detection of the mRNA expression of human angiotensin-converting enzyme 2 as a SARS coronavirus functional receptor in human femoral head]. Nan Fang Yi Ke Da Xue Xue Bao 2008, 28, 441–443. [Google Scholar]
- Djomkam, A.L.Z.; Olwal, C.O.; Sala, T.B.; Paemka, L. Commentary: SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Front. Oncol. 2020, 10, 1448. [Google Scholar] [CrossRef]
- Simmons, G.; Gosalia, D.N.; Rennekamp, A.J.; Reeves, J.D.; Diamond, S.L.; Bates, P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA 2005, 102, 11876–11881. [Google Scholar] [CrossRef] [PubMed]
- Simmons, G.; Reeves, J.D.; Rennekamp, A.J.; Amberg, S.M.; Piefer, A.J.; Bates, P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. USA 2004, 101, 4240–4245. [Google Scholar] [CrossRef] [PubMed]
- Teeling, E.C.; Vernes, S.C.; Davalos, L.M.; Ray, D.A.; Gilbert, M.T.P.; Myers, E.; Bat, K.C. Bat Biology, Genomes, and the Bat1K Project: To Generate Chromosome-Level Genomes for All Living Bat Species. Annu. Rev. Anim. Biosci. 2018, 6, 23–46. [Google Scholar] [CrossRef] [PubMed]
- Seim, I.; Fang, X.; Xiong, Z.; Lobanov, A.V.; Huang, Z.; Ma, S.; Feng, Y.; Turanov, A.A.; Zhu, Y.; Lenz, T.L.; et al. Genome analysis reveals insights into physiology and longevity of the Brandt’s bat Myotis brandtii. Nat. Commun. 2013, 4, 2212. [Google Scholar] [CrossRef]
- Thomas, S.P.; Suthers, R.A. The physiology and energetics of bat flight. J. Exp. Biol. 1972, 57, 317–335. [Google Scholar]
- Wilkinson, G.S.; South, J.M. Life history, ecology and longevity in bats. Aging Cell 2002, 1, 124–131. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, Q.; Wang, H.; Yao, X. Severe zoonotic viruses carried by different species of bats and their regional distribution. Clin. Microbiol. Infect. 2024, 30, 206–210. [Google Scholar] [CrossRef]
- Forni, D.; Cagliani, R.; Clerici, M.; Sironi, M. Molecular Evolution of Human Coronavirus Genomes. Trends Microbiol. 2017, 25, 35–48. [Google Scholar] [CrossRef]
- Hall, J.S.; Hofmeister, E.; Ip, H.S.; Nashold, S.W.; Leon, A.E.; Malave, C.M.; Falendysz, E.A.; Rocke, T.E.; Carossino, M.; Balasuriya, U.; et al. Experimental infection of Mexican free-tailed bats (Tadarida brasiliensis) with SARS-CoV-2. mSphere 2022, 8, e0026322. [Google Scholar] [CrossRef]
- Hu, B.; Zeng, L.P.; Yang, X.L.; Ge, X.Y.; Zhang, W.; Li, B.; Xie, J.Z.; Shen, X.R.; Zhang, Y.Z.; Wang, N.; et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017, 13, e1006698. [Google Scholar] [CrossRef]
- Li, W.; Shi, Z.; Yu, M.; Ren, W.; Smith, C.; Epstein, J.H.; Wang, H.; Crameri, G.; Hu, Z.; Zhang, H.; et al. Bats are natural reservoirs of SARS-like coronaviruses. Science 2005, 310, 676–679. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.; Lau, S.K.; Lam, C.S.; Lau, C.C.; Tsang, A.K.; Lau, J.H.; Bai, R.; Teng, J.L.; Tsang, C.C.; Wang, M.; et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012, 86, 3995–4008. [Google Scholar] [CrossRef] [PubMed]
- Kaza, B.; Aguilar, H.C. Pathogenicity and virulence of henipaviruses. Virulence 2023, 14, 2273684. [Google Scholar] [CrossRef]
- Kuang, G.; Xu, Z.; Wang, J.; Gao, Z.; Yang, W.; Wu, W.; Liang, G.; Shi, M.; Feng, Y. Nelson Bay Reovirus Isolated from Bats and Blood-Sucking Arthropods Collected in Yunnan Province, China. Microbiol. Spectr. 2023, 11, e0512222. [Google Scholar] [CrossRef]
- Towner, J.S.; Nyakarahuka, L.; Atimnedi, P. Bat-Borne Pathogens and Public Health in Rural African Artisanal Gold Mines. AMA J. Ethics 2024, 26, E109–E115. [Google Scholar] [CrossRef]
- Leendertz, S.A.J.; Gogarten, J.F.; Düx, A.; Calvignac-Spencer, S.; Leendertz, F.H. Assessing the Evidence Supporting Fruit Bats as the Primary Reservoirs for Ebola Viruses. EcoHealth 2016, 13, 18–25. [Google Scholar] [CrossRef]
- Chen, L.; Liu, B.; Yang, J.; Jin, Q. DBatVir: The database of bat-associated viruses. Database 2014, 2014, bau021. [Google Scholar] [CrossRef]
- Ruiz-Aravena, M.; McKee, C.; Gamble, A.; Lunn, T.; Morris, A.; Snedden, C.E.; Yinda, C.K.; Port, J.R.; Buchholz, D.W.; Yeo, Y.Y.; et al. Ecology, evolution and spillover of coronaviruses from bats. Nat. Rev. Microbiol. 2022, 20, 299–314. [Google Scholar] [CrossRef]
- Sanchez-Gomez, W.S.; Selem-Salas, C.I.; Cordova-Aldana, D.I.; Erales-Villamil, J.A. Common vampire bat (Desmodus rotundus) abundance and frequency of attacks to cattle in landscapes of Yucatan, Mexico. Trop. Anim. Health Prod. 2022, 54, 130. [Google Scholar] [CrossRef]
- Abundes-Gallegos, J.; Salas-Rojas, M.; Galvez-Romero, G.; Perea-Martínez, L.; Obregón-Morales, C.Y.; Morales-Malacara, J.B.; Chomel, B.B.; Stuckey, M.J.; Moreno-Sandoval, H.; García-Baltazar, A.; et al. Detection of Dengue Virus in Bat Flies (Diptera: Streblidae) of Common Vampire Bats, Desmodus rotundus, in Progreso, Hidalgo, Mexico. Vector Borne Zoonotic Dis. 2018, 18, 70–73. [Google Scholar] [CrossRef]
- Volokhov, D.V.; Becker, D.J.; Bergner, L.M.; Camus, M.S.; Orton, R.J.; Chizhikov, V.E.; Altizer, S.M.; Streicker, D.G. Novel hemotropic mycoplasmas are widespread and genetically diverse in vampire bats. Epidemiol. Infect. 2017, 145, 3154–3167. [Google Scholar] [CrossRef] [PubMed]
- Becker, D.J.; Bergner, L.M.; Bentz, A.B.; Orton, R.J.; Altizer, S.; Streicker, D.G. Genetic diversity, infection prevalence, and possible transmission routes of Bartonella spp. in vampire bats. PLoS Negl. Trop. Dis. 2018, 12, e0006786. [Google Scholar] [CrossRef] [PubMed]
- Arellano-Sota, C. Vampire bat-transmitted rabies in cattle. Rev. Infect. Dis. 1988, 10 (Suppl. 4), S707–S709. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Cordero, V.; Botello, F.; Magaña-Cota, G.; Iglesias, J. Vampire bats, Desmodus rotundus, feeding on white-tailed deer, Odocoileus virginianus. Mammalia 2011, 75, 91–92. [Google Scholar] [CrossRef]
- Kotwa, J.D.; Lobb, B.; Massé, A.; Gagnier, M.; Aftanas, P.; Banerjee, A.; Banete, A.; Blais-Savoie, J.; Bowman, J.; Buchanan, T.; et al. Genomic and transcriptomic characterization of delta SARS-CoV-2 infection in free-ranging white-tailed deer (Odocoileus virginianus). iScience 2023, 26, 108319. [Google Scholar] [CrossRef]
- Kuchipudi, S.V.; Surendran-Nair, M.; Ruden, R.M.; Yon, M.; Nissly, R.H.; Vandegrift, K.J.; Nelli, R.K.; Li, L.; Jayarao, B.M.; Maranas, C.D.; et al. Multiple spillovers from humans and onward transmission of SARS-CoV-2 in white-tailed deer. Proc. Natl. Acad. Sci. USA 2022, 119, e2121644119. [Google Scholar] [CrossRef]
- Martins, M.; Boggiatto, P.M.; Buckley, A.; Cassmann, E.D.; Falkenberg, S.; Caserta, L.C.; Fernandes, M.H.V.; Kanipe, C.; Lager, K.; Palmer, M.V.; et al. From Deer-to-Deer: SARS-CoV-2 is efficiently transmitted and presents broad tissue tropism and replication sites in white-tailed deer. PLoS Pathog. 2022, 18, e1010197. [Google Scholar] [CrossRef]
- McBride, D.S.; Garushyants, S.K.; Franks, J.; Magee, A.F.; Overend, S.H.; Huey, D.; Williams, A.M.; Faith, S.A.; Kandeil, A.; Trifkovic, S.; et al. Accelerated evolution of SARS-CoV-2 in free-ranging white-tailed deer. Nat. Commun. 2023, 14, 5105. [Google Scholar] [CrossRef]
- Willgert, K.; Didelot, X.; Surendran-Nair, M.; Kuchipudi, S.V.; Ruden, R.M.; Yon, M.; Nissly, R.H.; Vandegrift, K.J.; Nelli, R.K.; Li, L.; et al. Transmission history of SARS-CoV-2 in humans and white-tailed deer. Sci. Rep. 2022, 12, 12094. [Google Scholar] [CrossRef]
- Guito, J.C.; Prescott, J.B.; Arnold, C.E.; Amman, B.R.; Schuh, A.J.; Spengler, J.R.; Sealy, T.K.; Harmon, J.R.; Coleman-McCray, J.D.; Kulcsar, K.A.; et al. Asymptomatic Infection of Marburg Virus Reservoir Bats Is Explained by a Strategy of Immunoprotective Disease Tolerance. Curr. Biol. 2021, 31, 257–270.e5. [Google Scholar] [CrossRef]
- Lee, D.N.; Papeş, M.; Van den Bussche, R.A. Present and potential future distribution of common vampire bats in the Americas and the associated risk to cattle. PLoS ONE 2012, 7, e42466. [Google Scholar] [CrossRef]
- Hall, E.R. The Mammals of Northe America; John Wiley and Sons: Hoboken, NJ, USA, 1981. [Google Scholar]
- Leslie, M.J.; Messenger, S.; Rohde, R.E.; Smith, J.; Cheshier, R.; Hanlon, C.; Rupprecht, C.E. Bat-associated rabies virus in Skunks. Emerg. Infect. Dis. 2006, 12, 1274–1277. [Google Scholar] [CrossRef] [PubMed]
- Lack, J.B.; Wilkinson, J.E. Range-wide population genetic structure of the pallid bat (Antrozous pallidus)—Incongruent results from nuclear and mitochondrial DNA. Acta Chiropterologica 2010, 12, 401–413. [Google Scholar]
- Griffin, B.D.; Chan, M.; Tailor, N.; Mendoza, E.J.; Leung, A.; Warner, B.M.; Duggan, A.T.; Moffat, E.; He, S.; Garnett, L.; et al. SARS-CoV-2 infection and transmission in the North American deer mouse. Nat. Commun. 2021, 12, 3612. [Google Scholar] [CrossRef]
- Kapczynski, D.R.; Sweeney, R.; Spackman, E.; Pantin-Jackwood, M.; Suarez, D.L. Development of an in vitro model for animal species susceptibility to SARS-CoV-2 replication based on expression of ACE2 and TMPRSS2 in avian cells. Virology 2022, 569, 1–12. [Google Scholar] [CrossRef]
- Briggs, K.; Sweeney, R.; Blehert, D.S.; Spackman, E.; Suarez, D.L.; Kapczynski, D.R. SARS-CoV-2 utilization of ACE2 from different bat species allows for virus entry and replication in vitro. Virology 2023, 586, 122–129. [Google Scholar] [CrossRef]
- Gralinski, L.E.; Menachery, V.D. Return of the Coronavirus: 2019-nCoV. Viruses 2020, 12, 135. [Google Scholar] [CrossRef]
- Boshart, M.; Weber, F.; Jahn, G.; Dorsch-Hasler, K.; Fleckenstein, B.; Schaffner, W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell 1985, 41, 521–530. [Google Scholar] [CrossRef]
- Qin, J.Y.; Zhang, L.; Clift, K.L.; Hulur, I.; Xiang, A.P.; Ren, B.Z.; Lahn, B.T. Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS ONE 2010, 5, e10611. [Google Scholar] [CrossRef]
- Chazotte, B. Labeling nuclear DNA using DAPI. Cold Spring Harb. Protoc. 2011, 2011, pdb prot5556. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Higgins, D.G.; Thompson, J.D.; Gibson, T.J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996, 266, 383–402. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Sharma, A.K.; Aguilar, K.; Boghani, F.; Sarcan, S.; George, M.; Ramesh, J.; Van Der Eerden, J.; Panda, C.S.; Lopez, A.; et al. Simple virus-free mouse models of COVID-19 pathologies and oral therapeutic intervention. iScience 2024, 27, 109191. [Google Scholar] [CrossRef] [PubMed]
- Bi, Z.; Hong, W.; Yang, J.; Lu, S.; Peng, X. Animal models for SARS-CoV-2 infection and pathology. MedComm 2021, 2, 548–568. [Google Scholar] [CrossRef]
- Cleary, S.J.; Pitchford, S.C.; Amison, R.T.; Carrington, R.; Robaina Cabrera, C.L.; Magnen, M.; Looney, M.R.; Gray, E.; Page, C.P. Animal models of mechanisms of SARS-CoV-2 infection and COVID-19 pathology. Br. J. Pharmacol. 2020, 177, 4851–4865. [Google Scholar] [CrossRef]
- Dillard, J.A.; Martinez, S.A.; Dearing, J.J.; Montgomery, S.A.; Baxter, V.K. Animal Models for the Study of SARS-CoV-2-Induced Respiratory Disease and Pathology. Comp. Med. 2023, 73, 72–90. [Google Scholar] [CrossRef]
- Gabrielson, K.; Myers, S.; Yi, J.; Gabrielson, E.; Jimenez, I.A. Comparison of Cardiovascular Pathology In Animal Models of SARS-CoV-2 Infection: Recommendations Regarding Standardization of Research Methods. Comp. Med. 2023, 73, 58–71. [Google Scholar] [CrossRef]
- Martins, M.; Nooruzzaman, M.; Cunningham, J.L.; Ye, C.; Caserta, L.C.; Jackson, N.; Martinez-Sobrido, L.; Fang, Y.; Diel, D.G. The SARS-CoV-2 Spike is a virulence determinant and plays a major role on the attenuated phenotype of Omicron virus in a feline model of infection. J. Virol. 2024, 98, e0190223. [Google Scholar] [CrossRef]
- Taha, T.Y.; Chen, I.P.; Hayashi, J.M.; Tabata, T.; Walcott, K.; Kimmerly, G.R.; Syed, A.M.; Ciling, A.; Suryawanshi, R.K.; Martin, H.S.; et al. Rapid assembly of SARS-CoV-2 genomes reveals attenuation of the Omicron BA.1 variant through NSP6. Nat. Commun. 2023, 14, 2308. [Google Scholar] [CrossRef]
- Hui, K.P.Y.; Ho, J.C.W.; Cheung, M.C.; Ng, K.C.; Ching, R.H.H.; Lai, K.L.; Kam, T.T.; Gu, H.; Sit, K.Y.; Hsin, M.K.Y.; et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature 2022, 603, 715–720. [Google Scholar] [CrossRef] [PubMed]
- Meng, B.; Abdullahi, A.; Ferreira, I.; Goonawardane, N.; Saito, A.; Kimura, I.; Yamasoba, D.; Gerber, P.P.; Fatihi, S.; Rathore, S.; et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 2022, 603, 706–714. [Google Scholar] [CrossRef] [PubMed]
- Begum, M.M.; Ichihara, K.; Takahashi, T.; Nasser, H.; Jonathan, M.; Tokunaga, K.; Yoshida, I.; Nagashima, M.; Sadamasu, K.; Yoshimura, K.; et al. Virological characteristics correlating with SARS-CoV-2 spike protein fusogenicity. Front. Virol. 2024, 4, 1353661. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakre, A.; Sweeney, R.; Espinoza, E.; Suarez, D.L.; Kapczynski, D.R. The ACE2 Receptor from Common Vampire Bat (Desmodus rotundus) and Pallid Bat (Antrozous pallidus) Support Attachment and Limited Infection of SARS-CoV-2 Viruses in Cell Culture. Viruses 2025, 17, 507. https://doi.org/10.3390/v17040507
Bakre A, Sweeney R, Espinoza E, Suarez DL, Kapczynski DR. The ACE2 Receptor from Common Vampire Bat (Desmodus rotundus) and Pallid Bat (Antrozous pallidus) Support Attachment and Limited Infection of SARS-CoV-2 Viruses in Cell Culture. Viruses. 2025; 17(4):507. https://doi.org/10.3390/v17040507
Chicago/Turabian StyleBakre, Abhijeet, Ryan Sweeney, Edna Espinoza, David L. Suarez, and Darrell R. Kapczynski. 2025. "The ACE2 Receptor from Common Vampire Bat (Desmodus rotundus) and Pallid Bat (Antrozous pallidus) Support Attachment and Limited Infection of SARS-CoV-2 Viruses in Cell Culture" Viruses 17, no. 4: 507. https://doi.org/10.3390/v17040507
APA StyleBakre, A., Sweeney, R., Espinoza, E., Suarez, D. L., & Kapczynski, D. R. (2025). The ACE2 Receptor from Common Vampire Bat (Desmodus rotundus) and Pallid Bat (Antrozous pallidus) Support Attachment and Limited Infection of SARS-CoV-2 Viruses in Cell Culture. Viruses, 17(4), 507. https://doi.org/10.3390/v17040507






