2. Structural Identifiability of Phenomenological Models
Structural identifiability analysis is a theoretical framework used to determine whether the parameters of a model can be uniquely inferred from unlimited, error-free observations. This analysis is essential for ensuring the mathematical integrity of a model before applying it to real-world data, as it confirms that parameter estimates are both unique and reliable. Various techniques, such as differential algebra methods, the Taylor series expansion method, and input-output approaches, have been developed to evaluate structural identifiability [
10,
12].
In this study, we employ the differential algebra method, a powerful approach that systematically eliminates unobserved state variables and derives differential algebraic polynomials involving the observed variables and model parameters [
12,
22,
23]. To achieve this, we utilize the
StructuralIdentifiability.jl package in JULIA, which enables efficient derivation of these polynomials and facilitates rigorous identifiability analysis [
7].
We reformulate the model structure for models with non-integer power exponents by introducing additional state variables. This reformulation not only preserves the fundamental structure of the model but also extends the applicability of identifiability analysis to previously intractable cases. By applying this approach, we ensure that parameters and state variables remain identifiable across models (
3), (
6), (
9), (
11), (
13), (
15), broadening the utility of these models in practical epidemic forecasting.
Structural identifiability theory: To analyze the structural identifiability of models, we rewrite them in the following compact form:
where
denotes the vector of state variables,
represents the observations, and
p denotes the vector of parameters. A parameter
p in model (
1) is called structurally identifiable if its value can be uniquely determined from the observable
(assuming noise-free measurements). A similar property for states is typically referred to as observability, but in the context of the present paper, it will be convenient for us to use the term identifiability in both cases. In fact, we will define identifiability for an arbitrary function of parameters and states. We will give a simplified version of the definition to avoid unnecessary technicalities; a formalized version can be found in [
24] (Definition 2.5).
Definition 1. A function in the states and parameters of model (1) is said to be structurally globally identifiable if, for generic and any , the following implication holds: If all the parameters are identifiable, then we will say that the model is identifiable. Definition 2. A function in the states and parameters of model (1) is said to be structurally locally identifiable if, for generic , there exists a neighborhood such that for any , the following implication holds: While the definitions above reflect the intuitive notion of identifiability, when it comes to checking identifiability for specific models, they are not very convenient. One standard approach to assessing structural identifiability is via input-output equations (also referred to as the differential algebra approach). We will not describe input-output equations in full generality, only for the single-output model, since all the models considered in this paper belong to this class. For a single-output model, the input-output equation is the irreducible equation of minimal order satisfied by the output. We will normalize the equation by dividing by the leading coefficient (considered a polynomial in
y and its derivatives) to obtain a monic polynomial. Let
be the coefficients of this equation. Then, under a certain assumption on the Wronskian of the input-output equations (see, e.g., [
25] (Lemma 4.6)), the identifiable functions of the parameters are precisely the ones expressible in terms of
. In this study, we used the software
StructuralIdentifiability.jl version v1.10.2 to assess identifiability; the software automatically checks if the aforementioned condition on Wronskians is fulfilled.
Structural identifiability results for the generalized growth model
We derive input-output equations by eliminating unobserved state variables using the differential algebra method. This process generates algebraic relationships involving the observed variables and model parameters, enabling identifiability analysis. For example, the generalized growth model is defined by the following differential equation:
where
denotes incidences at time
t,
denotes the cumulative number of cases at time
t, and
denotes the growth rate of the infectious diseases such that
Since epidemic data are typically collected as incident case counts over discrete time intervals, we assume that the incidence,
, corresponds to the observed data. This assumption is made for methodological consistency and computational feasibility, as it simplifies the identifiability analysis by ensuring that our model structure directly aligns with real-world epidemic datasets. It also allows us to use a standardized observation function across different growth models, facilitating direct comparisons in both structural and practical identifiability assessments.
To facilitate this process, we introduce an additional state variable, which allows us to reformulate models with non-integer power exponents into a structure that can be analyzed using the differential algebra approach. For this purpose, we introduce an additional state variable
to eliminate the non-integer power exponent, resulting in the extended version of the model by letting
; then,
GGM becomes
In this study, we examine the scenario in which the observations are equal to the incidences; that is,
. The monic input-output equation obtained from the
StructuralIdentifiability.jl package in JULIA, with the observation
, is given by
In the context of structural identifiability, the input-output equation indicates that
can be identified with the observation
, so
is identifiable as well. Now, we can express
in terms of identifiable parameters and state variables. As a result,
becomes identifiable. This implies that the parameter
r is identifiable. Therefore, model (
2) is structurally identifiable with the observation
We state the following proposition:
Proposition 1. The generalized growth model GGM is structurally identifiable from the observation of incidences .
We would like to stress that the way the lifting to the extended model (
3) is performed is important to obtain correct identifiability results for the original model. Suppose that, instead of
, we would have introduced
. Then, we would obtain a different extended system:
In this model, neither
r nor
are identifiable because there exists an output-preserving transformation
for any nonzero number
. The discrepancy between the identifiability results for different extended models can be explained as follows. The trajectories of the original versions of model (
2) and of (
3) are parametrized by three numbers:
for the former, and
for the latter. On the other hand, the space of trajectories of (
4) is four-dimensional, parametrized by
. The images of the trajectories of (
2) among the trajectories of (
4) are constrained to a manifold
. Thus, this is the additional trajectories that (
4) possesses, which turn
r into nonidentifiable status.
Structural identifiability results for the generalized logistic growth model
The generalized logistic growth model is given by the following equation:
where
r is the generalized growth rate,
k is the final epidemic size,
denotes incidences at time
t,
denotes the cumulative number of cases at time
t, and the parameter
denotes the different growth scenarios; the constant incidents is
, sub-exponential growth is
, and exponential growth is
. To obtain the extended version of the model without a non-integer exponent, we substitute
, where
GLM becomes
Here, we consider the case where the observations
correspond to the incidences. The input-output equation obtained from JULIA is normalized, with the observation
given by
We will check that all the parameters can be expressed in terms of the coefficients of this equations, thus showing that they are identifiable. First,
can be expressed from
, so it is identifiable. Next,
k can be expressed from
and the coefficient
. This concludes that only parameters
k and
are identifiable.
By using the StructuralIdentifiability.jl package in JULIA, we obtained the state variable , and is identifiable (observable) from the given observation. Then, can be written as a combination of identifiable parameters and the state variables. Therefore, the parameter r is also identifiable. We assert the following proposition:
Proposition 2. The generalized logistic growth model (5) is structurally identifiable from the observation of incidences, . Structural identifiability results for the Richards model
The Richards model is given by the following equation:
We let
; the
Richards model becomes
Here, we consider the case where the observations,
, correspond to the incidences. The normalized input-output equation of the model (
9) is given by
The parameter
a can be expressed from the coefficient
. Next, parameter
r can be expressed from
a and coefficient
. Thus, both
a and
r are identifiable from the observation
.
Now, we check the identifiability of the state variables; both and are identifiable from the observation. Therefore, parameter a can be written as a combination of the identifiable parameters and state variables. Thus, a is identifiable. We state the following proposition:
Proposition 3. The Richards model (8) is structurally identifiable from the observation of incidences, . Structural identifiability results for the generalized Richards model
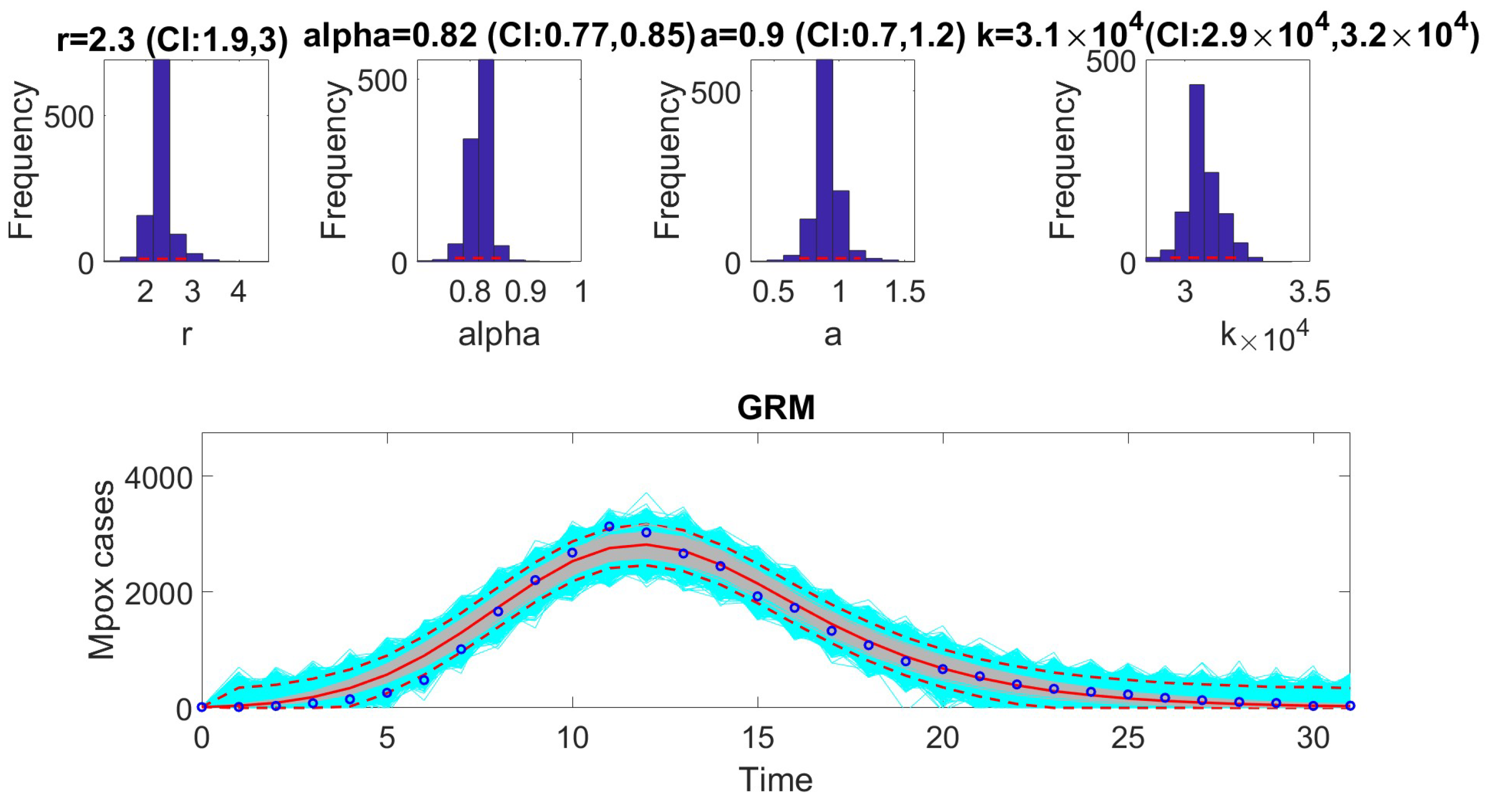
The model is represented by the following equation:
where
r is the generalized growth rate,
k is the final epidemic size,
denotes incidences at time
t,
denotes the cumulative number of cases at time
t, and the parameter
denotes the different growth scenarios; the constant incidents is
, sub-exponential growth is
, and exponential growth is
, and the exponent
a denotes the deviation from the symmetric s-shaped dynamics of the simple logistic curve.
To obtain the extended version without any non-integer power exponent, we let
, where
GRM becomes
By using StructuralIdentifiability.jl in JULIA, we determined that the parameters a and are locally identifiable, and the state variables and are also locally identifiable. Furthermore, the product and the summation are globally identifiable. Since a is positive, it follows that a is globally identifiable, ensuring that and are globally identifiable as well. To obtain the identifiability of parameters r and k, we express and in terms of identifiable states and parameters. Therefore, the parameters r and k are identifiable. We conclude the following proposition:
Proposition 4. The generalized Richards model (10) is structurally identifiable from the observation of incidences under the assumption of the positivity of a and k. Structural identifiability results for the Gompertz model
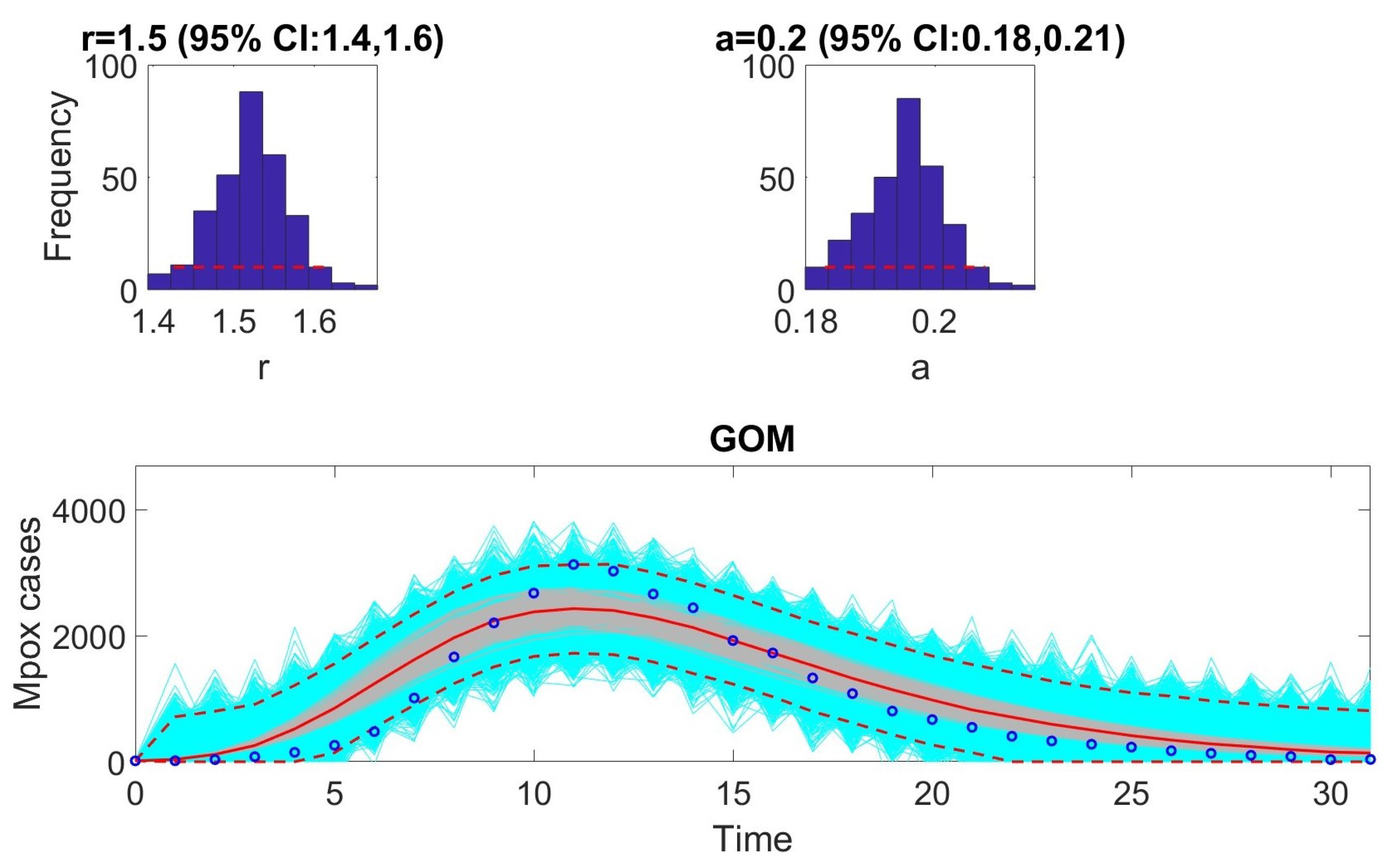
The Gompertz model is given by the following equation:
When letting
,
GOM becomes
Here, we consider the case where the observations, , correspond to the incidences,
The input-output equation of model (
13) is given by
Since
a appears among the coefficients, it is identifiable. The
StructuralIdentifiability package in JULIA shows that both
and
are identifiable from the observation
. Thus, the parameter
can be written as a product of identifiable parameters and state variables. Thus,
r is identifiable.
Proposition 5. The model (12) is structurally identifiable from the observation of incidences, . Structural identifiability results for the SEIR model with inhomogeneous mixing
The SEIR model with inhomogeneous mixing is given by the following system of equations:
Let
then
SEIR becomes
Here, we consider the case where the observations are
. By using
StructuralIdentifiability.jl in JULIA, we determined that the parameters
,
, and
are globally identifiable, whereas
N is not identifiable. However, since
N represents the total population size of a closed population, its value is known, and
. With
N being a known quantity, we treat it as an additional observation in the model. We then perform the identifiability analysis again and obtain the fact the state variable
is identifiable from observations
and
N. Thus, we can express
as a combination of identifiable parameters and state variables. Thus, model (
14) is structurally identifiable. The summary of the structural identifiability results for models (
3), (
6), (
9), (
11), (
13), and (
15), obtained from
StructuralIdentifiability.jl in JULIA, is given in
Table 1.
Cases with time-varying introduce additional degrees of freedom into the model, potentially leading to the unidentifiability of key parameters unless additional constraints or external data sources (e.g., demographic data) are available. Future work could explore methods such as including auxiliary equations for or assuming known functional forms to restore identifiability.
Another critical challenge in real-world applications is the uncertainty in initial conditions. Since initial values for , , , and are often unknown or estimated from limited data, they can significantly impact parameter identifiability and estimation accuracy. Given these considerations, we consider analyses with and without knowing the initial conditions of the systems.
Proposition 6. The model (14) is structurally identifiable from the observation of incidences, , with known initial conditions. 5. Discussion
This study evaluates the structural and practical identifiability of six commonly used phenomenological growth models in epidemiology. It demonstrates their robustness in parameter estimation and applicability across diverse datasets. To assess structural identifiability, we reformulated the models to address challenges posed by non-integer power exponents; we created an extended structure with fewer parameters and additional equations while preserving the original degrees of freedom. Our findings confirm that all reformulated models are structurally identifiable, even with unknown initial conditions. The original model formats were applied for data fitting and practical identifiability analyses, revealing that all models remained practically identifiable under varying noise levels, with performance differing across datasets.
The first step in validating these models involved assessing whether all unknown parameters were structurally identifiable, ensuring they could theoretically be uniquely determined from perfect, unlimited data. This step establishes a necessary foundation for reliable parameter estimation. We used the differential algebra method for this evaluation [
12,
22,
23]. Given the lack of dedicated software tools for analyzing systems of ordinary differential equations with non-integer exponents, we reformulated the models by introducing additional state variables. This reformulation enabled the use of the
StructuralIdentifiability.jl package in Julia [
7]. Our findings demonstrated that all parameters in the reformulated models were structurally identifiable, even with unknown initial conditions. By assessing the observability of state variables, we inferred the structural identifiability of the original models, bridging the gap between the reformulated and original structures [
8,
9].
Model validation was conducted using the
GrowthPredict MATLAB Toolbox, a specialized tool for fitting and forecasting time series trajectories based on phenomenological growth models. This toolbox facilitated comparisons across datasets and enabled the calculation of performance metrics such as AIC, MAE, MSE, and WIS. This toolbox was applied to three epidemiological datasets: weekly incidence data for monkeypox, COVID-19, and Ebola [
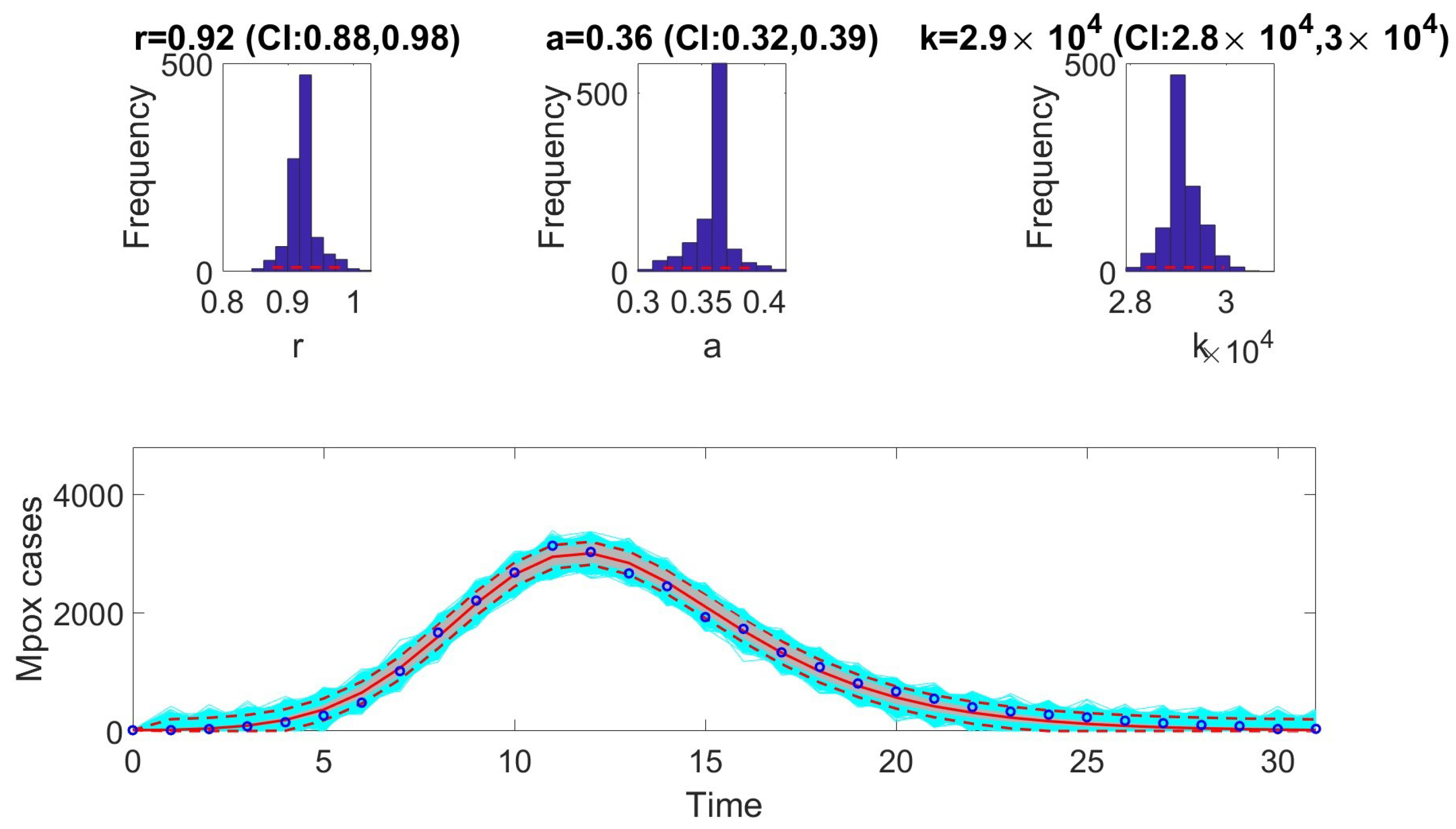
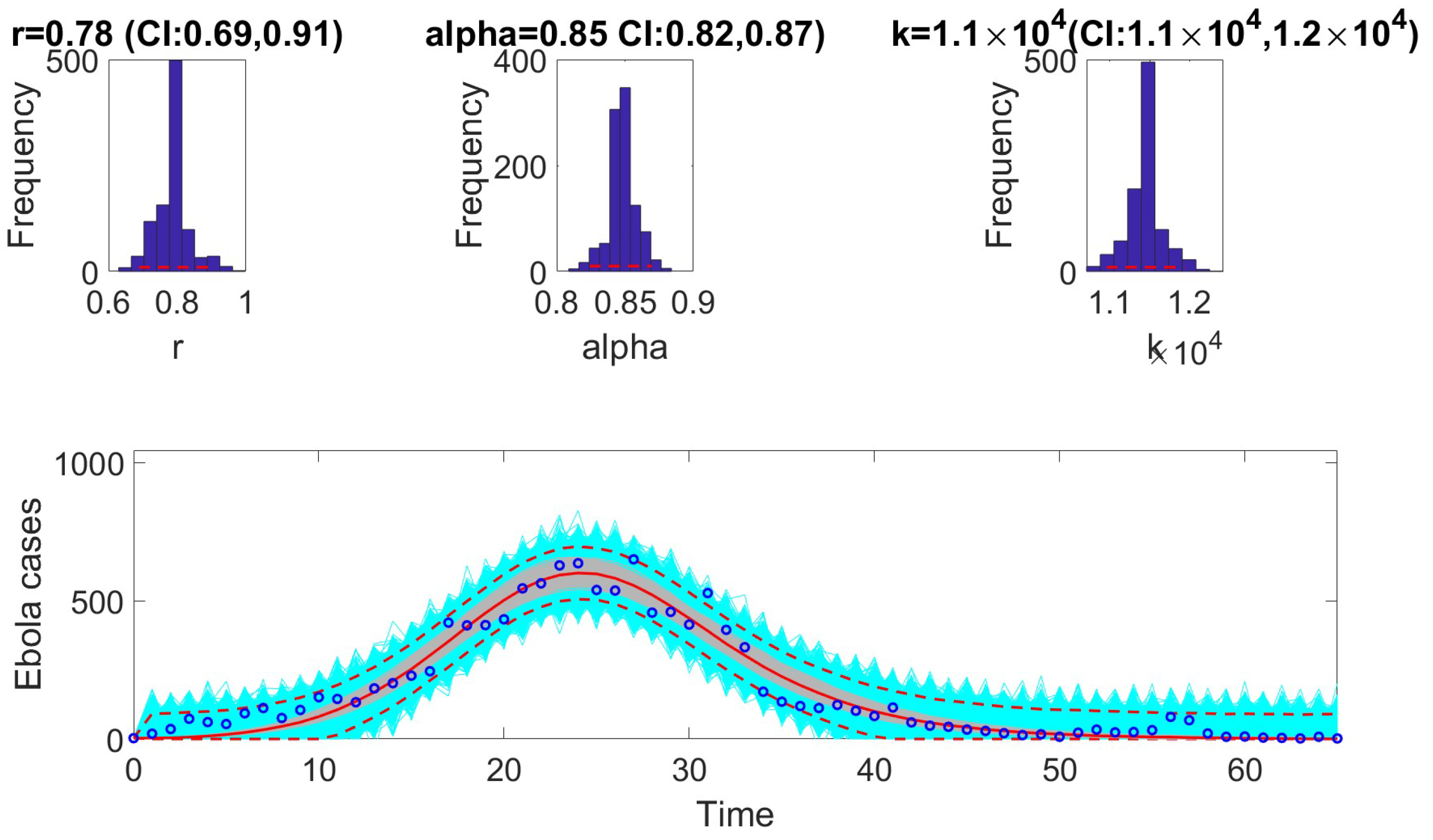
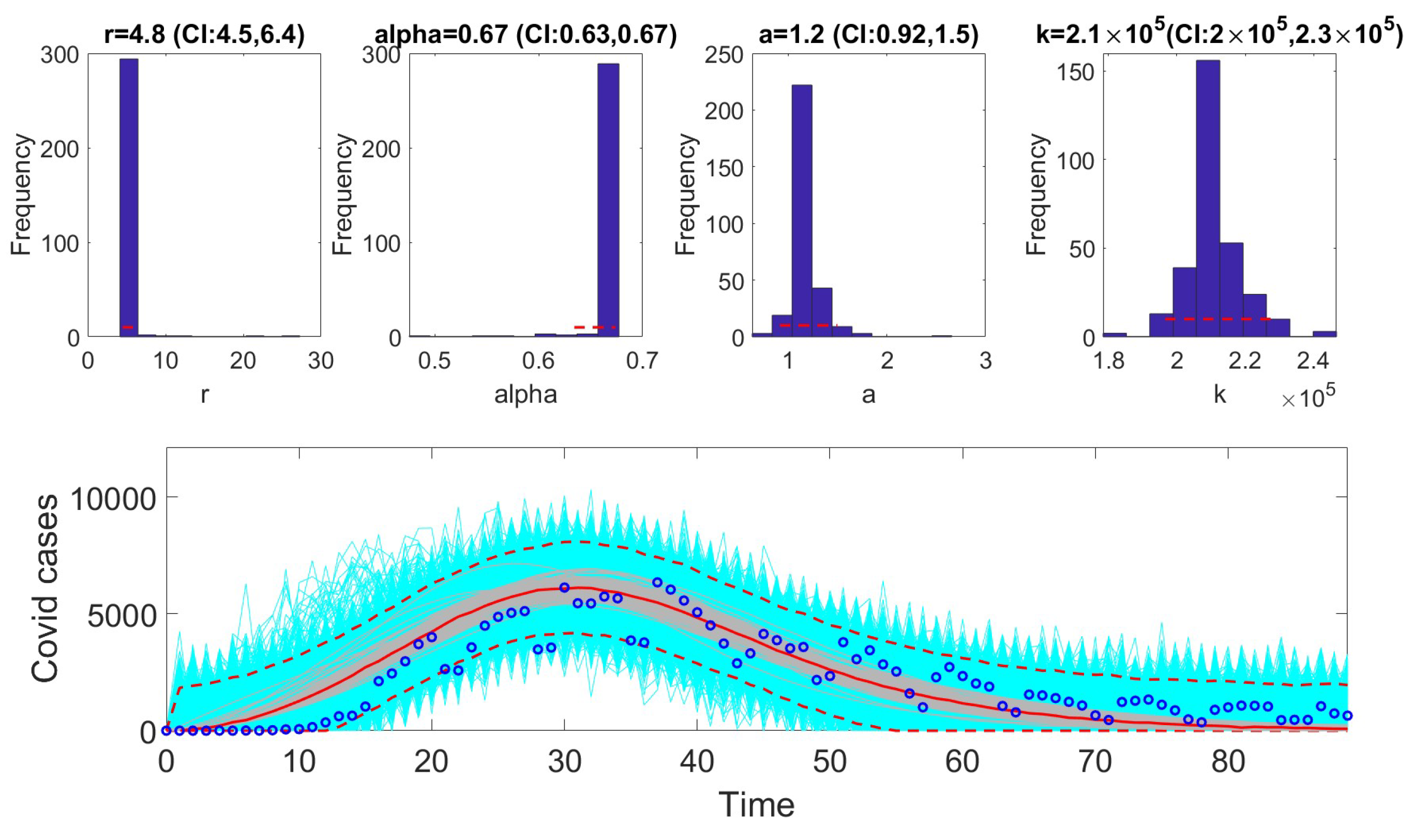
13]. To compare performance, metrics such as AIC, MAE, MSE, WIS, and coverage were calculated for each model. The selection of models based on AIC values revealed that certain models are better suited to specific epidemic contexts. For example, the Richards model provided the best fit for monkeypox data. In contrast, the GRM model excelled at fitting the COVID-19 data. The GLM model was most effective for the Ebola data.
In the next phase, we assessed practical identifiability by applying the models to datasets that produced the best fits, as indicated by the AIC values. We focused on the Richards, SEIR, and Gompertz models with the monkeypox dataset, the GLM with the Ebola dataset, and the GRM with the COVID-19 dataset. Practical identifiability was evaluated using Monte Carlo simulations, highlighting the robustness of parameter estimation under real-world conditions, where data are often noisy and limited. Other approaches, such as the Fisher Information Matrix (FIM), Profile Likelihood Method, and Bayesian methods, could complement this analysis [
1,
10,
12,
23]. Our results showed that all the models were practically identifiable for their respective datasets. However, parameter estimation accuracy varied across models and datasets, emphasizing the sensitivity of these models to data quality. These findings underscore the importance of including practical identifiability analysis in epidemiological studies to ensure robust model validation.
By addressing the challenge of non-integer power exponents and validating the models under practical conditions, this study broadens the applicability of phenomenological models. It strengthens confidence in their use for epidemic forecasting. However, several limitations must be acknowledged. First, while introducing additional state variables facilitates structural identifiability analysis, further research is needed to assess its impact on computational efficiency and model interpretability. Second, the dependency of parameter estimation accuracy on data quality remains a challenge, emphasizing the need for high-quality, well-calibrated datasets. Third, alternative approaches such as Bayesian methods and Fisher Information Matrix analysis should be explored to complement the Monte Carlo simulations performed here. Finally, future research should examine the role of time-varying parameters and unknown initial conditions in shaping identifiability outcomes.
Our analysis highlights the structural and practical identifiability of six phenomenological models across various epidemiological datasets. The results underscore the utility of these models in forecasting disease dynamics and their adaptability to diverse epidemic contexts. However, the accuracy of parameter estimates is highly dependent on data quality, emphasizing the critical need for careful data collection and the integration of practical identifiability evaluations in the model selection process.