The Clinical and Laboratory Landscape of COVID-19 During the Initial Period of the Pandemic and at the Beginning of the Omicron Era
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Study Participants and Samples
2.3. Molecular Genetic Analysis
2.4. Laboratory Data
2.5. Hemagglutination Inhibition Test (HI)
2.6. Statistical Analysis
3. Results
3.1. Molecular Genetic Analysis of the N Protein Gene of SARS-CoV-2 Antigenic Variants
3.2. The Main Data on the Observed Patient Cohorts
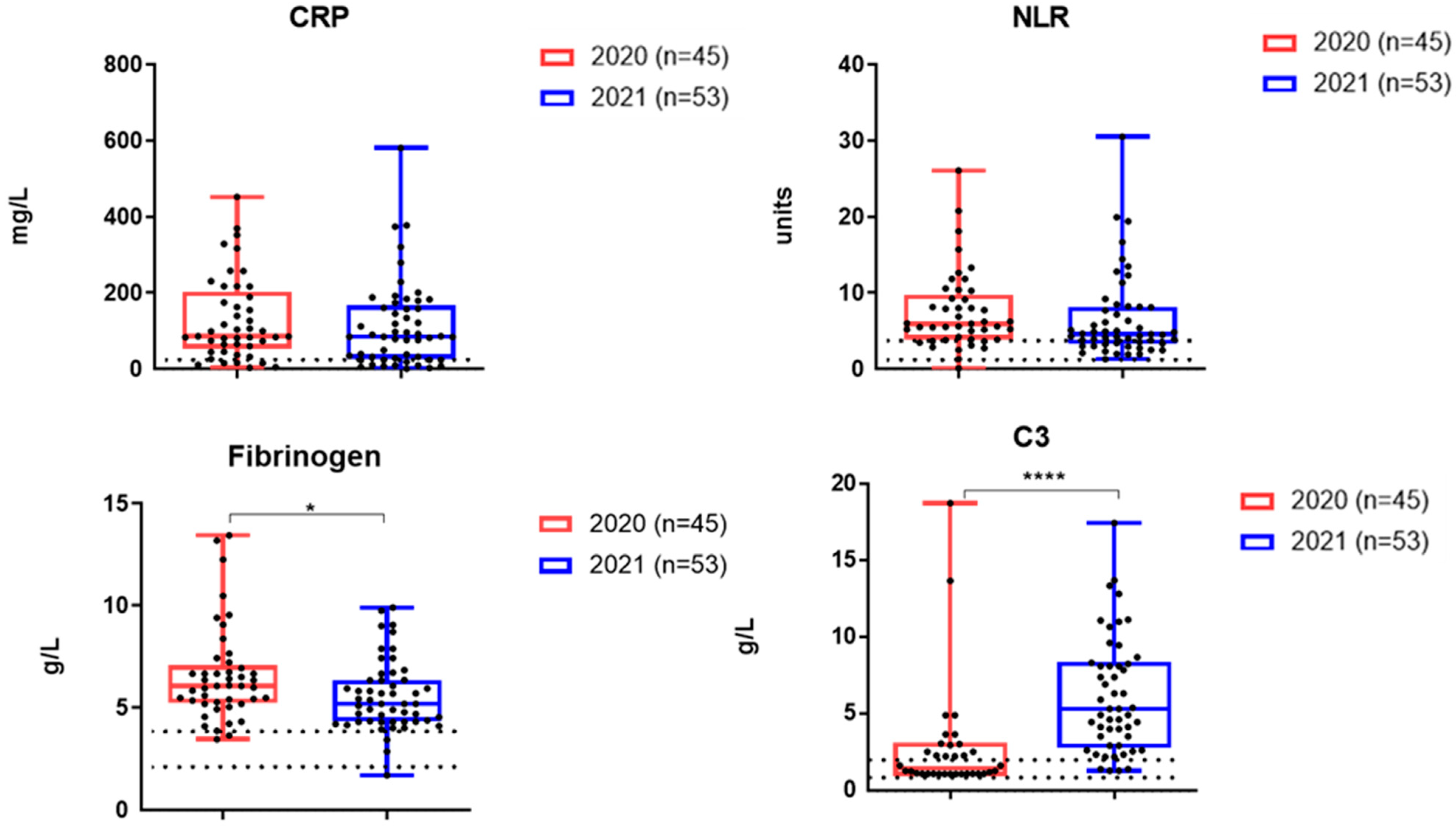
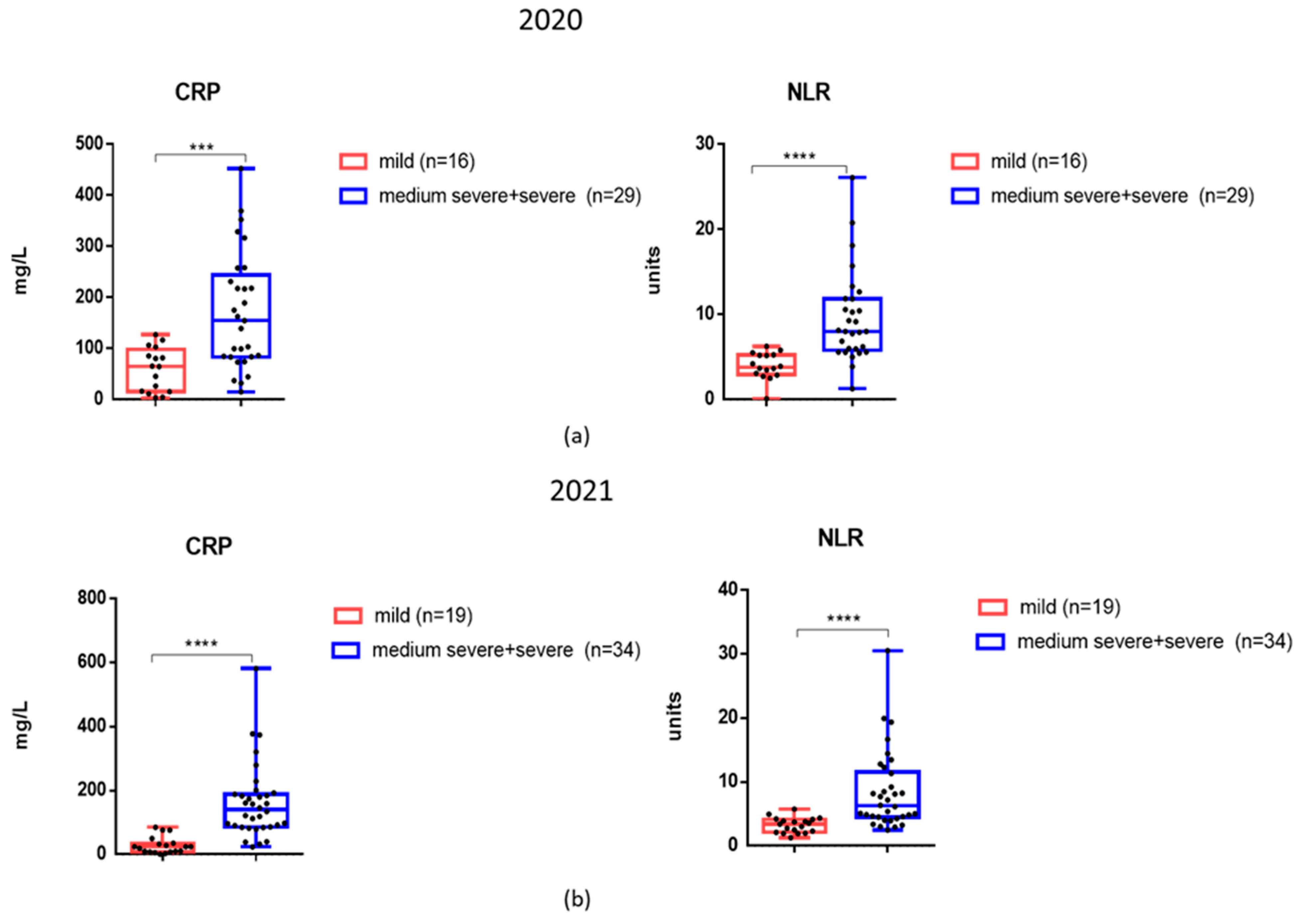
3.3. The Levels of Inflammatory Markers and Cytokines in the Serum Samples of Patients Examined
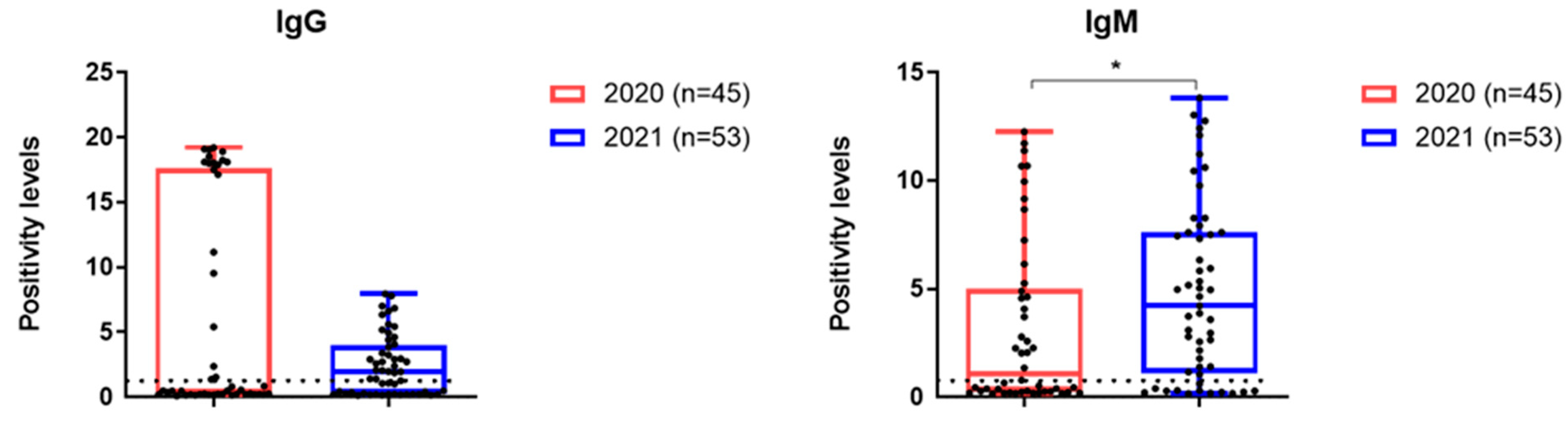
3.4. Antibodies to SARS-CoV-2 Analyzed Patient Cohorts
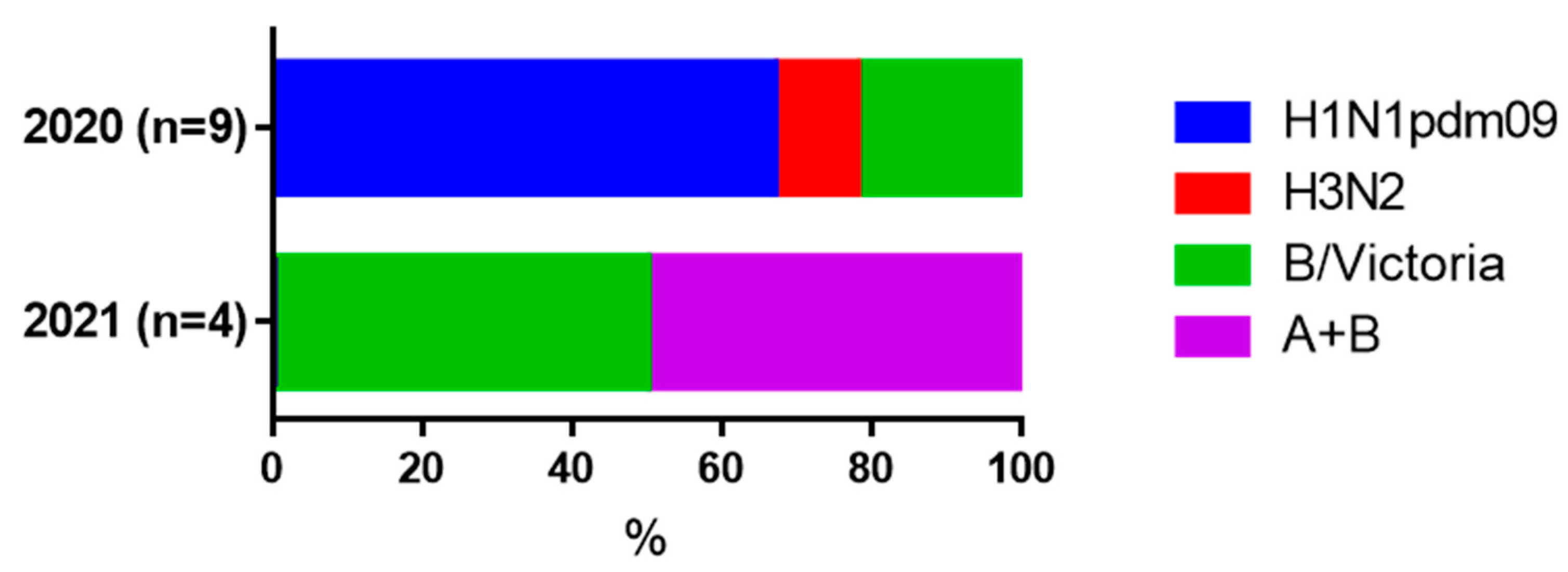
3.5. Increases in Serum HI Antibodies to Influenza Viruses in Paired Blood Sera
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| COVID-19 | coronavirus disease 19 |
| CRP | C-reactive protein |
| HI | hemagglutination inhibition |
| HRM | high-resolution melting |
| IFN-α | interferon 1 alpha |
| IL-6 | interleukin-6 |
| Me | medians |
| M-MulV RT | Moloney Murine Leukemia Virus Reverse Transcriptase |
| NLR | NLR neutrophil/lymphocyte ratio |
| Q1; Q3 | lower and upper quartiles |
| RBCs | red blood cells |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus 2 |
| TNF-α | tumor necrosis factor alpha |
References
- Kawamura, S.; Yamaguchi, F.; Kusakado, R.; Go, Y.; Nohmi, S.; Yoshizaki, C.; Suzuki, H. Changes in Clinical Features and Severity of COVID-19 with the Emergence of Omicron Variants: A Shift Towards a Common Disease. Infect. Drug Resist. 2024, 17, 5595–5603. [Google Scholar] [CrossRef] [PubMed]
- Akimkin, V. COVID-19 epidemic process and evolution of SARS-CoV-2 genetic variants in the Russian Federation. Microbiol. Res. 2024, 15, 213–224. [Google Scholar] [CrossRef]
- Meo, S.A.; Meo, A.S.; Al-Jassir, F.F.; Klonoff, D.C. Omicron SARS-CoV-2 new variant: Global prevalence and biological and clinical characteristics. Europ. Rev. Med. Pharmacol. Sci. 2021, 25, 8012–8018. [Google Scholar]
- Long, B.; Carius, B.M.; Chavez, S.; Liang, S.Y.; Brady, W.J.; Koyfman, A.; Gottlieb, M. Clinical update on covid-19 for the emergency clinician: Presentation and evaluation. Am. J. Emerg. Med. 2022, 54, 46–57. [Google Scholar] [CrossRef]
- Akimkin, V.G.; Popova, A.Y.; Khafizov, K.F.; Dubodelov, D.V.; Ugleva, S.V.; Semenenko, T.A.; Ploskireva, A.A.; Gorelov, A.V.; Pshenichnaya, N.Y.; Yezhlova, E.B.; et al. COVID-19: Evolution of the pandemic in Russia. Report II: Dynamics of the circulation of SARS-CoV-2 genetic variants. J. Microbiol. Epidemiol. Immunobiol. 2022, 99, 381–396. [Google Scholar] [CrossRef]
- Park, S.W.; Sun, K.; Abbott, S.; Sender, R.; Bar-On, Y.M.; Weitz, J.S.; Funk, S.; Grenfell, B.T.; Backer, J.A.; Wallinga, J.; et al. Inferring the differences in incubation-period and generation-interval distributions of the Delta and Omicron variants of SARS-CoV-2. Proc. Nat. Acad. Sci. USA 2023, 120, e2221887120. [Google Scholar] [CrossRef]
- Hui, K.P.Y.; Ho, J.C.W.; Cheung, M.C.; Ng, K.C.; Ching, R.H.H.; Lai, K.L.; Kam, T.T.; Gu, H.; Sit, K.-Y.; Hsin, M.K.Y.; et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature 2022, 603, 715–720. [Google Scholar] [CrossRef]
- Ren, S.Y.; Wang, W.B.; Gao, R.D.; Zhou, A.M. Omicron variant (B. 1.1. 529) of SARS-CoV-2: Mutation, infectivity, transmission, and vaccine resistance. World J. Clin. Cases 2022, 10, 1–11. [Google Scholar] [CrossRef]
- Xia, S.; Wang, L.; Zhu, Y.; Lu, L.; Jiang, S. Origin, virological features, immune evasion and intervention of SARS-CoV-2 Omicron sublineages. Signal Transduct. Target. Ther. 2022, 7, 241. [Google Scholar] [CrossRef]
- Liu, Y.; Huang, X.; Li, P.; Wang, D.; Yin, H.; Wang, N.; Sun, S. Clinical Profile and Outcome Analysis of Ear-Nose-Throat Symptoms in SARS-CoV-2 Omicron Subvariant Infections. Int. J. Public Health 2023, 68, 1606403. [Google Scholar] [CrossRef]
- Chemaitelly, H.; Ayoub, H.H.; Coyle, P.; Tang, P.; Hasan, M.R.; Yassine, H.M.; Abu-Raddad, L.J. Differential protection against SARS-CoV-2 reinfection pre-and post-Omicron. Nature 2025, 639, 1024–1031. [Google Scholar] [CrossRef]
- Ao, D.; Lan, T.; He, X.; Liu, J.; Chen, L.; Baptista-Hon, D.T.; Wei, X. SARS-CoV-2 Omicron variant: Immune escape and vaccine development. MedComm. 2022, 3, e126. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Bhattacharya, M.; Nag, S.; Dhama, K.; Chakraborty, C. A detailed overview of SARS-CoV-2 omicron: Its sub-variants, mutations and pathophysiology, clinical characteristics, immunological landscape, immune escape, and therapies. Viruses 2023, 15, 167. [Google Scholar] [CrossRef] [PubMed]
- Hussein, A.; Majeed, H.A.; Majeed, N.R.; Ali, N.H.; Saeed, D.A.; Ali, N.A. An Overview of Signs and Symptoms to Determine Coronavirus and Omicron Patients in Primary Care and Hospitals. Cihan Univ. Erbil Sci. J. 2024, 8, 8–17. [Google Scholar] [CrossRef]
- Hansen, P.R. Relative contagiousness of emerging virus variants: An analysis of the Alpha, Delta, and Omicron SARS-CoV-2 variants. Economet. J. 2022, 25, 739–761. [Google Scholar] [CrossRef]
- Malik, Y.A. COVID-19 variants: Impact on transmissibility and virulence. Malays. J. Path. 2022, 44, 387–396. [Google Scholar]
- Zabidi, N.Z.; Liew, H.L.; Farouk, I.A.; Puniyamurti, A.; Yip, A.J.W.; Wijesinghe, V.N.; Lal, S.K. Evolution of SARS-CoV-2 variants: Implications on immune escape, vaccination, therapeutic and diagnostic strategies. Viruses 2023, 15, 944. [Google Scholar] [CrossRef]
- Fan, H.; Tian, M.; Liu, S.; Ye, C.; Li, Z.; Wu, K.; Zhu, C. Strategies Used by SARS-CoV-2 to Evade the Innate Immune System in an Evolutionary Perspective. Pathogens 2024, 13, 1117. [Google Scholar] [CrossRef]
- Afridonova, Z.E.; Toptygina, A.P.; Mikhaylov, I.S. Humoral and cellular immune response to SARS-CoV-2 S and N proteins. Biochemistry 2024, 89, 872–882. [Google Scholar] [CrossRef]
- El-Maradny, Y.A.; Badawy, M.A.; Mohamed, K.I.; Ragab, R.F.; Moharm, H.M.; Abdallah, N.A.; Redwan, E.M. Unraveling the role of the nucleocapsid protein in SARS-CoV-2 pathogenesis: From viral life cycle to vaccine development. Intern. J. Bio. Macromol. 2024, 279, 135201. [Google Scholar] [CrossRef]
- Rahman, M.S.; Islam, M.R.; Alam, A.R.U.; Islam, I.; Hoque, M.N.; Akter, S.; Hossain, M.A. Evolutionary dynamics of SARS-CoV-2 nucleocapsid protein and its consequences. J. Med. Virol. 2021, 93, 2177–2195. [Google Scholar] [CrossRef] [PubMed]
- Alosaimi, B.; Naeem, A.; Hamed, M.E.; Alkadi, H.S.; Alanazi, T.; Al Rehily, S.S.; Zafar, A. Influenza co-infection associated with severity and mortality in COVID-19 patients. Virol. J. 2021, 18, 127. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Cai, Y.; Huang, X.; Yu, X.; Zhao, L.; Wang, F.; Zhan, Q. Co-infection with SARS-CoV-2 and influenza A virus in patient with pneumonia, China. Emerg. Infect. Dis. 2020, 26, 1324. [Google Scholar] [CrossRef]
- Cuadrado-Payán, E.; Montagud-Marrahi, E.; Torres-Elorza, M.; Bodro, M.; Blasco, M.; Poch, E.; Piñeiro, G.J. SARS-CoV-2 and influenza virus co-infection. Lancet 2020, 395, e84. [Google Scholar] [CrossRef]
- Shvedova, T.N.; Kopteva, O.S.; Kudar, P.A.; Lerner, A.A.; Desheva, Y.A. The role of coinfection with influenza viruses in the pathogenesis of severe infection in patients with COVID-19. Med. Ac. J. 2021, 21, 159–164. [Google Scholar] [CrossRef]
- Dhanasekaran, V.; Sullivan, S.; Edwards, K.M.; Xie, R.; Khvorov, A.; Valkenburg, S.A.; Barr, I.G. Human seasonal influenza under COVID-19 and the potential consequences of influenza lineage elimination. Nat. Commun. 2022, 13, 1721. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Desheva, Y.A.; Smolonogina, T.A.; Landgraf, G.O.; Rudenko, L.G. Development of the A/H6N1 influenza vaccine candidate based on A/Leningrad/134/17/57 (H2N2) master donor virus and the genome composition analysis using high resolution melting (HRM). Microbiol. Ind. Res. J. 2016, 3, 61–67. [Google Scholar] [CrossRef]
- Katz, J.M.; Hancock, K.; Xu, X. Serologic assays for influenza surveillance, diagnosis and vaccine evaluation. Expert. Rev. Anti-Infect. Ther. 2011, 9, 669–683. [Google Scholar] [CrossRef]
- Sharma, R.; Dubey, S.K. Computational Management of Alignment of Multiple Protein Sequences Using ClustalW. In Proceedings of the First International Conference on Sustainable Technologies for Computational Intelligence: Proceedings of ICTSCI, Rajasthan, India, 29–30 March 2019; Springer Nature: Wien, Austria, 2019; Volume 2020, pp. 177–186. [Google Scholar]
- O’Toole, Á.; Pybus, O.G.; Abram, M.E.; Kelly, E.J.; Rambaut, A. Pango lineage designation and assignment using SARS-CoV-2 spike gene nucleotide sequences. BMC Genom. 2022, 23, 121. [Google Scholar] [CrossRef]
- Liang, F. Quantitative Mutation Analysis of Genes and Proteins of Major SARS-CoV-2 Variants of Concern and Interest. Viruses 2023, 15, 1193. [Google Scholar] [CrossRef] [PubMed]
- Sui, J.; Noubouossie, D.F.; Gandotra, S.; Cao, L. Elevated plasma fibrinogen is associated with excessive inflammation and disease severity in COVID-19 patients. Front. Cell. Infect. Microbiol. 2021, 11, 734005. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.T.; Ellsworth, C.R.; Qin, X. Emerging role of complement in COVID-19 and other respiratory virus diseases. Cell. Mol. Life Sci. 2024, 81, 94. [Google Scholar] [CrossRef] [PubMed]
- Mellors, J.; Dhaliwal, R.; Longet, S.; Tipton, T.; Barnes, E.; Dunachie, S. Complement-Mediated Enhancement of SARS-CoV-2 Antibody Neutralisation Potency in Vaccinated Individuals. Nat. Commun. 2025, 16, 1–5. [Google Scholar] [CrossRef]
- Ellsworth, C.R.; Chen, Z.; Xiao, M.T.; Qian, C.; Wang, C.; Khatun, M.S.; Qin, X. Enhanced complement activation and MAC formation accelerates severe COVID-19. Cell. Mol. Life Sci. 2024, 81, 405. [Google Scholar] [CrossRef]
- Detsika, M.G.; Palamaris, K.; Dimopoulou, I.; Kotanidou, A.; Orfanos, S.E. The complement cascade in lung injury and disease. Respir. Res. 2024, 25, 20. [Google Scholar] [CrossRef]
- Dumitrescu, G.; Antovic, J.; Soutari, N.; Gran, C.; Antovic, A.; Al-Abani, K.; Taxiarchis, A. The role of complement and extracellular vesicles in the development of pulmonary embolism in severe COVID-19 cases. PLoS ONE 2024, 19, e0309112. [Google Scholar] [CrossRef]
- Yatsyshina, S.B.; Artamonova, A.A.; Elkina, M.A.; Valdokhina, A.V.; Bulanenko, V.P.; Berseneva, A.A.; Akimkin, V.G. Genetic characteristics of influenza A and B viruses circulating in Russia in 2019–2023. J. Microbiol. Epidemiol. Immunobiol. 2024, 101, 719–734. [Google Scholar] [CrossRef]
- Pawlowski, C.; Silvert, E.; O’Horo, J.C.; Lenehan, P.J.; Challener, D.; Gnass, E.; Soundararajan, V. SARS-CoV-2 and influenza coinfection throughout the COVID-19 pandemic: An assessment of coinfection rates, cohort characteristics, and clinical outcomes. PNAS Nexus 2022, 1, pgac071. [Google Scholar] [CrossRef]
- Varshney, K.; Pillay, P.; Mustafa, A.D.; Shen, D.; Adalbert, J.R.; Mahmood, M.Q. A systematic review of the clinical characteristics of influenza-COVID-19 co-infection. Clin. Exp. Med. 2023, 23, 3265–3275. [Google Scholar] [CrossRef]
- Debisarun, P.A.; Gössling, K.L.; Bulut, O.; Kilic, G.; Zoodsma, M.; Liu, Z.; Netea, M.G. Induction of trained immunity by influenza vaccination-impact on COVID-19. PLoS Pathog. 2021, 17, e1009928. [Google Scholar] [CrossRef]
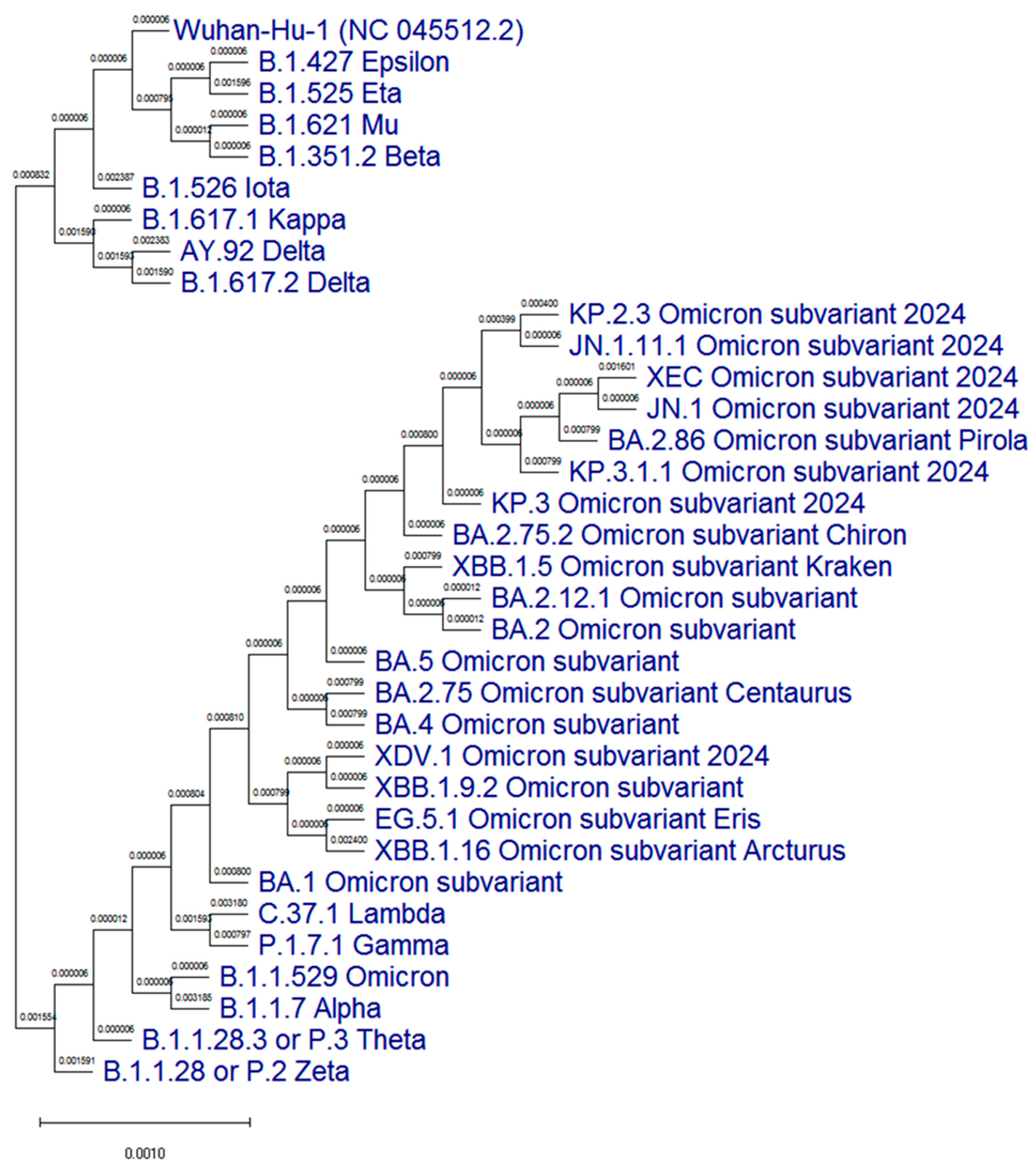
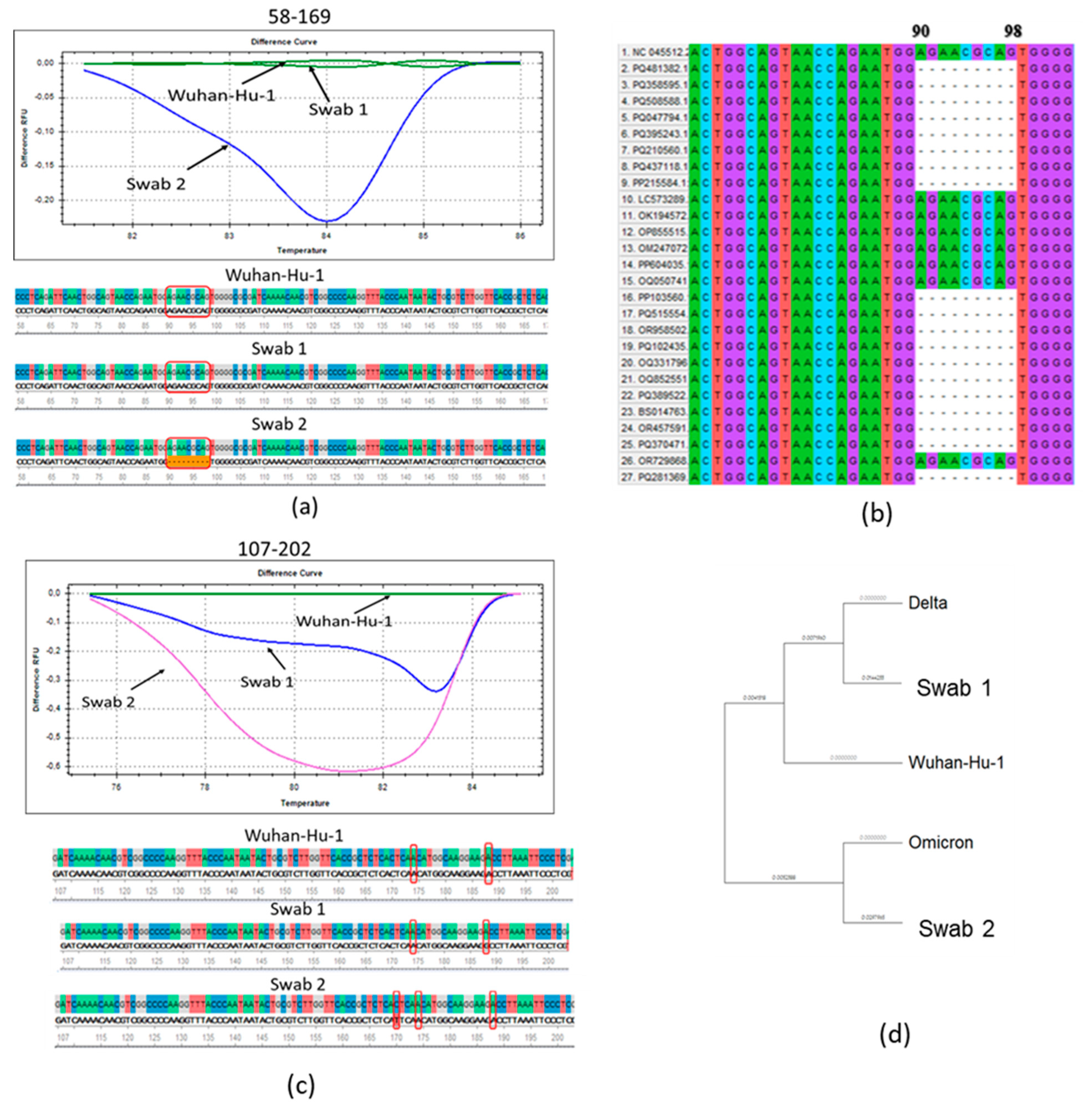

| Position in the Nucleotide Chain | Nucleotide Sequence (5′-3′) | Length of Fragment, bp | T Annealing, °C |
|---|---|---|---|
| F58 | CCCTCAGATTCAACTGGCAGT | 112 | |
| R169 | TGAAGAGCGGTGAACCAAGAC | 55 |
| Characteristic | 1st Cohort (2020), n = 45 | 2nd Cohort (2021), n = 53 | p= |
|---|---|---|---|
| Age, Me (Q1; Q3) | 62.00 (55.00; 70.00) | 67.00 (56.50; 76.00) | 0.15 |
| Males | 22 (48.9%) | 28 (52.8%) | 0.5 |
| Females | 23 (51.1%) | 25 (47.2%) | 0.5 |
| <65 years old | 28 (62.2%) | 23 (43.4%) | 0.43 |
| ≥65 years old | 17 (37.8%) | 30 (56.6%) | 0.43 |
| Mild COVID-19 | 16 (35.6%) | 19 (35.8%) | 0.57 |
| Medium severe + severe COVID-19 | 29 (64.4%) | 34 (64.2%) | 0.57 |
| Days from onset of illness, Me (Q1; Q3) | 7.00 (5.00; 9.00) | 4.00 (3.00; 5.00) | <0.0001 |
| Positive PCR test for SARS-CoV-2 on the day of hospitalization | 22 (48.9%) | 53 (100%) | <0.001 |
| Viremia (positive serum PCR-test) | 12 (26.7%) | 0 (0%) | <0.0001 |
| SARS-CoV-2 vaccination | 0 (0%) | 37 (69.8%) | <0.001 |
| Comorbidities: | |||
| Cardiovascular | 31 (68.8%) | 31 (58.5%) | 0.2 |
| Diabetes | 9 (20%) | 6 (11.3%) | 0.17 |
| Chronic pulmonary disorders | 5 (11.1%) | 3 (5.6%) | 0.27 |
| Bacterial coinfections | 18 (40.0%) | 22 (41.5%) | 0.52 |
| Lethal outcome | 13 (24.1%) | 0 (0%) | <0.001 |
| Positive for IgG | 15 (33.3%) | 31 (58.5) | 0.01 |
| Positive for IgM | 15 (33.3%) | 40 (45.5%) | <0.0001 |
| Influenza seroconversions among paired samples | 9 out of 28 (32.2%) | 5 out of 14 (35.7%) | 0.12 |
| Characteristic | 1st Cohort (2020) | 2nd Cohort (2021) | p= |
|---|---|---|---|
| Number of patients | 28 | 14 | |
| Mild | 9 (32.1%) | 5 (35.7%) | 0.54 |
| Medium severe + severe | 19 (67.9%) | 9 (64%) | 0.54 |
| Average period between the 1st and 2nd serum | 4 (3.0; 4.0) | 4 (3.0; 5.0) | 0.18 |
| Seroconversion to influenza in mild COVID-19 | 0 (0%) | 0 (0%) | n/a |
| Seroconversion to influenza in medium severe + severe COVID-19 | 9 (47.4%) 1 | 4 (44.4%) | 0.69 |
| Seroconversion to influenza viruses (HI) | |||
| A/H1N1pdm09 | 6 (21.4%) | 1 (7.1%) | 0.19 |
| A/H3N2 | 1 (3.6%) | 1 (7.1%) | 0.67 |
| B/Victoria | 2 (7.1%) | 4 (28.6%) | 0.08 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Desheva, Y.A.; Shvedova, T.N.; Kopteva, O.S.; Guzenkov, D.S.; Kudar, P.A.; Kotomina, T.S.; Petrachkova, D.S.; Grigorieva, E.P.; Lerner, A.A.; Ponkratov, S.V. The Clinical and Laboratory Landscape of COVID-19 During the Initial Period of the Pandemic and at the Beginning of the Omicron Era. Viruses 2025, 17, 481. https://doi.org/10.3390/v17040481
Desheva YA, Shvedova TN, Kopteva OS, Guzenkov DS, Kudar PA, Kotomina TS, Petrachkova DS, Grigorieva EP, Lerner AA, Ponkratov SV. The Clinical and Laboratory Landscape of COVID-19 During the Initial Period of the Pandemic and at the Beginning of the Omicron Era. Viruses. 2025; 17(4):481. https://doi.org/10.3390/v17040481
Chicago/Turabian StyleDesheva, Yulia A., Tamara N. Shvedova, Olga S. Kopteva, Danila S. Guzenkov, Polina A. Kudar, Tatiana S. Kotomina, Daria S. Petrachkova, Elena P. Grigorieva, Anna A. Lerner, and Stanislav V. Ponkratov. 2025. "The Clinical and Laboratory Landscape of COVID-19 During the Initial Period of the Pandemic and at the Beginning of the Omicron Era" Viruses 17, no. 4: 481. https://doi.org/10.3390/v17040481
APA StyleDesheva, Y. A., Shvedova, T. N., Kopteva, O. S., Guzenkov, D. S., Kudar, P. A., Kotomina, T. S., Petrachkova, D. S., Grigorieva, E. P., Lerner, A. A., & Ponkratov, S. V. (2025). The Clinical and Laboratory Landscape of COVID-19 During the Initial Period of the Pandemic and at the Beginning of the Omicron Era. Viruses, 17(4), 481. https://doi.org/10.3390/v17040481







