Abstract
Dichorhavirus is a recently accepted plant virus genus within the family Rhabdoviridae. Species assigned to the genus consist of bi-segmented, negative sense, single-stranded RNA viruses and are transmitted by Brevipalpus spp. Currently, there are five recognized species and two unclassified members in the genus Dichorhavirus. Four out of seven-orchid fleck virus (OFV), citrus leprosis virus N, citrus chlorotic spot virus, and citrus bright spot virus-can infect citrus and produce leprosis disease-like symptoms. The E-probe Diagnostic for Nucleic Acid Analysis (EDNA) was developed to reduce computational effort and then integrated within Microbe-Finder (MiFi®) online platform to design and evaluate e-probes in raw High Throughput Sequencing (HTS) data. During this study, Dichorhavirus genomes were downloaded from public databases and e-probes were designed using the MiProbe incorporated into the MiFi® platform. Three different sizes of e-probes, 40, 60, and 80 nucleotides, were developed and selected based on whole genome comparisons with near-neighbor genomes. For curation, each e-probe was searched in the NCBI nucleotide sequence database using BLASTn. All the e-probes that had hits with non-target species with ≥90% identities were removed. The sensitivity and specificity of Dichorhavirus genus, species, strain, and variant-specific e-probes were validated in vivo using HTS meta-transcriptomic libraries generated from Dichorhavirus-suspected citrus, orchid, and ornamentals. Through downstream analysis of HTS data, EDNA not only detected the known hosts of OFV but also discovered an unknown host leopard plant (Farfugium japonicum), and the possible existence of a new ornamental strain of OFV in nature.
1. Introduction
Rhabdoviridae is one of the families in the order Mononegavirales. Except for unassigned genera, all other taxonomically accepted rhabdoviruses are allocated into three distinct subfamilies: Alpharhabdovirinae, Betarhabdovirinae, and Gammarhabdovirinae. Out of three, the subfamily Betarhabdovirinae includes six virus genera infecting plant hosts and arthropod vectors. Genera assigned to the family Rhabdoviridae consist of mono- and bi-segmented, negative-sense, single-stranded RNA viruses [1,2]. Very recently, a data mining expedition discovered the first tri-segmented rhabdovirus genome from multiple hosts tentatively named “Trirhavirus” [3]. The first occurrence of Medicago trirhavirus 1 has been confirmed in commercial alfalfa fields in Washington State, USA [4]. There are two genera (Dichorhavirus and Varicosavirus) under the family Rhabdoviridae, having bi-segmented genomes and infecting plants only. Among these, only Dichorhavirus genus is transmitted by false spider mites, belonging to the genus Brevipalpus [2,5]. Since the discovery of orchid fleck virus (OFV), the type species of the genus Dichorhavirus, four more approved members, [coffee ring spot virus (CoRSV), citrus leprosis virus N (CiLV-N), citrus chlorotic spot virus (CiCSV), and clerodendrum chlorotic spot virus (ClCSV)], and two more unassigned species [citrus bright spot virus (CiBSV) and Dichorhavirus sp. ‘monocotyledonae’] were included (https://www.ncbi.nlm.nih.gov/taxonomy/?term=Dichorhavirus) (accessed on 29 January 2025). Genome segment 1 (RNA1; ~7.0 kb) of the dichorhaviruses consists of five open reading frames (ORFs) that encode the nucleocapsid protein (N), phosphoprotein (P), movement protein (MP), matrix protein (M), and glycoprotein (G). Segment 2 (RNA2; ~6.0 Kb) has only a single ORF that encodes for the large (L) protein, also known as RNA-dependent RNA polymerase (RdRp).
Dichorhaviruses have been detected in multiple crops such as citrus (Citrus spp.), coffee (Coffea sp.), and several ornamentals including orchids [1]. Dichorhavirus-like OFV replicates inside mites [6], but plant-infected tissues develop localized chlorotic and/or necrotic lesions as the viruses do not move systemically [7]. The relationship between Brevipalpus spp. and Dichorhavirus transmissions is often very complex, as multiple false spider species are involved. OFV is the only Brevipalpus-transmitted virus (BTV) with worldwide distribution, transmitted by B. californicus to orchids, citrus, and ornamentals [8,9]. However, outside the family Orchidaceae, OFV has been found naturally infecting Dieffenbachia sp. (Araceae), Swinglea glutinosa (Rutaceae) [9], lilyturf (Liriope spicata, Asparagaceae) [10], green ti plant (Cordyline terminalis, Asparagaceae) [1], common hollyhock (Alcea rosea, Malvaceace) (https://hdl.handle.net/2263/84240, accessed on 18 February 2025), and spike speedwell (Veronica spicata, Plantaginaceae) [11]. Recent findings on rough lemon and mandarin orange (Rutaceae) in Hawaii [12], greenbrier (Smilax auriculata, Smilacaceae) [13], cast-iron plant (Aspidistra elatior), lilyturf or monkey grass (Liriope muscari), aztec grass (Ophiopogon intermedius, Asparagaceae) [14], and pandan grass (Pandanus amaryllifolius, Pandanaceae) in Florida [15], and in orchids (Cymbidium sp., Dendrobium sp., and Dendrochilum magnum) in California pose a potential threat to billion-dollar US citrus industry [16]. Current literature describes that OFV has two orchids (OFV-Orc1 and OFV-Orc2) and two citrus strains (OFV-Cit1 and OFV-Cit2). At least one of these four OFV-variants has been implicated in causing CiL disease (CiLD) in Colombia, Mexico, Hawaii, and South Africa [12,17,18,19,20,21], but there has been no report of OFV-Cit infection in orchids or other host species.
Several diagnostic methods have been developed for detecting viruses, but a limited number of diagnostic assays are available for Dichorhavirus detection [22,23,24,25,26,27]. Plant viruses are routinely detected with serological and molecular techniques such as the enzyme-linked immunosorbent assay (ELISA) and polymerase chain reaction (PCR), respectively. At present, no Dichorhavirus antibody is commercially available for serological tests. Only developed conventional RT-PCR assays are utilized for the routine detection of dichorhaviruses [22,23,24,25,26,27].
The e-Probe Diagnostic Nucleic Acid Analysis (EDNA) is a computational pipeline that utilizes multiple short (40–120 nt) pathogen-specific curated sequences, known as e-probes, to detect and identify known pathogens (single or multiple) of interest from raw HTS datasets [28]. To overcome the HTS challenges, like time-consuming and laborious data analysis and variation in laboratory-specific cutoff values for diagnostic decisions, the Microbe-Finder (MiFi®) online platform was created. The easy-to-use MiFi® platform incorporates the EDNA pipeline to consolidate e-probe design and validation within a single interface [29]. There are two tools, MiProbe and MiDetect, incorporated in the MiFi® web application to identify curated e-probes and detect/identify target organism/s from HTS libraries, respectively. This technology has been utilized earlier for the detection and identification of plum pox virus [30], citrus tristeza virus [31], hop viruses, and viroids [32] and has proven its capability in the detection of oomycetes [33], fungi [34], bacteria Ralstonia solanacearum [35], and fastidious prokaryotes like ‘Candidatus Liberibacter asiaticus’ and Spiroplasma citri [31]. Very recently, the MiFi® metagenome analysis platform was used for the simultaneous detection of multiple pathogens associated with bovine respiratory disease (BRD) complex [36].
Here, we explored the EDNA technology integrated with the online MiFi® platform for the detection of dichorhaviruses infecting multiple plant species, with emphasis on strain differentiation of OFV. In this study, e-probes for the genus Dichorhavirus, and specific e-probes for its six species, OFV strains, and its variants were developed. The detection accuracy of the EDNA technology was validated by comparison of results with the existing gold standard molecular diagnostics (RT-qPCR) and by detecting Dichorhavirus towards strain level using RNASeq data from multiple OFV-infected plant species. In the validation study, the sensitivity and specificity of each set of curated e-probes of Dichorhavirus were evaluated using HTS meta-transcriptomic libraries infecting citrus, orchid, and multiple ornamental plant species. Newly developed Dichorhavirus e-probes successfully identified Farfugium japonicum (family Asteraceae) as a new host for OFV and exposed the possibility of a new strain of OFV in nature.
2. Materials and Methods
2.1. Phylogenetic Analysis of Dichorhaviruses Using Available Genome Sequences in GenBank
Three lineages of dichorhaviruses identified in the phylogenetic trees based on L-protein (RNA2) were called sub-groups 1, 2, and 3 [23]. The topology of the phylogenetic tree that likely reflects a Dichorhavirus and mite vector coevolutionary relationship is very important to visualize the subgroups of dichorhaviruses before designing the generic and specific e-probes for disease diagnostics. To verify the phylogenetic relationship among dichorhaviruses, 28 complete Dichorhavirus RNA1 and RNA2 genome sequences available in GenBank were downloaded and aligned using the in-built MUSCLE program in MEGA 11 [37]. For this study, phylogenetic trees for RNA1 and RNA2 were created utilizing the Neighbor-Joining method [38] supported by 1000 bootstrap replicates (next to the branches) [39] and the evolutionary distances were computed using the Maximum Composite Likelihood method [40] (Figure 1).

Figure 1.
Phylogenetic relationships of dichorhaviruses [orchid fleck virus (OFV), citrus leprosis virus N (CiLV-N), citrus chlorotic spot virus (CiCSV), citrus bright spot virus (CiBSV), clerodendrum chlorotic spot virus (ClCSV), and coffee ring spot virus (CoRSV)] using neighbor-joining methods based on (A) RNA1 and (B) RNA2 genome sequences. Phylogenies were supported by 1000 bootstrap replicates and bootstrap values greater than 50 are shown at the nodes. Genetic pieces of information associated with the NCBI accessions mentioned in the distinct subgroup clades were considered for e-probes design.
2.2. Buildup of the Dichorhavirus e-Probe Sequences Infecting Different Hosts
To generate e-probes for leprosis-related dichorhaviruses, genomic data was retrieved from three sources: (1) GenBank and published reports, (2) In-house collection of BTVs (kitaviruses- and Dichorhavirus-infected/suspected samples sequences from Florida, California, Hawaii, Colombia, Costa Rica, and Mexico, and (3) Sequence data shared by the researchers working on BTVs. Primarily, scientists from the United States Department of Agriculture (USDA)–Agricultural Research Service, USDA–Animal Plant Health Inspection Service; Plant Protection Quarantine, and Institute for Biosecurity and Microbial Forensic at Oklahoma State University worked together and initiated e-probes development for dichorhaviruses infecting citrus, coffee, Clerodendrum, and orchid [16].
2.3. Development, Curation, and in Silico Validation of Dichorhavirus e-Probes
To generate e-probes, two specific FASTA formatted files are needed as input into EDNA: (1) a target genomes file and (2) a near neighbors’ file. The target and near neighbor files were retrieved using https://www.ncbi.nlm.nih.gov/taxonomy (accessed on 15 November 2022) website by entering the search term ‘Dichorhavirus’. As an example, to retrieve a set of target genomes for Dichorhavirus leprosis (CiLV-N) associated with CiLD in Brazil, the Taxonomy ID: 2560386 was retrieved and then the NCBI database (https://www.ncbi.nlm.nih.gov, accessed on 15 November 2022) was searched for the term “txid2560386 [Organism:exp]”. A custom sequence length in the range of 5500 to 7000 bp was used to capture the full-length sequences of RNA1 and RNA2 of dichorhaviruses. Then, the target sequences were downloaded in FASTA format, and the raw e-probes were developed and curated for BLASTn analysis. This file is the target genome needed for input into EDNA. To create the near neighbors’ file for e-probe design, the taxonomy ID: 1913605 was retrieved for the Dichorhavirus genus. To retrieve the near neighbor genomes from NCBI in FASTA format, the search term “(txid1913605 [Organism:exp]) NOT txid2560386 [Organism:exp]” was entered into https://www.ncbi.nlm.nih.gov/ (accessed on 15 November 2022) where the NOT txid Boolean statement excludes the target. Raw e-probe sequences that did not match the target pathogen (e-value of ≤1 × 10−10 were removed from the final e-probe set to ensure diagnostic specificity [28,30,41]. The files were uploaded into MiFi®, and e-probes for 40, 60, and 80 nucleotides were created using the e-probe developer software (MiProbe v2) inside MiFi®. These probes were further manually curated using BLASTn to the nr/nt database, excluding Dichorhavirus leprosis (CiLV-N), with Taxonomy ID: 2560386. E-probes that hit a different organism with a percent identity and query coverage greater or equal to 90% were removed. Furthermore, the newly developed e-probes were filtered to remove any known hits to the host genome, using the same percent identity and query coverage as that used for the nt database. The manually curated probes were then re-uploaded to MiFi®. For each target and near neighbor FASTA file, the same procedure was repeated for Dichorhavirus citri (CiCSV), Dichorhavirus clerodendri (ClCSV), Dichorhavirus coffeae, (CoRSV), Dichorhavirus orchidaceae, (OFV or citrus necrotic spot virus; CiNSV), and unclassified Dichorhavirus (CiBSV). In addition, the sequence of Dichorhavirus sp. ‘monocotyledonae’, an unclassified member, was added for validation of curated Dichorhavirus e-probes.
Pairwise genome sequence alignments for all RNA-1 and -2 genome segments were carried out using nucmer (v4.0.0rc1) [42] to establish links displaying similar regions among the studied dichorhaviruses. Gene annotations were retrieved from their respective accessions in the NCBI nucleotide database and e-probe target regions were identified between Dichorhavirus genus vs. species-specific. GenBank accessions of all 28 Dichorhavirus genomes were listed in the Supplementary Table S1. The Circos plot with corresponding links and annotated features was generated using Circos (v0.69-8) [43]. Labels and formatting were processed using Inkscape 1.3.2.
2.4. Detection of Dichorhaviruses Using Real-Time RT-qPCR Assays
There are four dichorhaviruses (OFV, CiLV-N, CiCSV, and CiBSV) that infect citrus and produce leprosis-like symptoms. Out of four dichorhaviruses, only OFV has four distinct variants (OFV-Orc1, OFV-Orc2, OFV-Cit1, and OFV-Cit2) which infect citrus [9,12,17,21]. To detect the dichorhaviruses (CiLV-N, CiCSV, and OFV) associated with citrus leprosis-like symptoms, a multiplex real-time RT-PCR assay was used [44]. For RNA-quality check, plant internal control nad5 gene primers and probes were also included in the assays. Furthermore, to differentiate the strain of OFVs in the infected tissue, separate real-time quadruplex RT-qPCR assays were optimized [45]. To validate the outcome of the e-probe analysis in the HTS data, RT-qPCR was utilized as a screening tool, whereas the bioinformatic analysis of meta-transcriptomic data [46] was used as a confirmatory diagnostic tool. RT-qPCR was performed in a QuantStudioTM 5 Real-Time PCR System (Thermo Fisher Scientific Inc. Carlsbad, CA, USA) following the optimized manufacturer protocol [44,45].
2.5. High Throughput Sequencing of Dichorhaviruses from Different Hosts
For this study, an optimized Illumina ‘Ribo-Zero Total RNA’ HTS protocol [47] was utilized to extract total RNA from 31 samples consisting of 14 host species including C. aurantium, C. reticulata, Sapium sp., Aralia sp., orchids (Cymbidium sp., Dendrobium sp., and Dendrochilum magnum), and ornamentals (Hibiscus rosa-sinensis, Liriope sp., Aspidistra sp., Ophiopogan sp. Smilax auriculata, Pedilanthus tithymaloides, and F. japonicum) using the RNeasy Plant Mini Kit (Qiagen, Germantown, MD, USA) following a modification of the manufacturer protocol [46]. Before proceeding to the cDNA library preparation, the quality and concentration of extracted RNA were measured using a TapeStation (Agilent, Santa Clara, CA, USA). The Illumina TruSeqR Stranded Total RNA Library Prep Plant kit (Illumina, Inc., San Diego, CA, USA) was utilized to construct the 31 cDNA libraries following the modified Illumina ‘Ribo-Zero Total RNA’ protocol [47]. Library titers were quantified using the Qubit fluorometer (Invitrogen, Carlsbad, CA, USA) and quality was tested using TapeStation (Agilent, Santa Clara, CA, USA) as per the manufacturer’s instructions. Either single- or pair-end sequencing was conducted using a MiSeq platform or NextSeq 550 system with 2 × 75 bp, Hi-output Illumina sequencing reagent kits (Illumina, San Diego, CA, USA). A published bioinformatic pipeline [46] was utilized to determine the genomic sequence of known and novel viruses/strains if any, in the generated FASTQ sequence data files.
2.6. Dichorhavirus Detection Using MiFi® Platforms and Determination of the Diagnostic Sensitivity and Specificity of Its e-Probes
For the in vivo validation study, extracted total RNAs from 31 BTV suspected samples were evaluated before proceeding with HTS data generated by either MiSeq or Nextseq 550 Illumina platform. In total, 31 libraries (Table 1) were scanned using e-probes, and the outcome was compared with the real-time RT-qPCR results. For further confirmation, data was analyzed using an optimized bioinformatic pipeline [46]. The metagenome sequence output files (.fastq) were concatenated, compressed in gzip format, and uploaded into MiFi® Metagenomes section. The developed 40, 60, and 80 nts probes were searched against all the libraries created for known and unassigned Dichorhavirus sp. using the MiDetect v2 software in the MiFi® (https://bioinfo.okstate.edu, accessed on 14 March 2023) platform. To determine a positive alignment, an e-value ≤ 1 × 10−10 was used as the threshold. The analysis was based on comparing t test scores between the target (detection signal) and decoy (reverse e-probe sequence described as background signal) e-probes. True positives were called positive when the sequence library scanned using all three e-probes having different lengths (40, 60, and 80 nts) were predicted to be positive and matched with wet lab RT-qPCR assay to confirm that the sample contained the target pathogen. Likewise, true negatives were called negative when the sequence library scanned using e-probes (40, 60, and 80 nts) were predicted to be negative and matched laboratory confirmation (RT-qPCR-verified) that the sample was negative for the target pathogen. Contradictory suspicious, false negative and false positive e-probe diagnosis were further confirmed by bioinformatic analysis. The following formulae were used to determine the e-probe sensitivity (TP/TP + FN) and specificity (TN/TN + FP), where ‘TP’ = true positive, ‘TN’ = true negative, ‘FN’ = false negative, and ‘FP’ = false positive.

Table 1.
Three different lengths (40, 60, and 80 nt) of Dichorhavirus genus, species, strain, and variant-specific e-probes were designed using the e-probe developer software (MiProbe v2) inside the Microbe Finder (MiFi®) platform.
3. Results
3.1. e-Probe Development and Curation
A total of 28 Dichorhavirus RNA-1 and -2 genome segments sequences available in the GenBank, including 13 OFV (subgroup-1), 4 CiLV-N, 3 CiBSV (subgroup-2), 4 CiCSV, 3 ClCSV, and 1 CoRSV (subgroup-3), were included to generate the Dichorhavirus genus and species-specific e-probes (Figure 2). To determine the optimal size of Dichorhavirus e-probes, 40, 60, and 80 nt lengths of e-probes were designed (Table 1). The number of Dichorhavirus e-probe sequences was reduced after curation. In total, 684 Dichorhavirus e-probes were curated by comparing genome sequences against mono-segmented cytoplasmic and nuclear rhabdovirus genera assigned to the family Rhabdoviridae, with cileviruses belonging to the family Kitaviridae and other citrus infecting viruses. Furthermore, available host genome sequences of interest were also included in the BLASTn analysis to discard the sequences having identity with the host and retain the Dichorhavirus-specific e-probes of interest. All the Dichorhavirus e-probes were further categorized into the genus, species, strain, and variant-specific group. A total of 180 genus-specific; 394 species-specific (99, 114, and 181 e-probes for subgroup-1, -2, and -3, respectively); 10 OFV strain-specific (3 for OFV-Orc and 7 for OFV-Cit); 62 OFV-Orc (6 for Orc1 and 56 for Orc2); and 38 OFV-Cit (14 for Cit1 and 24 for Cit2) variant-specific e-probes were uploaded in the MiFi® platform (Table 1). Except for OFV, the number of Dichorhavirus e-probes for each species was increased when the nucleotide length of the e-probe was reduced from 80 to 40. Overall, 40 mer size e-probes produced the maximum number, followed by 60 and 80 mer e-probes (Table 1). We have successfully designed 40 nt e-probes for all the Dichorhavirus species but failed to curate 60 and 80 nt strain-specific OFV-Cit, OFV-Orc e-probes, and 80 nts e-probe for OFV-Orc2 variant as there is a limited number of sequences for select species in the public databases (Table 1). Moreover, the smaller cutoff (40 mer e-probe) as compared to the larger cutoff (60 or 80 mer) has a higher potential to generate more e-probes for smaller pathogen genomes by the MiFi® platform.
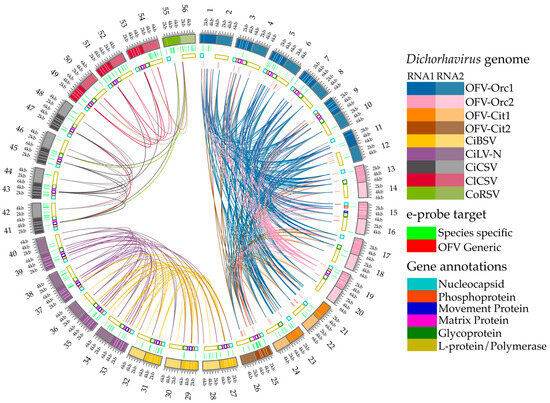
Figure 2.
Multiple pairwise whole genome alignment with gene annotations and highlights for e-probe target regions of the dichorhaviruses used in this study. Alignments performed with the MiProbe v2 software revealed regions of genomic similarity between the targeted dichorhaviruses, and the links between viruses display local high-identity pairwise alignments. The number showing outside the outermost ring represents three subgroups of dichorhavirus; 1–26 represent subgroup 1 (OFV), 27–40 represent subgroup 2 (CiBSV and CiLV-N), and 41–56 represent subgroup 3 (CiCSV, ClCSV, and CoRSV). Odd and even numbers outside the circle represent Dichorhavirus RNA1 and RNA2 genome segments, respectively. From the outermost to innermost rings: (outermost ring) nucleotide positions of RNA1 and RNA2 of genomes of dichorhaviruses; (inner ring 1) green highlights corresponding to e-probes targeting exclusively specific virus species; (inner ring 2) gene annotations retrieved from NCBI reports for all viruses for both RNA1 and RNA2 segments; (innermost half-circle) red highlights corresponding to e-probes designed for orchid fleck virus (OFV) generic regions shared by multiple targets. As stated by the latest NCBI annotations of the studied accessions, hypothetical proteins and orphan ORFs were not considered for gene annotation display.
3.2. Screening of Potential Dichorhavirus-Infected Samples Using RT-qPCR Assays
A total of 31 BTV suspected samples belonging to 14 host species were collected from four countries (12 from USA, 12 from Mexico, 4 from Colombia, and 3 from Costa Rica) and tested for Brevipalpus-transmitted Dichorhavirus infection utilizing Dichorhavirus species-specific (OFV, CiCSV, CiLV-N) and OFV strain-specific RT-qPCR assays. There are no RT-qPCR diagnostic assays available for generic Dichorhavirus, CoRSV, CiBSV, and ClCSV, and generic OFV-Orc and OFV-Cit strain detection. Out of 31, only 16 OFV-positive samples were identified from Mexico (Querétaro, Colima, and Jalisco) and the USA (California, Florida, and Hawaii) using species-specific RT-qPCR assay. Among 16 samples, 13 were positive for either single or two OFV strains/variants. The positive samples included single infections of OFV-Cit1 (n = 1), OFV-Cit2 (n = 1), OFV-Orc2 (n = 5), and mixed infections of OFV-Orc1 and -Orc2 (n = 6) but the strains associated with the remaining three samples (Liriope S1-TH, Aralia CO002, and Dendrobium CO005ac) were undetermined (Table 2).

Table 2.
Detection of Dichorhavirus species [orchid fleck virus (OFV), citrus leprosis virus N (CiLV-N), and citrus chlorotic spot virus (CiCSV)] and four variants of orchid fleck virus (OFV-Orc1, OFV-Orc2, OFV-Cit1, and OFV-Cit2) infecting citrus, orchids, and ornamentals in the USA, Mexico, Colombia, and Costa Rica using two separate Taqman RT-qPCR assays. No RT-qPCR diagnostic assays are available for citrus bright spot virus (CiBSV), clerodendrum chlorotic spot virus (ClCSV), coffee ring spot virus (CoRSV), and generic orchid (OFV-Orc) and citrus (OFV-Cit) strains of OFV. * Refers to the absence of RT-qPCR assay. NT refers to ‘not tested’ and UD refers to ‘undetermined’.
3.3. Detection of Dichorhavirus Genus, Species, OFV Strains, and Variants Utilizing e-Probes in HTS Data
To compare the results of RT-qPCR assays, all studied samples, including asymptomatic (healthy control), Dichorhavirus negative but symptomatic (negative control), and OFV positive (positive control), were processed for HTS, the meta-transcriptomic data uploaded into the Microbe Finder (MiFi®) platform and utilized for e-probe detection. The number of raw reads of RNASeq data generated by Illumina (MiSeq/NextSeq) sequencing platforms per sample varied from 9.5 to 89 million. MiFi detected Dichorhavirus in 15 RNASeq datasets using the e-probes designed for its species, strains, and variants whereas the remaining 16 samples were negative, even though one of the 16 samples (CO005ac) was suspicious for Dichorhavirus genus-specific e-probe diagnosis. Twelve out of fifteen were positive with all the generic Dichorhavirus e-probes sets (40, 60, and 80 nt) whereas the remaining three were positive with either 40 nt (FL7OHP) or 60 nt (CO002) length or both e-probes (ASP-FL) (Table 3). All the e-probe sets irrespective of length (40, 60, and 80 nts) successfully detected Dichorhavirus species OFV in 14 samples even though the total number of hits and the p values decreased as the e-probe length decreased (Table 3). Expected OFV strain-specific positive and unexpected false negative diagnostic calls were obtained for 12 and 2 OFV positive samples, respectively. Out of twelve, nine samples had a single infection with the OFV-Orc strain, two with the OFV-Cit strain, and one (sample CA2) with both strains. OFV strain-specific e-probes failed to clearly detect the presence of strains in the RNASeq data obtained from OFV-positive samples (FL7OHP and CA3) but successfully detected variant OFV-Orc1 or both the OFV-Orc variants in FL7OHP and CA3, respectively. Interestingly, the e-probe diagnosis was unsuccessful in determining the variant of OFV-Cit in the sample CA2. E-probe sets specific to OFV-Cit variants successfully detected OFV-Cit1 in the sample QR035 (F. japonicum) and OFV-Cit2 in JA022 (C. aurantium). Expected negative diagnostic calls were obtained for 15 RT-qPCR negative samples, including the healthy control (JA023) and other 14 negative samples (Table 2) with the three e-probe sets, indicating that pathogen e-probes were specific to the Dichorhavirus species and OFV strains and not cross-reacting with the citrus, orchid, and other ornamental hosts genome sequences (Table 3). Interestingly, the sample CO002 (Aralia sp.) gave a positive diagnostic call for Dichorhavirus only with 60 nt length e-probe (p-value 0.0416). However, a suspicious call for OFV species raised the question for further confirmation. RNASeq data for BTV-suspected sample CO005ac (Dendrobium sp.) gave potentially true negative diagnostic calls with suspicious p-value 0.0794 for 40 nt Dichorhavirus generic e-probe only (Table 3).

Table 3.
Detection of Dichorhavirus in RNASeq data originated from healthy and Brevipalpus transmitted virus suspected symptomatic leaf tissues utilizing e-probe sets designed for Dichorhavirus species [coffee ring spot virus (CoRSV), citrus leprosis virus N (CiLV-N), citrus chlorotic spot virus (CiCSV), clerodendrum chlorotic spot virus (ClCSV), citrus bright spot virus (CiBSV), and orchid fleck virus (OFV)], OFV strains (OFV-Orc and OFV-Cit), and variants (OFV-Orc1, OFV-Orc2, OFV-Cit1, and OFV-Cit2) available in the MiFi platform. Out of 31, 16 RT-qPCR positive, 1 healthy and 1 negative sample e-probe analysis data were included. Except for OFV, none of the Dichorhavirus species was identified in the listed samples, and negative results for CiLV-N, CiCSV, CoRSV, ClCSV, and CiBSV are not included in this table. The target and decoy scores were compared using a t-test. Three tiers of diagnostic calls were used in the statistical test, positive (p-value ≤ 0.05), suspect (0.05 > p-value ≤ 0.1), and negative (p-value > 0.1). No significant difference between the two sets (target and decoy) indicated no evidence for the presence of pathogen sequences, and the sample was designated negative for the Dichorhavirus. The negative and suspicious e-probe p-values were highlighted using orange and lavender colors, respectively. Blue shaded ‘NP’ refers to ‘no probe’ while shadeless ‘NC’ refers to ‘not computed’.
3.4. Diagnostic Sensitivity and Specificity for Curated Dichorhavirus e-Probes in RNA-Seq Data and Confirmation of e-Probe Diagnostics Using Bioinformatic Pipeline Analysis
Diagnostic results of the Dichorhavirus e-probes from the field samples were compared with the results of Dichorhavirus-specific RT-qPCR assays to calculate diagnostic performance metrics using known positive, negative, and Brevipalpus-transmitted Dichorhavirus-suspected samples. In this study, 31 samples including citrus, orchids, and ornamentals were included for sensitivity and specificity testing and utilized metagenome compressed raw HTS fastq files from available RT-qPCR positive (n = 16) and negative (n = 15) samples uploaded to the MiFi® platform. The EDNA diagnostic sensitivity and specificity analysis was calculated for curated e-probe sequences with 40-, 60-, and 80-nt e-probe lengths and evaluated for false-positive and -negative results (Table 4).

Table 4.
In vivo sensitivity and specificity testing of Dichorhavirus e-probes comparing the gold standard RT-qPCR data and bioinformatic pipeline analysis. ‘ND’ means ‘Not Determined’ and ‘NT’ means ‘Not Tested’.
The number of raw reads RNASeq data ranged from 9.5 to 89 million but the total Dichorhavirus reads varies from minimum 372 [23.91% of 1556 total virus reads (0.007%), out of 21,801,326 post-trim reads] in FL7OHP to a maximum 10,766,019 [75.59% of 14,241,854 total virus reads (56.37%), out of 25,266,822 post-trim reads] in the sample S79_VOrBTP (Table 5). In contrast, maximum total virus reads (62,935,224/77,029,412 × 100 = 81.70%) were obtained in the sample S78_VDPCU with only 5.97% Dichorhavirus reads. Out of 22 Dichorhavirus positive samples, 11 had >90% Dichorhavirus reads with other viruses in mixed infection. Total virus reads from those 11 samples varied from 0.617% (119,926 reads) to 13.63% (3,013,247 reads) but Dichorhavirus reads represented between 91.49% and 99.94% of viral reads (Table 5). The relative abundance of Dichorhavirus sequence reads in the RNASeq data (Figure 3) was calculated using a curated Dichorhavirus e-probe irrespective of the optimal e-probe length (Table 5).

Table 5.
In vivo validation of Dichorhavirus e-probe diagnostic in 31 RNASeq data by comparing RT-qPCR results and bioinformatic pipeline analysis.
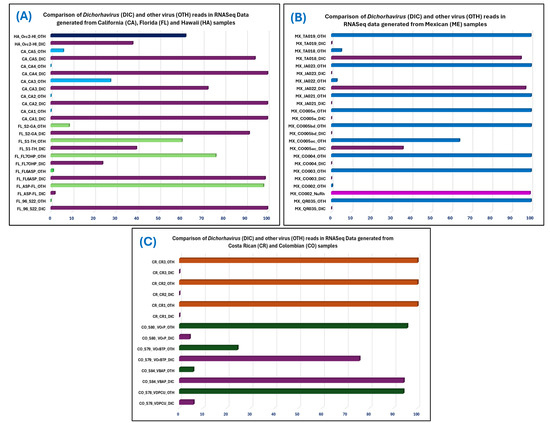
Figure 3.
Distribution of rhabdovirus and other virus reads (OTH) among total virus reads detected in 31 RNASeq data, which were obtained from Brevipalpus-transmitted virus-suspected samples from four countries: (A) USA, (B) Mexico (C) Colombia and Costa Rica. Samples from each country are represented by individual colors in the bar diagrams. Single-segment nucleo-rhabdovirus (NuRh) reads in the samples are represented by purple, bi-segment Dichorhavirus (DIC) is represented by plum color, whereas other viruses are represented by green (Florida), light blue (California), dark blue (Hawaii), in the USA samples, blue color in Mexican samples, dark green in Colombia samples, and orange color in Costa Rica samples.
Among 16 RT-qPCR-positive samples, 15 samples were positive for Dichorhavirus e-probe. None of the Dichorhavirus-positive samples were identified as negative in e-probe diagnosis except the sample CO002. Interestingly, bioinformatic analysis revealed that the sample CO002 was infected with a single-segmented novel nucleo-rhabdovirus belonging to the family Rhabdoviridae. Furthermore, novel Dichorhavirus sequences were detected in six RNAseq libraries (sample CO005ac, TA018 from Mexico and S78_VDPCU, S79_VOrBTP, S80_VOrP, and S84_VBAP from Colombia) (Figure 3, Table 5) but none of the Dichorhavirus sequences in those six samples were detected by Dichorhavirus genus-specific e-probes except CO005ac which was diagnosed as suspicious (p-value 0.0794) diagnostic sample for possible Dichorhavirus infection. Out of 16 RT-qPCR-positive samples, OFV species-specific e-probes (OFV-Gen) were detected in 14 positive samples and diagnosed CO002 as suspicious and CO005ac as negative samples. OFV species-specific e-probe results show doubtful infection of OFV in the sample CO002 (p-value 0.0805) and negative in CO005ac (p-value 0.1195) as infected with a novel nucleo-rhabdovirus and a Dichorhavirus, respectively. In both cases, the OFV-specific e-probes were bioinformatically cross-reacted to the distantly related sequences, and EDNA sensitivity for OFV species-specific e-probe improved from 87.5% (14/16) to 100% after evaluating the results of RT-qPCR and EDNA with bioinformatic analysis. OFV strain-specific RT-qPCR detected two OFV-Cit and 11 OFV-Orc variants among 16 OFV-positive samples whereas it failed to determine the type of OFV strain as well as the type of variant present in the samples S1–TH (Liriope sp.), CO002 (Aralia sp.), and CO005ac (Dendrobium sp.) (Table 2). Even though e-probe analysis detected an OFV-Orc strain-specific sequence (p-value 0.0001) in the sample S1–TH, bioinformatic analysis confirmed the existence of an unrevealed OFV strain. Furthermore, the OFV-Orc1 variant was detected in CO002 by e-probe diagnostic (p-value 0.0209–0.0415), but the true positive e-probe diagnostic result turned into false positive as no similar e-probe sequence was identified by bioinformatic pipeline. The calculated sensitivity of the OFV variant-specific probe is 93.75% (15/16). The sensitivity of variant-level e-probe detection can be improved further by adding the new strain sequence in the strain-specific e-probe curation folder in the MiFi® platform.
3.5. Discovery of a New Host Species of OFV and a Possible New OFV Strain in Known Hosts Utilizing e-Probe Diagnosis
Out of 14 host species studied for possible Dichorhavirus infection, 10 species (C. aurantium, C. reticulata, Cymbidium sp., Dendrobium sp., Dendrochilum magnum, H. rosa-sinensis, Liriope sp., Aspidistra sp., Ophiopogan sp., and S. auriculata) are known host for Dichorhavirus, whereas P. tithymaloides, Sapium sp., Aralia sp., and F. japonicum are not previously known as either natural or experimental hosts. Both preliminary RT-qPCR assays and the e-probe sets in the MiFi® platform successfully detected Dichorhavirus and OFV in 12 (10 known and 2 unknown) host tissues from total RNA and 16 RNASeq datasets, respectively. The remaining fifteen samples, including one P. tithymaloides and two Sapium samples, and corresponding RNASeq datasets were negative by both methods. Both RT-qPCR and EDNA methods successfully detected OFV infection up to variant level (OFV-Cit1) in the new host F. japonicum, whereas e-probe diagnostic failed to identify any known OFV variant in two Aspidistra samples (ASP-FL and FL6ASP) from Tallahassee, Florida. On the other hand, OFV-specific RT-qPCR positive with CT values 32.19 and 30.52 for Aralia (CO002) and Dendrobium (CO005ac) samples, respectively, were evaluated as false positive for Dichorhavirus infection by EDNA.
E-probe diagnostic results of dichorhaviruses’ presence or absence in symptomatic F. japonicum (QR035), Aspidistra sp. (ASP-FL and FL6ASP), Aralia sp. (CO002), and Dendrobium sp. (CO005ac) were validated using meta-transcriptomic data. During the bioinformatic analysis, the post-trim sequences were mapped to the Arabidopsis proteome and the available Dendrobium genome sequences in the NCBI database, and the identified host sequences were then removed. Out of 26,542,292 post-trims read in F. japonicum (QR035) cDNA library, 5,955,476 reads (23.44%) were mapped against plant viruses. The assembled contigs were blasted against the NCBI database and most of the total virus read matched with the genus Nepovirus (99.16%), Ophiovirus (0.54%), Mycovirus (0.23%), Cytorhabdovirus (0.045%), Dichorhavirus (0.0145%), and DNA plant virus sequences available in GenBank. In total, 11 Dichorhavirus-related contigs were obtained, with a maximum size of 2271 nt and a minimum of 197 nt. Assembled contigs cover 37.29% and 99.58% of RNA1 and RNA2 genome segments, respectively, and shared 99% nucleotide sequence identity with OFV-Cit1 (Acc. No. KF209275 and KF209276). Bioinformatics analysis confirmed the infection of OFV-Cit1 in F. japonicum obtained from El Pueblito in Querétaro, Mexico, and validated the e-probe diagnostic results.
Meta-transcriptomic analysis of two Aspidistra cDNA libraries (ASP-FL and FL6ASP) detected a total of 32,178 (0.037%) and 119,926 (0.62%) virus reads out of 87,154,532 and 19,424,805 post-trim reads, respectively. Furthermore, total virus read associated with ASP-FL and FL6ASP identified as DNA plant virus (8.11% and 0.46%), Mycovirus (89.96% and 0.72%), and Dichorhavirus (1.92% and 98.82%) read, respectively. In total, 11 Dichorhavirus-related contigs were obtained from ASP-FL RNASeq data, with a maximum size of 1245 nt and a minimum of 221 nt, whereas two large contigs (6027 and 6454) were obtained from FL6ASP RNASeq data. Assembled RNA1 and RNA2 genome contigs shared a maximum 90.63% and 98.43% nt identity with Dichorhavirus orchidaceae infected V. spicata (spike speedwell) in the United Kingdom (Acc. No. PP429912) and P. amaryllifolius (pandan grass) in Florida (OK624602), respectively. Data analysis confirmed the presence of a new OFV strain in the ornamental Aspidistra sp.
A total of 6,843,912 (31.12%) processed reads related to plant viruses were detected in the CO005ac library, which consists of mainly two virus genera: Potexvirus and Dichorhavirus. The 2nd highest percentage of viral reads (35.86%) were detected for Dichorhavirus after the Potexvirus reads (64.09%) (Table 2). The remaining 0.5% reads shared nucleotide identity either with Badnavirus or Tobamovirus sequences. The assembled contig (6827 nt) for Potexvirus was identified as cymbidium mosaic virus whereas the remaining two long contigs (6466 and 6053 nt) were identified as RNA 1 and RNA2 genome segment of an undescribed Dichorhavirus. BLASTn analysis of two contigs of Dichorhavirus (6466 and 6053 nt) genome segments shared 70.64% nt sequence identity with 34% genome coverage and 70.50% nt sequence identity with 67% genome coverage with OFV-Orc2 RNA1 (MW021482) and OFV-Cit2 RNA2 (MK578001), respectively, and identified as a novel species for Dichorhavirus, which is a distantly relative of OFV. Another novel Dichorhavirus species sequence was identified in the samples; S78_VDPCU, S79_VOrBTP, S80_VOrP, and S84_VBAP from Colombia via bioinformatic analysis but surprisingly none of the sample’s sequences computed any suspicious value for any generic or species-specific OFV e-probes.
Altogether 1,588,639 single-end Illumina reads recovered from the Aralia sp. (CO002) cDNA library were mapped to the Rhabdovirus, the highest proportion of viral reads (1,588,639/1,596,975 × 100 = 99.48%) in the sample, whereas the remaining 8336 viral reads were related to Badnavirus and endornavirus sequences (Table 4). Total Rhabdovirus reads were further categorized between two subfamilies, Betarhabdovirinae and Alpharhabdovirinae at the rate of 96.51% (1,533,153/1,588,639 × 100) and 3.49% (55,486/1,588,639 × 100), respectively. Two major contigs of 11,583 and 13,856 nt covering almost the entire single-segmented rhabdovirus genome were identified. The nucleotide sequence of contigs 1 and 2 shared 80.75% and 64% nt identities with query coverage of 98% and 8%, respectively, with the nucleotide sequences of datura yellow vein virus (DYVV, Betanucleorhabdovirius) (KM823531) infecting black-eyed Susan (Thunbergia alata) in Australia and eggplant mottled dwarf virus (EMDV) (OR613409) infecting eggplant (Solanum melongena) in Iran, respectively. Interestingly, no significant similarity was found in BLASTn analysis with the contig 1 query sequence and EMDV (Alphanucleorhabdovirius) genome sequence. However, the contig 1 sequence shared approximately 2% of genome coverage (1702 to 1801, 1956 to 1993, and 2378 to 2498 nts) and ~76% nt identity with OFV-Cit1 (KF209276), the member of the genus Dichorhavirus.
4. Discussion
Dichorhavirus infections mostly produce local chlorotic/necrotic lesions and/or chlorotic spots appearing on leaves and fruits in monocots as well as in dicots. The most economically important disease associated with Dichorhavirus infection is citrus leprosis, first described in Florida at the start of the 20th century [48], but its occurrence has not been observed since the mid-1960s. HTS of citrus-leprosis-like symptomatic herbarium samples from Florida revealed an association with a distant relative of OFV, referred to as CiLV-N0 [49]. The necrotic lesion symptoms observed in C. sinensis in Florida are associated with the Dichorhavirus, CiLV-N0 infection. Similar symptoms were observed in Citrus spp. in Colombia, Mexico, Hawaii, and South Africa but the identified pathogen in association with the leprosis-like symptom was OFV [9,12,17,18,19,21,50]. The economic consequences of Dichorhavirus infections, particularly CiLD, are substantial. The identification of new strains or hosts, as demonstrated in this study, could further complicate disease management strategies. The declining cost of HTS and the accessibility of computational biologists with advanced bioinformatics and computer programming knowledge have made it reasonable for many laboratories and plant regulatory agencies like USDA-APHIS-PPQ to implement this cutting-edge technology in diagnostics [47,51,52]. In the current research, we explored the EDNA technology [28] integrated with the online MiFi® platform [29] to validate the sensitivity and specificity of curated Dichorhavirus e-probes by utilizing meta-transcriptomic libraries of BTVs belonging to the families Kitaviridae and Rhabdoviridae, along with other viruses infecting citrus and virus-free citrus and other hosts.
This study aimed to develop and curate e-probes at the genus, species, strain, and variant level using the Dichorhavirus species OFV as a model by adopting the scope of EDNA detection [53]. To adapt the approach according to the specific goal, generic e-probes at least for two or more Dichorhavirus species were designed utilizing the conserved regions of all the 28 Dichorhavirus genome sequences available in the GenBank. In contrast, non-conserved regions of the target genomes of multiple OFV isolates further identified at the strains/variants level were included to increase the sensitivity and specificity of e-probes and curated against phylogenetically related neighbors.
Previously, analytical performance metrics were assessed in silico based on a limit of detection of high-quality hits of e-probes [34], but the current study is focused on in vivo validation. HTS data generated from 31 RT-qPCR tested field samples collected from Colombia, Costa Rica, Mexico, and three different geographical locations of the United States were utilized to confirm the broader specificity of curated e-probes. Field samples collected from 14 different hosts, which have apparent Brevipalpus or eriophyid mite-transmitted virus symptoms were sequenced and analyzed using curated Dichorhavirus e-probes, and the test results were confirmed by comparing RT-qPCR and bioinformatic analysis. Both Dichorhavirus species-specific and OFV strain-specific RT-qPCR assays were developed in-house and validated for their sensitivity and specificity [44,45]. Similarly, the bioinformatic pipeline optimized for virus detection and discovery was validated in multiple studies [46,47]. To use the bioinformatic tools for pathogen detection, a person should have the capability to analyze the output of the Sequence Alignment Map (SAM) format before mapping the reference genome and/or BLAST to the GenBank database. MiFi® eliminates the requirement of a dedicated bioinformatician as the user never creates any SAM or BLAST output files and provides a clear answer within 30 min.
Generally, the relative size of the pathogen and the number of raw e-probes are proportionally correlated as demonstrated for viroids, which are the smallest known pathogens and have the lowest number of e-probes [29,31]. To improve diagnostic specificity during Dichorhavirus e-probe curation, non-specific sequences were removed. In this study, we curated a total of 684 Dichorhavirus genus, species, strain, or variant-specific e-probes in the MiFi® platform (Table 1). We showed that the EDNA can be utilized to detect the presence or absence of Dichorhavirus, the discovery of a new natural host (F. japonicum), and the possible existence of a novel Rhabdovirus/Dichorhavirus or a new strain of OFV in nature. Three sizes (40, 60, and 80 nt) of e-probes were successfully designed to match only the Dichorhavirus of interest, and variant-specific e-probes for OFV-Cit and OFV-Orc strains, except 80 nt e-probe for OFV-Orc2 variant. To assess the analytical sensitivity of curated e-probes, either in silico or in vitro validation is required to determine if the benchmark needs any further improvement [29]. In vitro validation of e-probe diagnostic must be accompanied by gold standard real-time PCR results of the same samples being sequenced. Here, we evaluated the sensitivity and specificity of all the genus, species, OFV strains, and its variant-specific e-probes utilizing EDNA incorporated in the MiFi® platform. The performance of the MiFi® platform was determined when EDNA results were compared and validated with the gold-standard RT-qPCR assays. In total, three cases of OFV species-specific RT-qPCR positive samples (Liriope sp. S1-TH, Aralia sp. CO002, and Dendrobium sp. CO005ac) failed to determine the type of variant by OFV strain-specific RT-qPCR assay (Table 2). Moreover, RT-qPCR assays were not conducted for generic Dichorhavirus, CoRSV, CiBSV, and ClCSV species, and generic OFV-Orc and OFV-Cit strain detection. Therefore, bioinformatic pipeline analysis was included as a 3rd method for further confirmation of e-probe diagnostic results. The RT-qPCR diagnostic was wrong in six cases when the e-probe diagnosis was verified by bioinformatic analysis. Out of 25 contradictory e-probe diagnoses, 17 were the correct diagnosis, whereas 8 were incorrect (Table 6). Among eight incorrect e-probe diagnoses, analysis results were either false positive (n = 5) or false negative (n = 3) as compared to RT-qPCR and bioinformatic analysis. Interestingly, two out of eight false diagnoses occurred in the samples CO002 and CO005ac, which were negative to listed dichorhaviruses in the study, but were later determined to have a novel nucleo-rhabdovirus in CO002 and a novel dichorhavirus in CO005ac present in RNASeq libraries. False-negative results when using e-probes are often caused by the inability of the t-test to compare the target with decoy e-probes and identify variance. This issue arises from either no hits or an excessive number of hits that reach the maximum limit of 250 hits permitted by EDNA. Typically, this can be addressed by adding internal control e-probes as used previously to calculate the variance with other citrus pathogens [31]. Internal control provides a small amount of background variance that assures that the statistical analysis does not have zero variance in the algorithm denominator, which results in a non-computed (NC) result. False positive detection of OFV-Orc1 in the sample CO002 indicated that the e-probe was cross-reacted with a novel nucleo-rhabdovirus sequence, and this is an expected behavior for e-probes since they are designed based on available data in public databases and are not meant to detect new virus species (Table 6). Overall, e-probe diagnosis incorrectly identified the OFV-Cit strain in the sample CA2 without identifying any variant, suggesting that the OFV strain-specific e-probes might not be 100% specific or bioinformatic analysis failed to pick up the low read OFV-Cit variant sequence in the CA2 RNASeq library. Moreover, the discovery of new OFV strain sequences in the samples Liriope sp. (S1-TH) and Aspidistra sp. (FL6ASP) RNASeq libraries also supports the RT-qPCR data and clarifies the reason behind the false detection of the presence or absence of different OFV-variant sequences through e-probe diagnosis (Table 6). E-probe synthesis is limited by the availability of target and near-neighbor genomic information and generally improves as more related sequences become available. Recently deposited four more OFV complete genome sequences (Acc. PP429909-10, PP429912-13, LC771578-79, and LC846649-50) and a new dichorhavirus species vinca chlorotic spot virus sequence (OR372158-59) in GenBank further extended the scope for more generic or specific dichorhavirus e-probe development.

Table 6.
Validation of contradictory results of Dichorhavirus e-probe diagnostic utilizing confirmatory bioinformatic pipeline analysis. ‘UD’ refers to ‘undetermined’, which means that in all the cycles of the RT-qPCR reaction the signal from the target cDNA did not pass the threshold level. ‘NT’ refers to ‘not tested’ due to absence of RT-qPCR assay.
Calculation of sequencing depth is an important parameter that can improve the chances of finding the desired pathogen reads in a complex metagenome, but no correlation was found between total viruses read and the ratio of Dichorhavirus and other virus read number in the studied RNASeq data (Figure 3, Table 5). The unbiased computer search will find the read if it is intact and present in the meta-transcriptomic data. At least one pathogen (OFV) was detected by MiFi® in 14 out of 16 real-time PCR-positive (Ct values ≤ 34) samples. This indicates that the titers of OFV during an active disease process were high enough to accomplish metagenome-based e-probe detection. The relative abundance of Dichorhavirus reads is highest in the sample CA1 metagenome (0.13607), and lowest in the sample ASP-FL (7.09 × 10−6) (Table 5). The detection time depends on computer power, but if the sample is readily detected with curated e-probe in the MiFi® platform, then hits will happen within minutes instead of 20–30 min required when pathogen reads are few among the total number of reads in the metagenome (Table 5). In conclusion, meta-transcriptomic-based Dichorhavirus detection with MiFi® was successfully tested; most of the tested samples (29/31) were correctly diagnosed with minimum error and led to the discovery of a new nucleo-rhabdovirus and a novel Dichorhavirus species. Deposition of novel Dichorhavirus species sequences and additional OFV strain and variant sequences in the GenBank database will further improve the sensitivity and specificity of curated Dichorhavirus e-probes available at https://bioinfo.okstate.edu (accessed on 25 February 2025).
The advantages of the metagenomic-based bioinformatic approach to pathogen detection are further accelerated by utilizing the MiFi®-based sensitive, specific, and time-efficient e-probe diagnostic method application in the discovery of a novel host (F. japonicum). The broader implication of the MiFi® is that the platform follows a rules-based approach (e-probe size, BLAST parameters, and scores) and more narrow search space to selecting the appropriate query sequence (e-probes) in an efficient manner since it eliminates irrelevant sequences that are later used to detect the pathogen in metagenomes compared to a traditional approach of screening metagenomes manually by using BLASTn directly against the NCBI’s more general databases such as nr/nt. Additionally, adjusting the e-value threshold and choosing “general” Dichorhavirus e-probes allowed for the search and discovery of a novel rhabdovirus and Dichorhavirus in meta-transcriptomic data that are not the specific target organism. Further addition of multiple OFV genome sequences from different hosts could improve the diagnostic rate for indistinct variants in the current database. Thus, the EDNA system [28] integrated into the MiFi® platform [29] can be adjusted or designed to address a range of applications and/or scientific needs which will allow the research scientist to address multifaceted experiments where monitoring and surveillance, detection, and diagnosis of pathogens infecting plant, animal, and human are required. The potential limitation of the MiFi® platform and e-probe curation is its dependency on sequences from the public domain, such as the near-neighbor sequence selection. It could take an expert to weed out any spurious sequences. Additionally, it takes a specialist to understand the current and usually esoteric classification system of the organism and create the probes in the proper context. For example, a person should have a thorough knowledge of taxonomy to create probes for viruses specific to the genus, species, strain, or variant. It would also be interesting to observe how e-probe diagnosis performs between the HTS data generated from the same cDNA library utilizing higher and lower error rates of the nanopore and Illumina sequencing platforms.
5. Conclusions
In this study, we explored millions of reads of cDNA sequences generated from 31 suspected plant samples for their ability to match with the newly curated short sequence/s (e-probe) specific to Dichorhavirus genus, species, OFV strains, and variants. A platform (MiFi®) integrated with multiple complex software was utilized to detect the targeted short-specific Dichorhavirus sequence from the tested dataset. Newly developed e-probes for dichorhaviruses correctly diagnosed 29 tested samples with minimum error and led to the discovery of a new host (leopard plant, F. japonicum), new ornamental OFV strain, and novel rhabdoviruses. The results of the EDNA method were validated with the gold standard RT-qPCR diagnostic methods and confirmed by bioinformatic pipeline analysis. Overall, the EDNA method is very specific and sensitive and can detect the targeted pathogen within one to 30 min, depending on the total number of pathogens reads in the sequencing dataset. The same protocol can be used by other scientists to develop, curate, and validate e-probes related to any virus genera and in the discovery of new hosts, new virus species, new strains, or variants. The findings of this study further pave the way for future investigations into the Dichorhavirus research. A focus on the evolutionary dynamics of Dichorhavirus would aid in prediction and help stakeholders manage their potential impact on agriculture and natural ecosystems. Implementation of the EDNA method and updated host information for Dichorhavirus diagnosis should assist regulatory agencies in surveillance activities to monitor the distribution pattern of citrus leprosis-associated dichorhaviruses in alternate hosts in countries where it is present and to prevent dissemination into citrus growing countries where there is no report of these viruses.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v17030441/s1, Table S1: GenBank accession numbers of 28 dichorhaviruses consisting of five species (OFV, CiLV-N, CiCSV, ClCSV, and CoRSV) and an unassigned member (CiBSV) were downloaded to design Dichorhavirus species-specific and OFV generic e-probes.
Author Contributions
Conceptualization, A.R. and W.L.S.; sample collection, G.O.-C. and A.R.; methodology, A.R., A.S.E., J.S. and D.R.L.; software, J.S., A.S.E., D.R.L. and A.R.; validation, A.R., J.S., A.S.E. and K.C.; formal analysis, A.R., J.S. and A.S.E.; investigation, A.R., J.S., A.S.E., Y.R., V.A.M., M.K.N. and K.C.; resources, A.R., A.S.E., K.C., Y.R., V.A.M. and M.K.N.; data curation, J.S., A.S.E. and A.R.; writing—original draft preparation, A.R. and J.S.; writing—review and editing, A.R., A.S.E., J.S.; G.O.-C. and W.L.S.; supervision, A.R.; project administration, A.R.; funding acquisition, A.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported and funded by the United States Department of Agriculture’s Animal and Plant Health Inspection Service (USDA-APHIS) Plant Protection Act Section 7721 Project No. 2018-03.0557, USDA-APHIS PPQ Citrus Health Response Program (CHRP) Project No. PPQ-013258, and USDA-ARS in-House Appropriated Project NP303-8042-22000-319-000-D. This material was made possible, in part, by Cooperative Agreements from the USDA-APHIS. It may not necessarily express APHIS’ views.
Institutional Review Board Statement
Not Applicable.
Informed Consent Statement
Not Applicable.
Data Availability Statement
The SRA data used in the study were deposited in the NCBI repository at https://www.ncbi.nlm.nih.gov/, under BioProject ID accession number PRJNA1158807, Temporary Submission ID: SUB14716102, scheduled to be released immediately after publication.
Acknowledgments
We thank the following people: (i) Juan Carlos Campos Pinzón, Guillermo Leon Martinez, and Yeisson Gutierrez Lopez, AGROSAVIA, Centro de Investigación La Libertad; (ii) Pamela Murillo-Rojas, University of Costa Rica; (iii) Michael Melzer, University of Hawaii, Honolulu; (iv) Kishore Dey and Austin Fife, Florida Department of Agriculture and Consumer Services, Gainesville, Florida; and (v) Tian Tongyan, California Department of Food and Agriculture, for providing the Brevipalpus-transmitted, virus-suspected, OFV-positive tissue samples. We also thank Chellappan Padmanabhan, Schyler Nunziata, USDA-APHIS-PPQ, PPCDL, and Sam Grinstead, USDA-ARS, MPPL, for providing technical support; John Hammond, USDA-ARS, Floral and Nursery Plants Research Unit, Beltsville, MD, for critical comments on the manuscript; and Rosemarie Hammond, USDA-ARS, MPPL acting research leader, for administrative support.
Conflicts of Interest
The authors declare no conflicts of interest, as the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Dietzgen, R.G.; Freitas-Astúa, J.; Chabi-Jesus, C.; Ramos-González, P.L.; Goodin, M.M.; Kondo, H.; Tassi, A.D.; Kitajima, E.W. Dichorhaviruses in Their Host Plants and Mite Vectors. In Advances in Virus Research; Academic Press Inc.: Cambridge, MA, USA, 2018; Volume 102, pp. 119–148. ISBN 9780128151945. [Google Scholar]
- Walker, P.J.; Freitas-Astua, J.; Bejerman, N.; Blasdell, K.R.; Breyta, R.; Dietzgen, R.G.; Fooks, A.R.; Kondo, H.; Kurath, G.; Kuzmin, I.V.; et al. ICTV Virus Taxonomy Profile: Rhabdoviridae 2022. J. Gen. Virol. 2022, 103, 001689. [Google Scholar] [CrossRef] [PubMed]
- Bejerman, N.; Dietzgen, R.; Debat, H. Novel Tri-Segmented Rhabdoviruses: A Data Mining Expedition Unveils the Cryptic Diversity of Cytorhabdoviruses. Viruses 2023, 15, 2402. [Google Scholar] [CrossRef] [PubMed]
- Nemchinov, L.G.; Irish, B.M.; Grinstead, S. First report of Medicago trirhavirus 1 infecting alfalfa in Washington State, USA. Plant Dis. 2024. Epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Dietzgen, R.G.; Kuhn, J.H.; Clawson, A.N.; Freitas-Astúa, J.; Goodin, M.M.; Kitajima, E.W.; Kondo, H.; Wetzel, T.; Whitfield, A.E. Dichorhavirus: A Proposed New Genus for Brevipalpus Mite-Transmitted, Nuclear, Bacilliform, Bipartite, Negative-Strand RNA Plant Viruses. Arch. Virol. 2014, 159, 607–619. [Google Scholar] [CrossRef]
- Kondo, H.; Fujita, M.; Telengech, P.; Maruyam, K.; Hyodo, K.; Tassi, A.D.; Ochoa, R.; Andika, I.B.; Suzuki, N. Evidence for the replication of a plant rhabdovirus in its arthropod mite vector. Virus Res. 2025, 351, 199522. [Google Scholar] [CrossRef] [PubMed]
- de Lillo, E.; Freitas-Astúa, J.; Kitajima, E.W.; Ramos-González, P.L.; Simoni, S.; Tassi, A.D.; Valenzano, D. Phytophagous Mites Transmitting Plant Viruses: Update and Perspectives. Entomol. Gen. 2021, 41, 439–462. [Google Scholar] [CrossRef]
- Kondo, H.; Maeda, T.; Tamada, T. Orchid Fleck Virus: Brevipalpus californicus Mite Transmission, Biological Properties and Genome Structure. Exp. Appl. Acarol. 2003, 30, 215–223. [Google Scholar] [CrossRef]
- Roy, A.; Hartung, J.S.; Schneider, W.L.; Shao, J.; Leon, M.G.; Melzer, M.J.; Beard, J.J.; Otero-Colina, G.; Bauchan, G.R.; Ochoa, R.; et al. Role Bending: Complex Relationships between Viruses, Hosts, and Vectors Related to Citrus Leprosis, an Emerging Disease. Phytopathology 2015, 105, 872–884. [Google Scholar] [CrossRef]
- Mei, Y.; Bejerman, N.; Crew, K.S.; McCaffrey, N.; Dietzgen, R.G. First report of orchid fleck virus in Lilyturf (Liriope spicata) in Australia. Plant Dis. 2016, 100, 1028. [Google Scholar] [CrossRef]
- Harju, V.; Fowkes, A.R.; Skelton, A.; Adams, I.P.; Mcgreig, S.; Forde, S.M.D.; Pufal, H.; Conyers, C.; Frew, L.; Fox, A. First detection of orchid fleck virus on Veronica spicata and Dendrochilum magnum. New Dis. Rep. 2024, 50, e12312. [Google Scholar] [CrossRef]
- Olmedo Velarde, A.; Roy, A.; Padmanabhan, C.; Nunziata, S.; Nakhla, M.K.; Melzer, M.J. First Report of Orchid Fleck Virus Associated with Citrus Leprosis Symptoms in Rough Lemon (Citrus jambhiri) and Mandarin (C. reticulata) the United States. Plant Dis. 2021, 105, 2258. [Google Scholar] [CrossRef] [PubMed]
- Dey, K.; Velez-Climent, M.; Padmanabhan, C.; Nunziata, S.; Rivera, Y.; McVay, J.; Roy, A. Smilax auriculata: A new host for Orchid fleck dichorhavirus identified in Florida, USA. Plant Dis. 2021, 106, 2271. [Google Scholar] [CrossRef] [PubMed]
- Fife, A.; Carrillo, D.; Knox, G.; Iriarte, F.; Dey, K.; Roy, A.; Ochoa, R.; Bauchan, G.; Paret, M.; Martini, X. Brevipalpus-transmitted Orchid Fleck Virus infecting three new ornamental hosts in Florida. J. Integr. Pest Manag. 2021, 12, 43. [Google Scholar] [CrossRef]
- Alvarez-Quinto, R.A.; Grinstead, S.; Rott, P.; Mollov, D. Genome characterization and complete sequence of a new badnavirus from Pandanus amaryllifolius. Arch Virol. 2022, 167, 1717–1720. [Google Scholar] [CrossRef]
- Roy, A.; Espindola, A.; Shao JNunziata, S.; Rivera, Y.; Mavrodieva, V.A.; Nakhla, M.K.; Cardwell, K.F. Evaluation of electronic probes for In-silico detection of the Dichorhavirus in High-Throughput Sequencing data using Microbe Finder (MiFi®). Phytopathology 2023, S3, 149. [Google Scholar]
- Cook, G.; Kirkman, W.; Clase, R.; Steyn, C.; Basson, E.; Fourie, P.H.; Moore, S.D.; Carstens, T.G.E.; Hattingh, V. Orchid Fleck Virus Associated with the First Case of Citrus Leprosis-N in South Africa. Plant Pathol. 2019, 155, 1373–1379. [Google Scholar] [CrossRef]
- Cruz-Jaramillo, J.L.; Ruiz-Medrano, R.; Rojas-Morales, L.; Lopez-Buenfil, J.A.; Morales- Galvan, O.; Chavarin-Palacio, C.; Ramírez-Pool, J.A.; Xoconostle-Cázares, B. Characterization of a proposed Dichorhavirus associated with the citrus leprosis disease and analysis of the host response. Viruses 2014, 6, 2602–2622. [Google Scholar] [CrossRef]
- Roy, A.; Leon, M.G.; Stone, A.L.; Schneider, W.L.; Hartung, J.S.; Brlansky, R.H. First report of citrus leprosis virus nuclear type in sweet orange in Colombia. Plant Dis. 2014, 98, 1162. [Google Scholar] [CrossRef]
- Roy, A.; Stone, A.L.; Shao, J.; Otero-Colina, G.; Wei, G.; Choudhary, N.; Achor, D.; Levy, L.; Nakhla, M.K.; Hartung, J.S.; et al. Identification and molecular characterization of nuclear citrus leprosis virus, a member of the proposed Dichorhavirus genus infecting multiple Citrus species in Mexico. Phytopathology. 2015, 105, 564–575. [Google Scholar] [CrossRef]
- Roy, A.; Stone, A.L.; Otero-Colina, G.; Wei, G.; Brlansky, R.H.; Ochoa, R. Reassortment of genome segments creates stable lineages among strains of orchid fleck virus infecting citrus in Mexico. Phytopathology 2020, 110, 106–120. [Google Scholar] [CrossRef]
- Chabi-Jesus, C.; Ramos-González, P.L.; Tassi, A.D.; Guerra-Peraza, O.; Kitajima, E.W.; Harakava, R.; Beserra, J.E.A.; Salaroli, R.B.; Freitas-Astúa, J. Identification and Characterization of Citrus Chlorotic Spot Virus, a New Dichorhavirus Associated with Citrus Leprosis-like Symptoms. Plant Dis. 2018, 102, 1588–1598. [Google Scholar] [CrossRef] [PubMed]
- Chabi-Jesus, C.; Ramos-González, P.L.; Tassi, A.D.; Rossetto Pereira, L.; Bastianel, M.; Lau, D.; Canale, M.C.; Harakava, R.; Novelli, V.M.; Kitajima, E.W.; et al. Citrus Bright Spot Virus: A New Dichorhavirus, Transmitted by Brevipalpus azores, Causing Citrus Leprosis Disease in Brazil. Plants 2023, 12, 1371. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, E.W.; Chagas, C.M.; Braghini, M.T.; Fazuoli, L.C.; Locali-Fabris, E.C.; Salaroli, R.B. Natural Infection of Several Coffea Species and Hybrids and Psilanthus ebracteolatus by the Coffee Ringspot Virus (CoRSV). Sci. Agric. 2011, 68, 503–507. [Google Scholar] [CrossRef]
- Kubo, K.S.; Freitas-Astúa, J.; Machado, M.A.; Kitajima, E.W. Orchid Fleck Symptoms May Be Caused Naturally by Two Different Viruses Transmitted by Brevipalpus. J. Gen. Plant Pathol. 2009, 75, 250–255. [Google Scholar] [CrossRef]
- Kubo, K.S.; Novelli, V.M.; Bastianel, M.; Locali-Fabris, E.C.; Antonioli-Luizon, R.; Machado, M.A.; Freitas-Astúa, J. Detection of Brevipalpus-Transmitted Viruses in Their Mite Vectors by RT–PCR. Exp. Appl. Acarol. 2011, 54, 33–39. [Google Scholar] [CrossRef]
- Ramos-González, P.L.; Chabi-Jesus, C.; Guerra-Peraza, O.; Tassi, A.D.; Kitajima, E.W.; Harakava, R.; Salaroli, R.B.; Freitas-Astúa, J. Citrus Leprosis Virus N: A New Dichorhavirus Causing Citrus Leprosis Disease. Phytopathology 2017, 107, 963–976. [Google Scholar] [CrossRef]
- Stobbe, A.H.; Daniels, J.; Espindola, A.S.; Verma, R.; Melcher, U.; Ochoa-Corona, F.; Garzon, C.; Fletcher, J.; Schneider, W. E-probe Diagnostic Nucleic acid Analysis (EDNA): A theoretical approach for handling of next generation sequencing data for diagnostics. J. Microbiol. Methods 2013, 94, 356–366. [Google Scholar] [CrossRef]
- Espindola, A.S.; Cardwell, K.F. Microbe Finder (MiFi®): Implementation of an Interactive Pathogen Detection Tool in Metagenomic Sequence Data. Plants 2021, 10, 250. [Google Scholar] [CrossRef]
- Stobbe, A.H.; Schneider, W.L.; Hoyt, P.R.; Melcher, U. Screening metagenomic data for viruses using the e-probe diagnostic nucleic acid assay. Phytopathology 2014, 104, 1125–1129. [Google Scholar] [CrossRef]
- Dang, T.; Wang, H.; Espíndola, A.S.; Habiger, J.; Vidalakis, G.; Cardwell, K. Development and Statistical Validation of e-Probe Diagnostic Nucleic Acid Analysis (EDNA) Detection Assays for the Detection of Citrus Pathogens from Raw High Throughput Sequencing Data. PhytoFrontiers 2023, 3, 113–123. [Google Scholar] [CrossRef]
- Pasha, A.; Espindola, A.S.; Ziebell, H.; Ochoa-Corona, F.M. Highly Curated and Reliable E-Probes for Detection of Viral Pathogens in Unassembled HTS Datasets from Hops. PhytoFrontiers 2025. [CrossRef]
- Proaño-Cuenca, F.; Espindola, A.S.; Garzon, C.D. Detection of Phytophthora, Pythium, Globisporangium, Hyaloperonospora and Plasmopara species in High-Throughput Sequencing data by in silico and in vitro analysis using Microbe Finder (MiFi®). PhytoFrontiers 2023, 3, 124–136. [Google Scholar] [CrossRef]
- Espindola, A.; Schneider, W.; Hoyt, P.R.; Marek, S.M.; Garzon, C. A new approach for detecting fungal and oomycete plant pathogens in next generation sequencing metagenome data utilizing electronic probes. Int. J. Data Min. Bioinform 2015, 12, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Bocsanczy, A.M.; Espindola, A.S.; Cardwell, K.; Norman, D.J. Development and Validation of E-Probes with the MiFi System for Detection of Ralstonia solanacearum Species Complex in Blueberries. PhytoFrontiersTM 2023, 3, 137–147. [Google Scholar] [CrossRef]
- Narayanan, S.; Espindola, A.S.; Malayer, J.; Cardwell, K.; Ramachandran, A. Development and evaluation of Microbe Finder (MiFi)®: A novel in silico diagnostic platform for pathogen detection from metagenomic data. J. Med. Microbiol. 2023, 72, 001720. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef]
- Espindola, A.S.; Cardwell, K.; Martin, F.N.; Hoyt, P.R.; Marek, S.M.; Schneider, W.; Garzon, C.D. A step towards validation of high-throughput sequencing for the identification of plant pathogenic oomycetes. Phytopathology 2022, 112, 1859–1866. [Google Scholar] [CrossRef]
- Marçais, G.; Delcher, A.L.; Phillippy, A.M.; Coston, R.; Salzberg, S.L.; Zimin, A. MUMmer4: A fast and versatile genome alignment system. PLoS Comput Biol. 2018, 14, e1005944. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, C.; Rivera, Y.; Mavrodieva, V.A.; Roy, A. Development and Validation of TWO quadruplex TaqMan Real Time RT-PCR Assays for the Detection of the Citrus Leprosis Virus Disease Complex. Plant Health 2024, Organized by APS on July 27–30 at Memphis, Tennessee. 2024. Available online: https://www.apsnet.org/meetings/annual/meetingarchives/PH2024/Program/Pages/MemberPlayer.aspx?vid=1011140596 (accessed on 18 February 2025).
- Roy, A.; Padmanabhan, C.; Otero-Colina, G.; Rivera, Y.; Mavrodieva, V.A.; Nakhla, M.K. A multiplex real-time PCR assay for the universal detection of orchid fleck virus and differentiation among its four strains infecting multiple hosts (Abs). In Proceedings of the 12th International Conference of Plant Pathology (ICPP), Lyon, France, 2–25 August 2023; Abstract book; p. 230. [Google Scholar]
- Roy, A.; Grinstead, S.; Leon Martínez, G.; Pinzón, J.C.C.; Nunziata, S.O.; Padmanabhan, C.; Hammond, J. Meta-Transcriptomic Analysis Uncovers the Presence of Four Novel Viruses and Multiple Known Virus Genera in a Single Hibiscus rosa-sinensis Plant in Colombia. Viruses 2024, 16, 267. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, C.; Nunziata, S.; Leon, M.G.; Rivera, Y.; Mavrodieva, V.A.; Nakhla, M.K.; Roy, A. High-throughput sequencing application in the detection and discovery of viruses associated with the regulated citrus leprosis disease complex. Front. Plant Sci. 2023, 13, 1058847. [Google Scholar] [CrossRef]
- Fawcett, H.S. Scaly bark or nail-head rust of citrus. Fla. Agric. Exp. Stn. Bull. 1911, 106, 1–41. [Google Scholar]
- Hartung, J.S.; Roy, A.; Fu, S.; Shao, J.; Schneider, W.L.; Brlansky, R.H. History and diversity of citrus leprosis virus recorded in herbarium specimens. Phytopathology 2015, 105, 1277–1284. [Google Scholar] [CrossRef]
- Amarasinghe, G.K.; Bao, Y.; Basler, C.F.; Bavari, S.; Beer, M.; Bejerman, N.; Blasdell, K.R. Taxonomy of the order Mononegavirales: Update 2017. Arch. Virol. 2017, 162, 2493–2504. [Google Scholar] [CrossRef]
- Adams, I.P.; Glover, R.H.; Monger, W.A.; Mumford, R.; Jackeviciene, E.; Navalinskiene, M.; Samuitiene, M.; Boonham, N. Nextgeneration sequencing and metagenomic analysis: A universal diagnostic tool in plant virology. Mol. Plant Pathol. 2009, 10, 537–545. [Google Scholar] [CrossRef]
- Shendure, J.; Ji, H. Next-generation DNA sequencing. Nat. Biotechnol. 2008, 26, 1135–1145. [Google Scholar] [CrossRef]
- Espindola, A.S. Eukaryotic Plant Pathogen Detection Through High Throughput DNA/RNA Sequencing Data Analysis. Master’s Thesis, Oklahoma State University, Stillwater, OK, USA, 2016. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).