A Comprehensive Review of the Neglected and Emerging Oropouche Virus
Abstract
1. Oropouche Virus
2. OROV Replication in the Host Cell
3. Viral Transmission
4. Midge and Mosquito Vectors
5. Control Measures for Midges
6. Clinical Symptoms
7. Diagnostics of OROV Infection
8. Pathogenesis
9. Vaccine and Antiviral Development
10. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Sakkas, H.; Bozidis, P.; Franks, A.; Papadopoulou, C. Oropouche Fever: A Review. Viruses 2018, 10, 175. [Google Scholar] [CrossRef] [PubMed]
- Da Rosa, J.F.; De Souza, W.M.; de Paula Pinheiro, F.; Figueiredo, M.L.; Cardoso, J.F.; Acrani, G.O.; Nunes, M.R. Oropouche Virus: Clinical, Epidemiological, and Molecular Aspects of a Neglected Orthobunyavirus. Am. J. Trop. Med. Hyg. 2017, 96, 1019–1030. [Google Scholar] [CrossRef] [PubMed]
- WHO. Oropouche Virus Disease—Region of the Americas. 2024. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2024-DON530 (accessed on 17 February 2025).
- Deiana, M.; Malagò, S.; Mori, A.; Accordini, S.; Matucci, A.; Mantovani, R.P.; Gianesini, N.; Huits, R.; Piubelli, C.; Gobbi, F.G.; et al. Full Genome Characterization of the First Oropouche Virus Isolate Imported in Europe from Cuba. Viruses 2024, 16, 1586. [Google Scholar] [CrossRef]
- CDC. CDC Oropouche Current Year Data 2025. Available online: https://www.cdc.gov/oropouche/data-maps/current-year-data.html (accessed on 17 February 2025).
- Morrison, A.; White, J.L.; Hughes, H.R.; Guagliardo, S.A.J.; Velez, J.O.; Fitzpatrick, K.A.; Davis, E.H.; Stanek, D.; Kopp, E.; Dumoulin, P.; et al. Oropouche Virus Disease Among U.S. Travelers—United States. Morb. Mortal. Wkly. Rep. 2024, 73, 769–773. [Google Scholar] [CrossRef]
- Files, M.A.; Hansen, C.A.; Herrera, V.C.; Schindewolf, C.; Barrett, A.D.T.; Beasley, D.W.C.; Bourne, N.; Milligan, G.N. Baseline mapping of Oropouche virology, epidemiology, therapeutics, and vaccine research and development. NPJ Vaccines 2022, 7, 38. [Google Scholar] [CrossRef] [PubMed]
- Nunes, M.R.T.; Martins, L.C.; Rodrigues, S.G.; Chiang, J.O.; Azevedo, R.D.S.d.S.; da Rosa, A.P.T.; Vasconcelos, P.F.d.C. Oropouche virus isolation, southeast Brazil. Emerg. Infect. Dis. 2005, 11, 1610–1613. [Google Scholar] [CrossRef]
- Tilston-Lunel, N.L.; Hughes, J.; Acrani, G.O.; da Silva, D.E.A.; Azevedo, R.S.S.; Rodrigues, S.G.; Vasconcelos, P.F.C.; Nunes, M.R.T.; Elliott, R.M. Genetic analysis of members of the species Oropouche virus and identification of a novel M segment sequence. J. Gen. Virol. 2015, 96 Pt 7, 1636–1650. [Google Scholar] [CrossRef]
- Acrani, G.O.; Tilston-Lunel, N.L.; Spiegel, M.; Weidmann, M.; Dilcher, M.; da Silva, D.E.A.; Nunes, M.R.T.; Elliott, R.M. Establishment of a minigenome system for Oropouche virus reveals the S genome segment to be significantly longer than reported previously. J. Gen. Virol. 2015, 96 Pt 3, 513–523. [Google Scholar] [CrossRef]
- Tilston-Lunel, N.L.; Acrani, G.O.; Randall, R.E.; Elliott, R.M. Generation of Recombinant Oropouche Viruses Lacking the Nonstructural Protein NSm or NSs. J. Virol. 2016, 90, 2616–2627. [Google Scholar] [CrossRef]
- Bowden, T.A.; Bitto, D.; McLees, A.; Yeromonahos, C.; Elliott, R.M.; Huiskonen, J.T. Orthobunyavirus ultrastructure and the curious tripodal glycoprotein spike. PLoS Pathog. 2013, 9, e1003374. [Google Scholar] [CrossRef]
- Hellert, J.; Aebischer, A.; Wernike, K.; Haouz, A.; Brocchi, E.; Reiche, S.; Guardado-Calvo, P.; Beer, M.; Rey, F.A. Orthobunyavirus spike architecture and recognition by neutralizing antibodies. Nat. Commun. 2019, 10, 879. [Google Scholar] [CrossRef]
- Obijeski, J.F.; Bishop, D.H.; A Murphy, F.; Palmer, E.L. Structural proteins of La Crosse virus. J. Virol. 1976, 19, 985–997. [Google Scholar] [CrossRef] [PubMed]
- Aquino, V.H.; Moreli, M.L.; Figueiredo, L.T.M. Analysis of oropouche virus L protein amino acid sequence showed the presence of an additional conserved region that could harbour an important role for the polymerase activity. Arch. Virol. 2003, 148, 19–28. [Google Scholar] [CrossRef]
- Elliott, R.M. Molecular biology of the Bunyaviridae. J. Gen. Virol. 1990, 71 Pt 3, 501–522. [Google Scholar] [CrossRef] [PubMed]
- Murillo, J.L.; Cabral, A.D.; Uehara, M.; da Silva, V.M.; dos Santos, J.V.; Muniz, J.R.C.; Estrozi, L.F.; Fenel, D.; Garcia, W.; Sperança, M.A. Nucleoprotein from the unique human infecting Orthobunyavirus of Simbu serogroup (Oropouche virus) forms higher order oligomers in complex with nucleic acids in vitro. Amino Acids 2018, 50, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Elliott, R.M. Orthobunyaviruses: Recent genetic and structural insights. Nat. Rev. Microbiol. 2014, 12, 673–685. [Google Scholar] [CrossRef]
- Naveca, F.G.; de Almeida, T.A.P.; Souza, V.; Nascimento, V.; Silva, D.; Nascimento, F.; Mejía, M.; de Oliveira, Y.S.; Rocha, L.; Xavier, N.; et al. Human outbreaks of a novel reassortant Oropouche virus in the Brazilian Amazon region. Nat. Med. 2024, 30, 3509–3521. [Google Scholar] [CrossRef]
- Schwartz, D.A. Novel Reassortants of Oropouche Virus (OROV) Are Causing Maternal-Fetal Infection During Pregnancy, Stillbirth, Congenital Microcephaly and Malformation Syndromes. Genes 2025, 16, 87. [Google Scholar] [CrossRef]
- Santos, R.I.; Rodrigues, A.H.; Silva, M.L.; Mortara, R.A.; Rossi, M.A.; Jamur, M.C.; Oliver, C.; Arruda, E. Oropouche virus entry into HeLa cells involves clathrin and requires endosomal acidification. Virus Res. 2008, 138, 139–143. [Google Scholar] [CrossRef]
- Schwarz, M.M.; Price, D.A.; Ganaie, S.S.; Feng, A.; Mishra, N.; Hoehl, R.M.; Fatma, F.; Stubbs, S.H.; Whelan, S.P.J.; Cui, X.; et al. Oropouche orthobunyavirus infection is mediated by the cellular host factor Lrp1. Proc. Natl. Acad. Sci. USA 2022, 119, e2204706119. [Google Scholar] [CrossRef]
- Ganaie, S.S.; Schwarz, M.M.; McMillen, C.M.; Price, D.A.; Feng, A.X.; Albe, J.R.; Wang, W.; Miersch, S.; Orvedahl, A.; Cole, A.R.; et al. Lrp1 is a host entry factor for Rift Valley fever virus. Cell 2021, 184, 5163–5178 e24. [Google Scholar] [CrossRef] [PubMed]
- Olschewski, S.; Cusack, S.; Rosenthal, M. The Cap-Snatching Mechanism of Bunyaviruses. Trends Microbiol. 2020, 28, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Amroun, A.; Priet, S.; de Lamballerie, X.; Quérat, G. Bunyaviridae RdRps: Structure, motifs, and RNA synthesis machinery. Crit. Rev. Microbiol. 2017, 43, 753–778. [Google Scholar] [CrossRef]
- Barbosa, N.S.; Concha, J.O.; Dasilva, L.L.P.; Crump, C.M.; Graham, S.C. Oropouche Virus Glycoprotein Topology and Cellular Requirements for Glycoprotein Secretion. J. Virol. 2023, 97, e0133122. [Google Scholar] [CrossRef]
- Barbosa, N.S.; Mendonça, L.R.; Dias, M.V.S.; Pontelli, M.C.; da Silva, E.Z.M.; Criado, M.F.; da Silva-Januário, M.E.; Schindler, M.; Jamur, M.C.; Oliver, C.; et al. ESCRT machinery components are required for Orthobunyavirus particle production in Golgi compartments. PLoS Pathog. 2018, 14, e1007047. [Google Scholar] [CrossRef]
- Concha, J.O.; Gutierrez, K.; Barbosa, N.; Rodrigues, R.L.; de Carvalho, A.N.; Tavares, L.A.; Rudd, J.S.; Costa, C.S.; Andrade, B.Y.G.; Espreafico, E.M.; et al. Rab27a GTPase and its effector Myosin Va are host factors required for efficient Oropouche virus cell egress. PLoS Pathog. 2024, 20, e1012504. [Google Scholar] [CrossRef]
- Sciancalepore, S.; Schneider, M.C.; Kim, J.; Galan, D.I.; Riviere-Cinnamond, A. Presence and multi-species spatial distribution of oropouche virus in Brazil within the one health framework. Trop. Med. Infect. Dis. 2022, 7, 111. [Google Scholar] [CrossRef] [PubMed]
- CDC. Clinical Overview of Oropouche Virus Disease. 2024. Available online: https://www.cdc.gov/oropouche/hcp/clinical-overview/index.html (accessed on 28 January 2025).
- Cardoso, B.F.; Serra, O.P.; Heinen, L.B.D.S.; Zuchi, N.; de Souza, V.C.; Naveca, F.G.; Dos Santos, M.A.M.; Slhessarenko, R.D. Detection of Oropouche virus segment S in patients and inCulex quinquefasciatus in the state of Mato Grosso, Brazil. Memórias Inst. Oswaldo Cruz 2015, 110, 745–754. [Google Scholar] [CrossRef]
- Pinheiro, F.P.; da Rosa, A.T.; Vasconcelos, P.F.D.C. Oropouche fever. In Textbook of Pediatric Infectious Diseases; CABI Digital Library: Wallingford, CT, USA, 2004; pp. 2418–2423. [Google Scholar]
- Almeida, G.M.; Souza, J.P.; Mendes, N.D.; Pontelli, M.C.; Pinheiro, N.R.; Nogueira, G.O.; Cardoso, R.S.; Paiva, I.M.; Ferrari, G.D.; Veras, F.P.; et al. Neural Infection by Oropouche Virus in Adult Human Brain Slices Induces an Inflammatory and Toxic Response. Front. Neurosci. 2021, 15, 674576. [Google Scholar] [CrossRef]
- Martins, F.E.d.N.; Chiang, J.O.; Nunes, B.T.D.; Ribeiro, B.d.F.R.; Martins, L.C.; Casseb, L.M.N.; Henriques, D.F.; de Oliveira, C.S.; Maciel, E.L.N.; Cravo, L.d.C.C.; et al. Newborns with microcephaly in Brazil and potential vertical transmission of Oropouche virus: A case series. Lancet Infect. Dis. 2025, 25, 155–165. [Google Scholar] [CrossRef]
- Dashraath, P.; Nielsen-Saines, K.; Schwartz, D.A.; Musso, D.; Baud, D. Vertical transmission potential of Oropouche virus infection in human pregnancies. AJOG Glob. Rep. 2025, 5, 100431. [Google Scholar] [CrossRef]
- O’connor, T.W.; Hick, P.M.; Finlaison, D.S.; Kirkland, P.D.; Toribio, J.-A.L. Revisiting the Importance of Orthobunyaviruses for Animal Health: A Scoping Review of Livestock Disease, Diagnostic Tests, and Surveillance Strategies for the Simbu Serogroup. Viruses 2024, 16, 294. [Google Scholar] [CrossRef] [PubMed]
- Castilletti, C.; Huits, R.; Mantovani, R.P.; Accordini, S.; Alladio, F.; Gobbi, F. Replication-Competent Oropouche Virus in Semen of Traveler Returning to Italy from Cuba, 2024. Emerg. Infect. Dis. 2024, 30, 2684–2686. [Google Scholar] [CrossRef] [PubMed]
- Counotte, M.J.; Kim, C.R.; Wang, J.; Bernstein, K.; Deal, C.D.; Broutet, N.J.N.; Low, N. Sexual transmission of Zika virus and other flaviviruses: A living systematic review. PLoS Med. 2018, 15, e1002611. [Google Scholar] [CrossRef]
- Sherley, M.; Ong, C.-W. Sexual transmission of Zika virus: A literature review. Sex Health 2018, 15, 183–199. [Google Scholar] [CrossRef]
- Yee, D.A.; Bermond, C.D.; Reyes-Torres, L.J.; Fijman, N.S.; Scavo, N.A.; Nelsen, J.; Yee, S.H. Robust network stability of mosquitoes and human pathogens of medical importance. Parasit Vectors 2022, 15, 216. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.R.; Spence, L.; Downs, W.G.; Aitken, T.H.G. Oropouche virus: A new human disease agent from Trinidad, West Indies. Am. J. Trop. Med. Hyg. 1961, 10, 574–578. [Google Scholar] [CrossRef]
- McGregor, B.L.; Connelly, C.R.; Kenney, J.L. Infection, Dissemination, and Transmission Potential of North American Culex quinquefasciatus, Culex tarsalis, and Culicoides sonorensis for Oropouche Virus. Viruses 2021, 13, 226. [Google Scholar] [CrossRef]
- Pinheiro, F.P.; Da Rosa, A.P.T.; Da Rosa, J.F.T.; Ishak, R.; Freitas, R.B.; Gomes, M.L.; LeDuc, J.W.; Oliva, O.F. Oropouche virus. I. A review of clinical, epidemiological, and ecological findings. Am. J. Trop. Med. Hyg. 1981, 30, 149–160. [Google Scholar] [CrossRef]
- Smith, G.C.; Francy, D.B. Laboratory studies of a Brazilian strain of Aedes albopictus as a potential vector of Mayaro and Oropouche viruses. J. Am. Mosq. Control Assoc. 1991, 7, 89–93. [Google Scholar]
- Roberts, D.R.; Pinheiro, F.D.; Hoch, A.L.; LeDuc, J.W.; Peterson, N.E.; Santos, M.V.; Western, K.A. Vectors and Natural Reservoirs of Oropouche Virus in the Amazon Region; US Army Medical Research and Development Command: Washington, DC, USA, 1977. [Google Scholar]
- Pinheiro, F.P.; Hoch, A.L.; Gomes, M.D.; Roberts, O.R. Oropouche virus. IV. Laboratory transmission by Culicoides paraensis. Am. J. Trop. Med. Hyg. 1981, 30, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, F.P.; da Rosa, A.P.T.; Gomes, M.L.; LeDuc, J.W.; Hoch, A.L. Transmission of Oropouche virus from man to hamster by the midge Culicoides paraensis. Science 1982, 215, 1251–1253. [Google Scholar] [CrossRef] [PubMed]
- Vasconcelos, P.F.; Travassos Da Rosa, J.F.; Guerreiro, S.C.; Dégallier, N.; Travassos Da Rosa, E.S.; Travassos Da Rosa, A.P. 1st register of an epidemic caused by Oropouche virus in the states of Maranhao and Goias, Brazil. Rev. Inst. Med. Trop. Sao Paulo 1989, 31, 271–278. [Google Scholar] [CrossRef]
- Bandeira, A.C.; da Silva, A.C.; Souza, M.; da Costa Saavedra, R.; Pereira, F.M.; de Oliveira Santos, S.P.; Leal, A.; de Mello, S.; da Purificação, S.M.; de Souza, D.R.; et al. Clinical profile of Oropouche fever in Bahia, Brazil: Unexpected fatal cases. SciELO Prepr. 2024. [Google Scholar]
- Thorp, J.H.; Rogers, D.C. Family Ceratopogonidae. In Thorp and Covich’s Freshwater Invertebrates; Elsevier: Amsterdam, The Netherlands, 2018; pp. 625–659. [Google Scholar]
- Purse, B.V.; Carpenter, S.; Venter, G.J.; Bellis, G.; Mullens, B.A. Bionomics of temperate and tropical Culicoides midges: Knowledge gaps and consequences for transmission of Culicoides-borne viruses. Annu. Rev. Entomol. 2015, 60, 373–392. [Google Scholar] [CrossRef]
- Sick, F.; Beer, M.; Kampen, H.; Wernike, K. Culicoides Biting Midges-Underestimated Vectors for Arboviruses of Public Health and Veterinary Importance. Viruses 2019, 11, 376. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, X.; Wu, Z.; Feng, S.; Lu, K.; Zhu, W.; Sun, H.; Niu, G. Oropouche virus: A neglected global arboviral threat. Virus Res. 2024, 341, 199318. [Google Scholar] [CrossRef]
- Mellor, P.S.; Boorman, J.; Baylis, M. Culicoides biting midges: Their role as arbovirus vectors. Ann. Rev. Entomol. 2000, 45, 307–340. [Google Scholar] [CrossRef]
- Feitoza, L.H.M.; de Carvalho, L.P.C.; da Silva, L.R.; Meireles, A.C.A.; Rios, F.G.F.; Silva, G.S.; de Paulo, P.F.M.; Pessoa, F.A.C.; de Medeiros, J.F.; Julião, G.R. Influence of meteorological and seasonal parameters on the activity of Culicoides paraensis (Diptera: Ceratopogonidae), an annoying anthropophilic biting midge and putative vector of Oropouche Virus in Rondonia, Brazilian Amazon. Acta Trop. 2023, 243, 106928. [Google Scholar] [CrossRef]
- Hoch, A.; Roberts, D.R.; Pinheiro, F. Host-seeking behavior and seasonal abundance of Culicoides paraensis (Diptera: Ceratopogonidae) in Brazil. J. Am. Mosq. Control Assoc. 1990, 6, 110–114. [Google Scholar]
- Benelli, G.; Buttazzoni, L.; Canale, A.; D’Andrea, A.; Del Serrone, P.; Delrio, G.; Foxi, C.; Mariani, S.; Savini, G.; Vadivalagan, C.; et al. Bluetongue outbreaks: Looking for effective control strategies against Culicoides vectors. Res. Vet. Sci. 2017, 115, 263–270. [Google Scholar] [CrossRef]
- Carpenter, S.; Groschup, M.H.; Garros, C.; Felippe-Bauer, M.L.; Purse, B.V. Culicoides biting midges, arboviruses and public health in Europe. Antivir. Res. 2013, 100, 102–113. [Google Scholar] [CrossRef]
- Humblet, M.-F.; Losson, B.; Saegerman, C. Integrated management of blood-feeding arthropods in veterinary teaching facilities —Part 2: Overview of control methods against adults and immature stages. Rev. Sci. Tech. 2020, 39, 757–777. [Google Scholar] [CrossRef]
- Barrera, E.L.P.; Reales-González, J.; Salas, D.; Santamaría, E.R.; Bello, S.; Rico, A.; Pardo, L.; Parra, E.; Rodriguez, K.; Alarcon, Z.; et al. Fatal acute undifferentiated febrile illness among clinically suspected leptospirosis cases in Colombia, 2016–2019. PLoS Negl. Trop. Dis. 2023, 17, e0011683. [Google Scholar] [CrossRef] [PubMed]
- Culquichicón, C.; Cardona-Ospina, J.A.; Patiño-Barbosa, A.M.; Rodriguez-Morales, A.J. Bibliometric analysis of Oropouche research: Impact on the surveillance of emerging arboviruses in Latin America. F1000Research 2017, 6, 194. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, A.H.; Santos, R.I.; Arisi, G.M.; Bernardes, E.S.; Silva, M.L.; Rossi, M.A.; Lopes, M.B.S.; Arruda, E. Oropouche virus experimental infection in the golden hamster (Mesocrisetus auratus). Virus Res. 2011, 155, 35–41. [Google Scholar] [CrossRef]
- Martins-Luna, J.; del Valle-Mendoza, J.; Silva-Caso, W.; Sandoval, I.; del Valle, L.J.; Palomares-Reyes, C.; Carrillo-Ng, H.; Peña-Tuesta, I.; Aguilar-Luis, M.A. Oropouche infection a neglected arbovirus in patients with acute febrile illness from the Peruvian coast. BMC Res. Notes 2020, 13, 67. [Google Scholar] [CrossRef] [PubMed]
- Silva-Caso, W.; Aguilar-Luis, M.A.; Palomares-Reyes, C.; Mazulis, F.; Weilg, C.; del Valle, L.J.; Espejo-Evaristo, J.; Soto-Febres, F.; Martins-Luna, J.; del Valle-Mendoza, J. First outbreak of Oropouche Fever reported in a non-endemic western region of the Peruvian Amazon: Molecular diagnosis and clinical characteristics. Int. J. Infect. Dis. 2019, 83, 139–144. [Google Scholar] [CrossRef]
- Diseases, V.I. Oropouche Virus and Oropouche Fever. 2019. Available online: https://vivo.colostate.edu/hbooks/infectious/viruses/bunya/oropouche.html (accessed on 17 February 2025).
- Chiang, J.O.; Azevedo, R.S.; Justino, M.C.; Matos, H.J.; Cabeça, H.L.; Silva, S.P.; Henriques, D.F.; Silva, E.V.; Andrade, G.S.; Vasconcelos, P.F.; et al. Neurological disease caused by Oropouche virus in northern Brazil: Should it be included in the scope of clinical neurological diseases? J. Neurovirol. 2021, 27, 626–630. [Google Scholar] [CrossRef]
- Gaillet, M.; Pichard, C.; Restrepo, J.; Lavergne, A.; Perez, L.; Enfissi, A.; Abboud, P.; Lambert, Y.; Ma, L.; Monot, M.; et al. Outbreak of Oropouche Virus in French Guiana. Emerg. Infect. Dis. 2021, 27, 2711–2714. [Google Scholar] [CrossRef]
- Pavon, J.A.R.; Neves, N.A.d.S.; Silva, L.C.F.; de Azevedo, F.K.; Junior, J.A.B.d.F.; Nunes, M.R.T.; Slhessarenko, R.D. Neurological infection by chikungunya and a triple Arbovirus co-infection in Mato Grosso, Central Western Brazil during 2019. J. Clin. Virol. 2022, 146, 105056. [Google Scholar] [CrossRef]
- Vernal, S.; Martini, C.C.R.; da Fonseca, B.A.L. Oropouche Virus-Associated Aseptic Meningoencephalitis, Southeastern Brazil. Emerg. Infect Dis. 2019, 25, 380–382. [Google Scholar] [CrossRef] [PubMed]
- de Souza Bastos, M.; Figueiredo, L.T.; Naveca, F.G.; Monte, R.L.; Lessa, N.; de Figueiredo, R.M.; de Lima Gimaque, J.B.; Joao, G.P.; Ramasawmy, R.; Mourao, M.P. Identification of Oropouche Orthobunyavirus in the cerebrospinal fluid of three patients in the Amazonas, Brazil. Am. J. Trop. Med. Hyg. 2012, 86, 732–735. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, D.A.; Dashraath, P.; Baud, D. Oropouche Virus (OROV) in Pregnancy: An Emerging Cause of Placental and Fetal Infection Associated with Stillbirth and Microcephaly following Vertical Transmission. Viruses 2024, 16, 1435. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, E.; Azevedo, R.D.; Coelho-dos-Reis, J.G.; Antonelli, L.R.; Ferreira, M.S.; Campi-Azevedo, A.C.; Costa-Silva, M.F.; Martins, L.C.; Chiang, J.O.; Teixeira-Carvalho, A.; et al. IFN-α as a time-sensitive biomarker during Oropouche virus infection in early and late seroconverters. Sci. Rep. 2019, 9, 17924. [Google Scholar] [CrossRef]
- Nascimento, V.A.D.; Santos, J.H.A.; Monteiro, D.C.D.S.; Pessoa, K.P.; Cardoso, A.J.L.; de Souza, V.C.; Abdalla, L.F.; Naveca, F.G. Oropouche virus detection in saliva and urine. Memórias Inst. Oswaldo Cruz 2020, 115, e190338. [Google Scholar] [CrossRef]
- Weidmann, M.; Rudaz, V.; Nunes, M.R.T.; Vasconcelos, P.F.C.; Hufert, F.T. Rapid detection of human pathogenic orthobunyaviruses. J. Clin. Microbiol. 2003, 41, 3299–3305. [Google Scholar] [CrossRef]
- Casseb, A.D.; Nunes, M.R.; Rodrigues, S.G.; Travassos da Rosa, E.S.; Casseb, L.M.; Casseb, S.M.; da Silva, S.P.; Rodrigues, É.D.; Vasconcelos, P.F. Diagnosis of arboviruses using indirect sandwich IgG ELISA in horses from the Brazilian Amazon. J. Venom. Anim. Toxins Incl. Trop. Dis. 2014, 20, 29. [Google Scholar] [CrossRef]
- Reiber, H.; Lange, P. Quantification of virus-specific antibodies in cerebrospinal fluid and serum: Sensitive and specific detection of antibody synthesis in brain. Clin. Chem. 1991, 37, 1153–1160. [Google Scholar] [CrossRef]
- Tilston-Lunel, N.L. Oropouche Virus: An Emerging Orthobunyavirus. J. Gen. Virol. 2024, 105, 002027. [Google Scholar] [CrossRef]
- Batista, P.M.; Andreotti, R.; de Almeida, P.S.; Marques, A.C.; Rodrigues, S.G.; Chiang, J.O.; Vasconcelos, P.F.d.C. Detection of arboviruses of public health interest in free-living New World primates (Sapajus spp.; Alouatta caraya) captured in Mato Grosso do Sul, Brazil. Rev. Soc. Bras. Med. Trop. 2013, 46, 684–690. [Google Scholar] [CrossRef]
- Luna, L.K.D.S.; Rodrigues, A.H.; Santos, R.I.M.; Sesti-Costa, R.; Criado, M.F.; Martins, R.B.; Silva, M.L.; Delcaro, L.S.; Proenca-Modena, J.L.; Figueiredo, L.T.M.; et al. Oropouche virus is detected in peripheral blood leukocytes from patients. J. Med. Virol. 2017, 89, 1108–1111. [Google Scholar] [CrossRef] [PubMed]
- Wise, E.L.; Márquez, S.; Mellors, J.; Paz, V.; Atkinson, B.; Gutierrez, B.; Zapata, S.; Coloma, J.; Pybus, O.G.; Jackson, S.K.; et al. Oropouche virus cases identified in Ecuador using an optimised qRT-PCR informed by metagenomic sequencing. PLoS Negl. Trop. Dis. 2020, 14, e0007897. [Google Scholar] [CrossRef]
- Santos, R.I.; Almeida, M.F.; Paula, F.E.; Rodrigues, A.H.; Saranzo, A.M.; Paula, A.E.; Silva, M.L.; Correa, V.M.A.; Acrani, G.O.; Neder, L.; et al. Experimental infection of suckling mice by subcutaneous inoculation with Oropouche virus. Virus Res. 2012, 170, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Proenca-Modena, J.L.; Sesti-Costa, R.; Pinto, A.K.; Richner, J.M.; Lazear, H.M.; Lucas, T.; Hyde, J.L.; Diamond, M.S. Oropouche virus infection and pathogenesis are restricted by MAVS, IRF-3, IRF-7, and type I interferon signaling pathways in nonmyeloid cells. J. Virol. 2015, 89, 4720–4737. [Google Scholar] [CrossRef] [PubMed]
- Proenca-Modena, J.L.; Hyde, J.L.; Sesti-Costa, R.; Lucas, T.; Pinto, A.K.; Richner, J.M.; Gorman, M.J.; Lazear, H.M.; Diamond, M.S. Interferon-Regulatory Factor 5-Dependent Signaling Restricts Orthobunyavirus Dissemination to the Central Nervous System. J. Virol. 2016, 90, 189–205. [Google Scholar] [CrossRef]
- Ribeiro Amorim, M.; Cornejo Pontelli, M.; Fabiano de Souza, G.; Primon Muraro, S.; Toledo-Teixeira, D.A.; Forato, J.; Bispo-dos-Santos, K.; Barbosa, N.S.; Cavalheiro Martini, M.; Lorencini Parise, P.; et al. Oropouche Virus Infects, Persists and Induces IFN Response in Human Peripheral Blood Mononuclear Cells as Identified by RNA PrimeFlow and qRT-PCR Assays. Viruses 2020, 12, 785. [Google Scholar] [CrossRef]
- Pinto, A.K.; Williams, G.D.; Szretter, K.J.; White, J.P.; Proença-Módena, J.L.; Liu, G.; Olejnik, J.; Brien, J.D.; Ebihara, H.; Mühlberger, E.; et al. Human and Murine IFIT1 Proteins Do Not Restrict Infection of Negative-Sense RNA Viruses of the Orthomyxoviridae, Bunyaviridae, and Filoviridae Families. J. Virol. 2015, 89, 9465–9476. [Google Scholar] [CrossRef]
- Kang, S.; Brown, H.M.; Hwang, S. Direct Antiviral Mechanisms of Interferon-Gamma. Immune Netw. 2018, 18, e33. [Google Scholar] [CrossRef]
- Santos, R.I.; Bueno-Júnior, L.S.; Ruggiero, R.N.; Almeida, M.F.; Silva, M.L.; Paula, F.E.; Correa, V.M.A.; Arruda, E. Spread of Oropouche virus into the central nervous system in mouse. Viruses 2014, 6, 3827–3836. [Google Scholar] [CrossRef]
- Mlakar, J.; Korva, M.; Tul, N.; Popović, M.; Poljšak-Prijatelj, M.; Mraz, J.; Kolenc, M.; Resman Rus, K.; Vesnaver Vipotnik, T.; Fabjan Vodušek, V.; et al. Zika Virus Associated with Microcephaly. N. Engl. J. Med. 2016, 374, 951–958. [Google Scholar] [CrossRef]
- Paul, A.M.; Acharya, D.; Neupane, B.; Thompson, E.A.; Gonzalez-Fernandez, G.; Copeland, K.M.; Garrett, M.; Liu, H.; Lopez, M.E.; de Cruz, M.; et al. Congenital Zika Virus Infection in Immunocompetent Mice Causes Postnatal Growth Impediment and Neurobehavioral Deficits. Front. Microbiol. 2018, 9, 2028. [Google Scholar] [CrossRef] [PubMed]
- Cola, J.P.; dos Santos, A.P.B.; Zanotti, R.L.; Costa, A.E.d.S.D.; Del Carro, K.B.; Coelho, L.d.A.L.; Miranda, A.E.; Vicente, C.R. Maternal and Fetal Implications of Oropouche Fever, Espírito Santo State, Brazil, 2024. Emerg. Infect. Dis. 2025, 31. [Google Scholar] [CrossRef] [PubMed]
- Ribas Freitas, A.R.; Schwartz, D.A.; Lima Neto, A.S.; Rodrigues, R.; Cavalcanti, L.P.G.; Alarcón-Elbal, P.M. Oropouche Virus (OROV): Expanding Threats, Shifting Patterns, and the Urgent Need for Collaborative Research in Latin America. Viruses 2025, 17, 353. [Google Scholar] [CrossRef]
- Schwartz, D.A.; Baud, D.; Dashraath, P. A potential mechanism of transplacental transmission of Oropouche virus in pregnancy. Lancet Microbe 2025. [Google Scholar] [CrossRef]
- Felix, A.; Hallet, E.; Favre, A.; Kom-Tchameni, R.; Defo, A.; Fléchelles, O.; Rosenthal, J.-M.; Douine, M.; Nacher, M.; Elenga, N. Cerebral injuries associated with Zika virus in utero exposure in children without birth defects in French Guiana. Medicine 2017, 96, e9178. [Google Scholar] [CrossRef]
- Aragao, M.; Holanda, A.; Brainer-Lima, A.; Petribu, N.; Castillo, M.; van der Linden, V.; Serpa, S.; Tenório, A.; Travassos, P.; Cordeiro, M.; et al. Nonmicrocephalic Infants with Congenital Zika Syndrome Suspected Only after Neuroimaging Evaluation Compared with Those with Microcephaly at Birth and Postnatally: How Large Is the Zika Virus “Iceberg”? Am. J. Neuroradiol. 2017, 38, 1427–1434. [Google Scholar] [CrossRef]
- Stubbs, S.H.; Pontelli, M.C.; Mishra, N.; Zhou, C.; Souza, J.d.P.; Viana, R.M.M.; Lipkin, W.I.; Knipe, D.M.; Arruda, E.; Whelan, S.P.J. Vesicular Stomatitis Virus Chimeras Expressing the Oropouche Virus Glycoproteins Elicit Protective Immune Responses in Mice. mBio 2021, 12, e0046321. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, U.K.; Tayebi, M.; Rahman, M.M. Immunoinformatics Approach for Epitope-Based Peptide Vaccine Design and Active Site Prediction against Polyprotein of Emerging Oropouche Virus. J. Immunol. Res. 2018, 2018, 6718083. [Google Scholar] [CrossRef]
- Afonso, A.; Conraths, F. Schmallenberg virus. Prev. Vet. Med. 2014, 116, 337–338. [Google Scholar] [CrossRef]
- Kim, Y.-H.; Kweon, C.-H.; Tark, D.-S.; Lim, S.I.; Yang, D.-K.; Hyun, B.-H.; Song, J.-Y.; Hur, W.; Park, S.C. Development of inactivated trivalent vaccine for the teratogenic Aino, Akabane and Chuzan viruses. Biologicals 2011, 39, 152–157. [Google Scholar] [CrossRef]
- Association, B.V. VMD authorises SBV vaccine for use in the UK. Vet Rec. 2013, 172, 543. [Google Scholar]
- Ogawa, Y.; Eguchi, M.; Shimoji, Y. Two Akabane virus glycoprotein Gc domains induce neutralizing antibodies in mice. J. Vet. Med. Sci. 2022, 84, 538–542. [Google Scholar] [CrossRef] [PubMed]
- Kraatz, F.; Wernike, K.; Hechinger, S.; König, P.; Granzow, H.; Reimann, I.; Beer, M. Deletion mutants of Schmallenberg virus are avirulent and protect from virus challenge. J. Virol. 2015, 89, 1825–1837. [Google Scholar] [CrossRef]
- Geddes, V.E.V.; de Oliveira, A.S.; Tanuri, A.; Arruda, E.; Ribeiro-Alves, M.; Aguiar, R.S. MicroRNA and cellular targets profiling reveal miR-217 and miR-576-3p as proviral factors during Oropouche infection. PLoS Negl. Trop. Dis. 2018, 12, e0006508. [Google Scholar] [CrossRef] [PubMed]
- Saivish, M.V.; Menezes, G.d.L.; da Silva, R.A.; de Assis, L.R.; Teixeira, I.d.S.; Fulco, U.L.; Avilla, C.M.S.; Eberle, R.J.; Santos, I.d.A.; Korostov, K.; et al. Acridones as promising drug candidates against Oropouche virus. Curr. Res. Microb. Sci. 2024, 6, 100217. [Google Scholar] [CrossRef] [PubMed]
- Mandova, T.; Saivish, M.V.; Menezes, G.d.L.; Bezerra, K.S.; Fulco, U.L.; da Silva, R.A.; Da Costa, F.B.; Nogueira, M.L. Antiviral Activity and Molecular Dynamics Simulation of Hops Compounds against Oropouche Virus (Peribunyaviridae). Pharmaceutics 2023, 15, 2769. [Google Scholar] [CrossRef]
- Peinado, R.d.S.; Saivish, M.V.; Menezes, G.d.L.; Fulco, U.L.; da Silva, R.A.; Korostov, K.; Eberle, R.J.; Melo, P.A.; Nogueira, M.L.; Pacca, C.C.; et al. The search for an antiviral lead molecule to combat the neglected emerging Oropouche virus. Curr. Res. Microb. Sci. 2024, 6, 100238. [Google Scholar] [CrossRef]
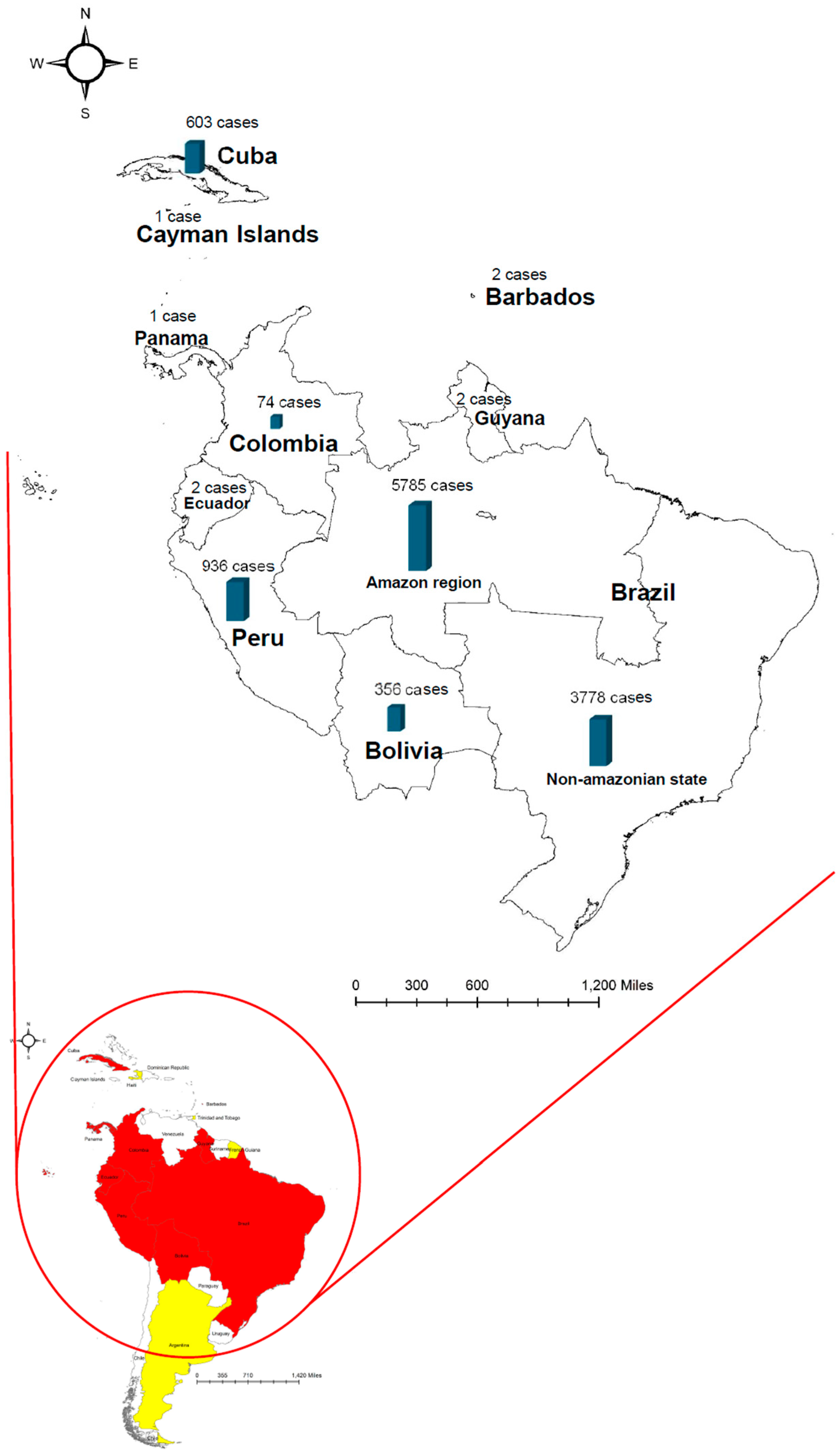
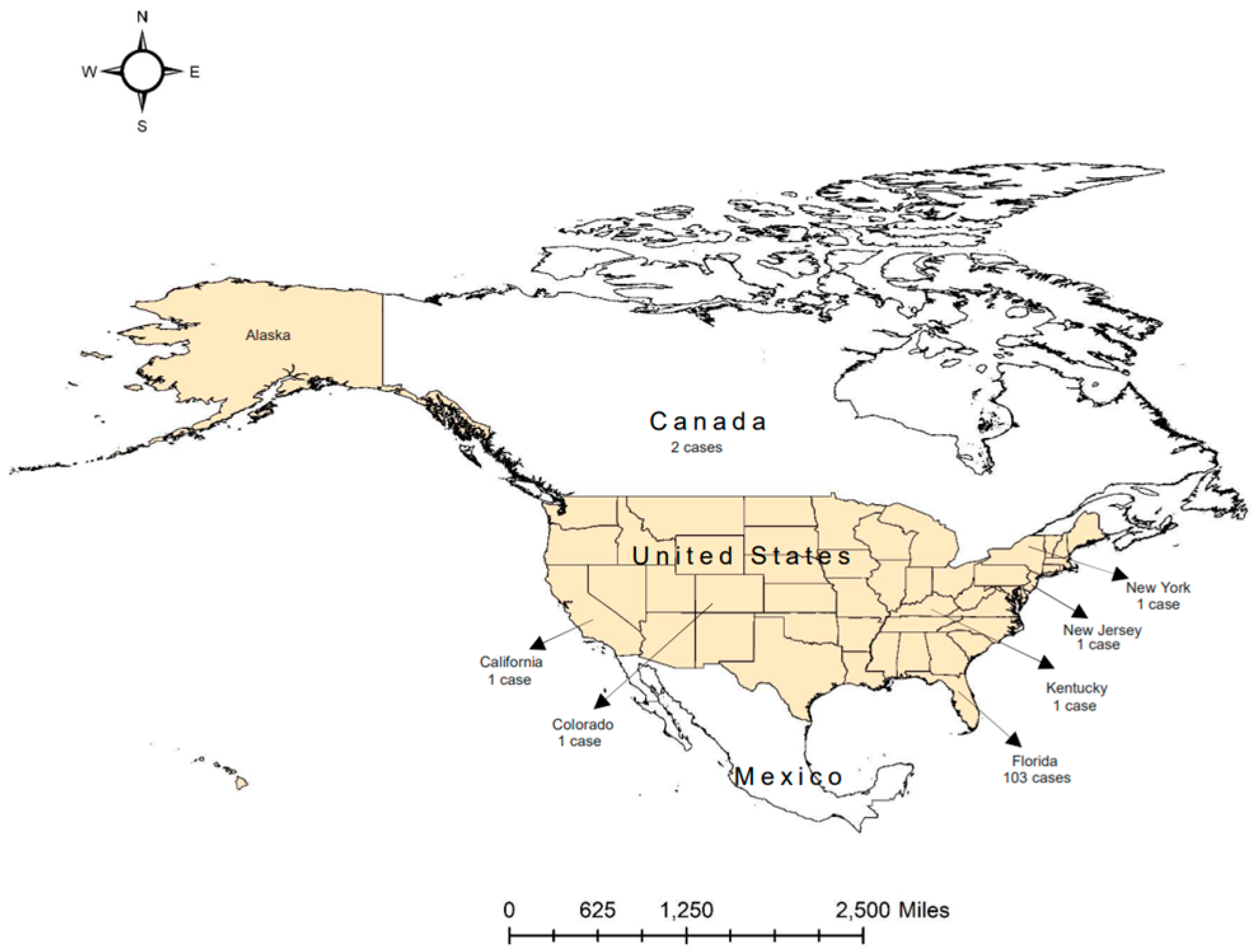
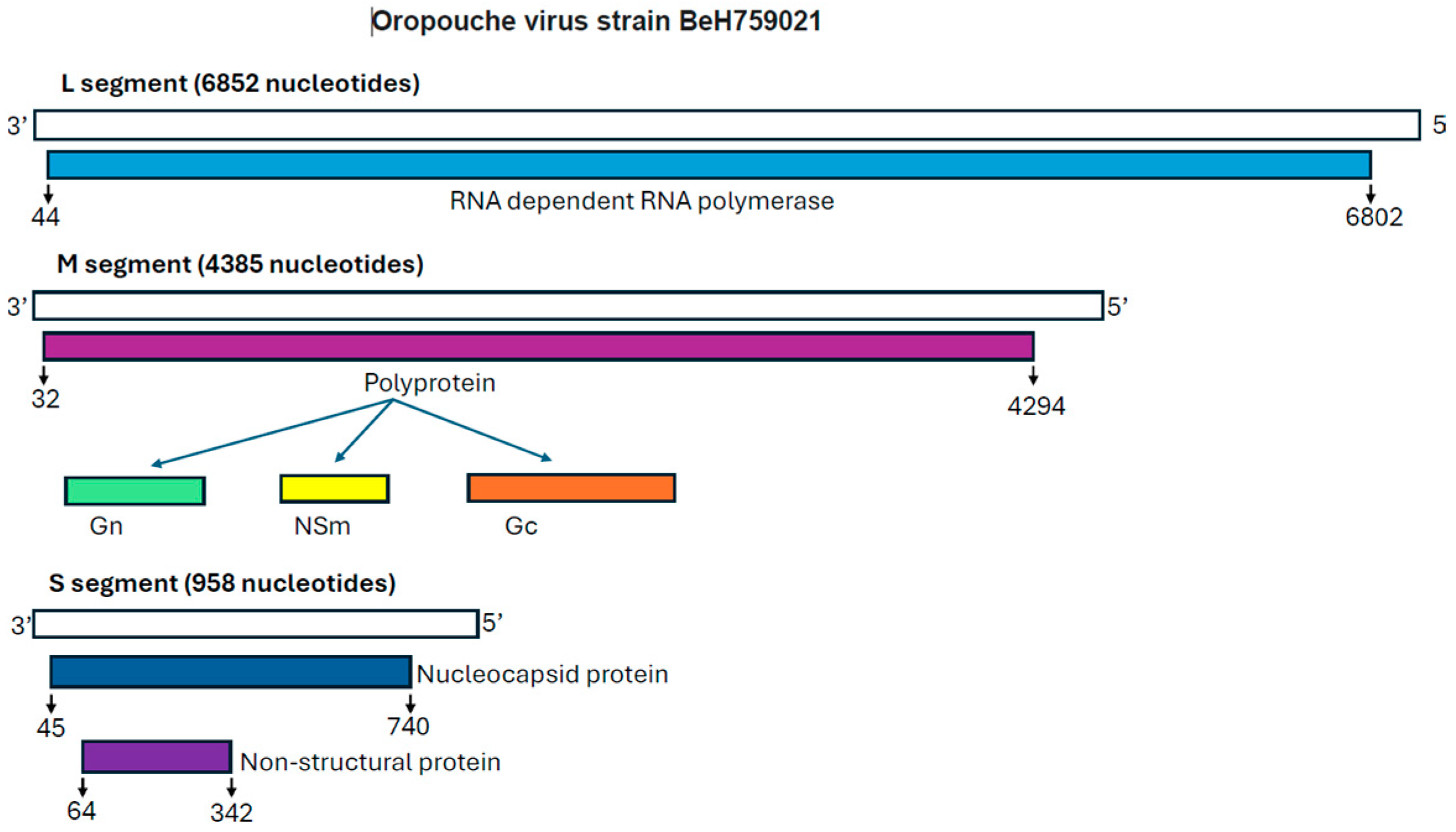
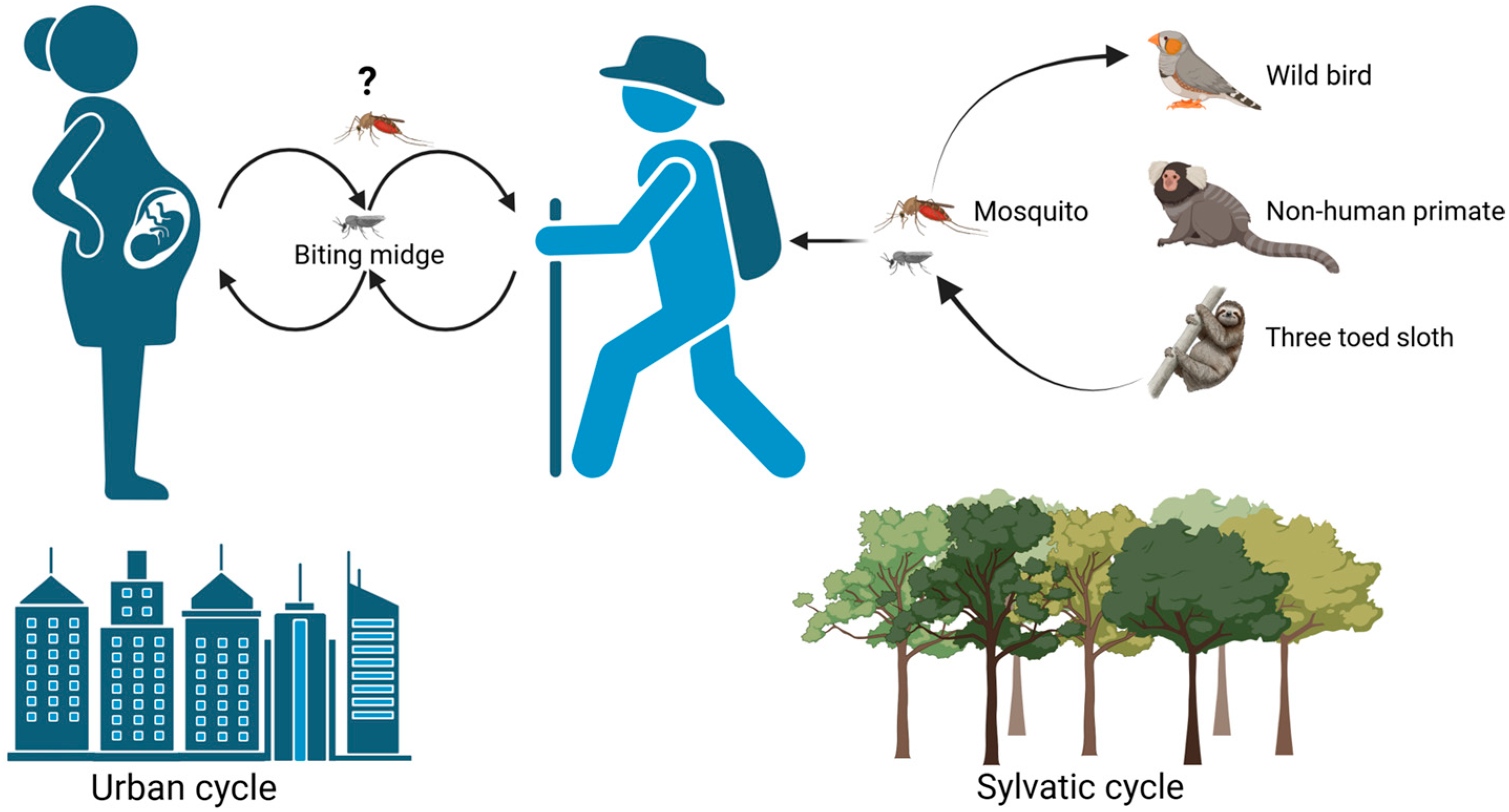
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, F.; Denyoh, P.M.D.; Urquhart, C.; Shrestha, S.; Yee, D.A. A Comprehensive Review of the Neglected and Emerging Oropouche Virus. Viruses 2025, 17, 439. https://doi.org/10.3390/v17030439
Bai F, Denyoh PMD, Urquhart C, Shrestha S, Yee DA. A Comprehensive Review of the Neglected and Emerging Oropouche Virus. Viruses. 2025; 17(3):439. https://doi.org/10.3390/v17030439
Chicago/Turabian StyleBai, Fengwei, Prince M. D. Denyoh, Cassandra Urquhart, Sabin Shrestha, and Donald A. Yee. 2025. "A Comprehensive Review of the Neglected and Emerging Oropouche Virus" Viruses 17, no. 3: 439. https://doi.org/10.3390/v17030439
APA StyleBai, F., Denyoh, P. M. D., Urquhart, C., Shrestha, S., & Yee, D. A. (2025). A Comprehensive Review of the Neglected and Emerging Oropouche Virus. Viruses, 17(3), 439. https://doi.org/10.3390/v17030439








