Integrating Viral Infection and Correlation Analysis in Passiflora edulis and Surrounding Weeds to Enhance Sustainable Agriculture in Republic of Korea
Abstract
1. Introduction
2. Materials and Methods
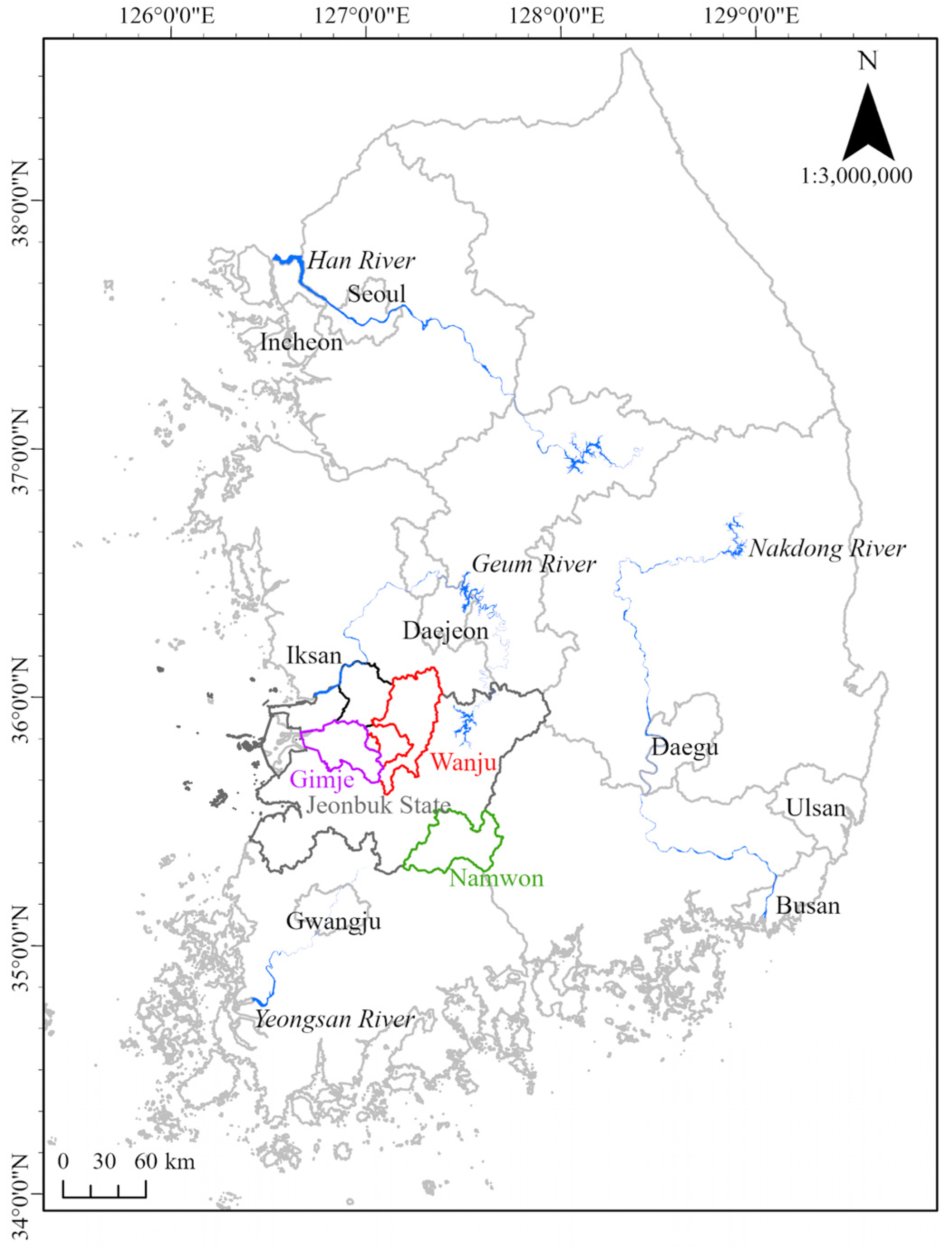
2.1. Selection of Study Area and P. edulis Sample Collection
2.2. PCR/RT-PCR Assays for Detection and Characterization of Viral Infection Status of P. edulis
2.3. Analysis of Viral Infection Status of Weeds Surrounding the P. edulis Cultivation Sites and Infection Status by Weed Species
2.4. Analysis of Correlation Between the Viruses Identified in P. edulis Cultivation Sites and the Infecting Weeds Surrounding the Sites
3. Results and Discussion
3.1. Status of Viral Infection of P. edulis
3.2. Status of Viral Infection of Weeds Surrounding the P. edulis Cultivation Sites
3.3. Current Status of Viral Infection of Weeds Surrounding the P. edulis Cultivation Sites by Weed Species
3.4. Correlation Between Viruses Infecting P. edulis in the Cultivation Sites and Infecting Weeds Surrounding the Cultivation Sites
4. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bernacci, L.C.; Soares-Scott, M.D.; Junqueira, N.T.V.; Passos, I.R.D.S.; Meletti, L.M.M. Passiflora edulis Sims: The correct taxonomic way to cite the yellow passion fruit (and of other colors). Rev. Bras. Frutic. 2008, 30, 566–576. [Google Scholar] [CrossRef]
- Deng, J.; Zhou, Y.; Bai, M.; Li, H.; Li, L. Anxiolytic and sedative activities of Passiflora edulis f. flavicarpa. J. Ethnopharmacol. 2010, 128, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Minor Tropical Fruits. Available online: https://www.fao.org/fileadmin/templates/est/COMM_MARKETS_MONITORING/Tropical_Fruits/Documents/Minor_Tropical_Fruits_FoodOutlook_1_2018.pdf (accessed on 2 February 2025).
- Huang, A.; Gu, P.; Ding, M.; Wang, Y. First report of Euphorbia leaf curl virus and Papaya leaf curl Guangdong virus on passion fruit in mainland China. J. Plant Pathol. 2021, 103, 1367. [Google Scholar] [CrossRef]
- Jeon, M.K.; An, H.J.; Lim, C.K.; Kim, S.A.; Jang, Y.J.; Chung, S.W. Incidence of Viral Disease on Purple Passion Fruit (Passiflora edulis). J. Agri. Life Sci. 2022, 56, 29–35, (In Korean with English Abstract). [Google Scholar] [CrossRef]
- Park, J.O.; Lee, S.M.; Kwon, H.Y.; Kim, H.J.; Lee, B.B.; Cho, K.C.; Jeong, H.J.; Cho, H.S. Development of Techniques for Improving Fruit High Quality and Overwintering Cultivation in Passionfruit (Passiflora edulis SIMS) Grown in the Plastic Film House; Rural Development Administration: Jeonju, Republic of Korea, 2021; p. 1, (In Korean with English summary). [Google Scholar]
- Rural Development Administration. 2023 Rural Development Administration Training Manual for Public Officials: Subtropical Crops; Korean Association for the Disabled in Sports and Recreation: Jeonju, Republic of Korea, 2023; pp. 7–9. (In Korean) [Google Scholar]
- Wu, W.; Ma, F.; Zhang, X.; Tan, Y.; Han, T.; Ding, J.; Xing, W.; Wu, B.; Huang, D.; Zhang, S.; et al. Research Progress on Viruses of Passiflora edulis. Biology 2024, 13, 839. [Google Scholar] [CrossRef]
- Iwai, H.; Yamashita, Y.; Nishi, N.; Nakamura, M. The potyvirus associated with the dappled fruit of Passiflora edulis in Kagoshima prefecture, Japan is the third strain of the proposed new species East Asian Passiflora virus (EAPV) phylogenetically distinguished from strains of Passion fruit woodiness virus. Arch. Virol. 2006, 151, 811–818. [Google Scholar] [CrossRef]
- Chiemsombat, P.; Prammanee, S.; Pipattanawong, N. Occurrence of Telosma mosaic virus causing fruit severe mosaic disease in Thailand and immunostrip test for rapid virus detection. Crop Prot. 2014, 63, 41–47. [Google Scholar] [CrossRef]
- Sokhandan, N.; Gillings, M.R.; Bowyer, J.W. Polymerase chain reaction detection and assessment of genetic variation in New South Wales isolates of passion fruit woodiness potyvirus. Australas. Plant Pathol. 1997, 26, 155–164. [Google Scholar] [CrossRef]
- Spiegel, S.; Zeidan, M.; Sobolev, I.; Beckelman, Y.; Holdengreber, V.; Tam, Y.; Bar Joseph, M.; Lipsker, Z.; Gera, A. The complete nucleotide sequence of Passifora latent virus and its phylogenetic relationship to other carlaviruses. Arch. Virol. 2007, 152, 181–189. [Google Scholar] [CrossRef]
- Ferreira, S.S.; Barros, D.R.; De Almeida, M.R.; Zerbini, F.M. Characterization of Passionfruit severe leaf distortion virus, a novel begomovirus infecting passionfruit in Brazil, reveals a close relationship with tomato-infecting begomoviruses. Plant Pathol. 2010, 59, 221–230. [Google Scholar] [CrossRef]
- Cheng, Y.H.; Deng, T.C.; Chen, C.C.; Chiang, C.H.; Chang, C.A. First Report of Euphorbia leaf curl virus and Papaya leaf curl Guangdong virus on Passion Fruit in Taiwan. Plant Dis. 2014, 98, 1746. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.J.; Ding, M.; Cao, M.J.; Zhang, S.; Zhong, L.Q.; Wang, Y. First Report of Papaya Leaf Curl China Virus on Passion Fruit in China. Plant Dis. 2020, 104, 1265. [Google Scholar] [CrossRef]
- Zhang, S.; Huang, A.J.; Zhou, X.; Li, Z.H.; Dietzgen, R.G.; Zhou, C.Y.; Cao, M.J. Natural Defect of a Plant Rhabdovirus Glycoprotein Gene: A Case Study of Virus–Plant Coevolution. Phytopatholoy 2021, 111, 227–236. [Google Scholar] [CrossRef]
- Huang, A.; Gu, P.; Wang, Y.; Li, J.; Yang, Z.; Yi, L. Simultaneous detection and differentiation of four viruses in passion fruit plants by a multiplex RT-PCR. Trop. Plant Pathol. 2023, 48, 23–29. [Google Scholar] [CrossRef]
- Hong, S.B. Development of a Multiplex PCR System for Diagnosis of Major Passionfruit Viruses and Their Epidemiology in Korea. Master’s Thesis, Jeonbuk National University, Jeonju, Republic of Korea, 22 February 2017. (In Korean with English Abstract). [Google Scholar]
- Roossinck, M.J. Evolutionary History of Cucumber Mosaic Virus Deduced by Phylogenetic Analyses. J. Virol. 2002, 76, 3382–3387. [Google Scholar] [CrossRef]
- Chen, L.; Sun, D.; Zhang, X.; Shao, D.; Lu, Y.; An, Y. Transcriptome analysis of yellow passion fruit in response to cucumber mosaic virus infection. PLoS ONE 2021, 16, e0247127. [Google Scholar] [CrossRef]
- Fischer, I.H.; Rezende, J.A.M. Diseases of Passion Flower (Passiflora spp.). Pest Technol. 2008, 2, 1–19. [Google Scholar]
- Do, D.H.; Chong, Y.H.; Ha, V.C.; Cheng, H.W.; Chen, Y.K.; Bui, T.N.L.; Nguyen, T.B.N.; Yeh, S.D. Characterization and Detection of Passiflora Mottle Virus and Two Other Potyviruses Causing Passionfruit Woodiness Disease in Vietnam. Phytopathology 2021, 111, 1675–1685. [Google Scholar] [CrossRef]
- Bao, S.; Ge, F.; Li, X.; Bo, B. First Report of Passiflora Latent Virus in Passion Fruit (Passiflora edulis) in China. Plant Dis. 2023, 107, 3326. [Google Scholar] [CrossRef]
- Choi, M.K.; Ju, H.J. First Report of Passiflora Latent Virus Infecting Passion Fruit (Passiflora edulis) in South Korea. Plant Dis. 2023, 107, 2893. [Google Scholar] [CrossRef]
- Roy, A.; Leon M, G.; Nunziata, S.; Padmanabhan, C.; Rivera, Y.; Brlansky, R.H.; Hartung, J.S. First Report of Passion Fruit Green Spot Virus in Yellow Passion Fruit (Passiflora edulis f. flavicarpa) in Casanare, Colombia. Plant Dis. 2023, 107, 2270. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Jiang, J.; Yang, Q.; Niu, L.; Wang, Y.; Long, X.; Malichan, S.; Xie, X. Occurrence and Distribution of Major Viruses Infecting Passion Fruit in Guizhou Province, China, and Molecular Characterization of Two Potyviruses. Plant Dis. 2023, 107, 2307–2312. [Google Scholar] [CrossRef] [PubMed]
- Junco, M.C.; Silva, C.D.C.; do Carmo, C.M.; Kotsubo, R.Y.; de Novaes, T.G.; Molina, R.D.O. Identification of potential hosts plants of Cowpea aphid-borne mosaic virus. J. Phytopathol. 2021, 169, 45–51. [Google Scholar] [CrossRef]
- Odum, E.P. Fundamentals of Ecology; W.B. Saunders Company: Philadelphia, PA, USA, 1971; p. 574. [Google Scholar]
- National Institute of Agricultural Sciences. Handbook of Useful Weeds; 21st Century Book Publishing Company: Paju, Republic of Korea, 2017; pp. 1–455. (In Korean) [Google Scholar]
- Kim, M.; Hong, S.B.; Kim, J.; Kwak, H.R.; Choi, H.S.; Kim, C.K.; Ko, S.J.; Seo, J.K.; Ju, H.J. First Report of Papaya Leaf Curl Guandong Virus in Bell Pepper in Korea. Plant Dis. 2018, 102, 2046. [Google Scholar] [CrossRef]
- Virus Diseases of Tropical Crops. Available online: https://www.researchgate.net/profile/John-Randles/publication/277693828_Virus_Diseases_of_Tropical_Crops/links/5a8e04eaa6fdcc808c0f112b/Virus-Diseases-of-Tropical-Crops.pdf (accessed on 2 February 2025).
- Kim, H.J.; Song, M.A.; Song, J.H.; Ko, Y.J.; Park, J.H.; Heo, T.H. Incidence and occurrence pattern of viruses in passion fruit on Jeju Island. In Proceedings of the Korean Society of Pesticide Science of the Conference, Kwangju, Republic of Korea, 24–25 October 2018; p. 297, (In Korean with English Abstract). [Google Scholar]
- Chong, Y.H.; Cheng, Y.H.; Cheng, H.W.; Huwang, Y.C.; Yeh, S.D. The virus causing passionfruit woodiness disease in Taiwan is reclassified as East Asian passiflora virus. J. Gen. Plant Pathol. 2018, 84, 208–220. [Google Scholar] [CrossRef]
- Palukaitis, P.; Roossinck, M.J.; Dietzgen, R.G.; Francki, R.I.B. Cucumber MOSAIC Virus. Adv. Virus Res. 1992, 41, 281–348. [Google Scholar] [CrossRef]
- Tomlinson, J.A. Epidemiology and control of virus diseases of vegetables. Ann. Appl. Biol. 1987, 110, 661–681. [Google Scholar] [CrossRef]
- Gioria, R.; Espinha, L.M.; Rezendea, J.A.M.; Gasparband, J.O.; Kitajima, E. Limited movement of Cucumber mosaic virus (CMV) in yellow passion flower in Brazil. Plant Pathol. 2002, 51, 127–133. [Google Scholar] [CrossRef][Green Version]
- Syller, J.; Grupa, A. The effects of co-infection by different Potato virus Y (PVY) isolates on virus concentration in solanaceous hosts and efficiency of transmission. Plant Pathol. 2014, 63, 466–475. [Google Scholar] [CrossRef]
- East Asian Passiflora Virus. Available online: https://www.cabidigitallibrary.org/doi/10.1079/cabicompendium.119879 (accessed on 2 February 2025).
- Iwai, H.; Ohmori, T.; Kurokawa, Y.; Muta, T.; Arai, K. New Record of Passionfruit Woodiness Virus in Japan. Jpn. J. Phytopathol. 1996, 62, 459–465. [Google Scholar] [CrossRef]
- Lee, I.Y.; Oh, Y.J.; Hong, S.H.; Choi, J.K.; Heo, S.J.; Lee, C.Y.; Hwang, K.S.; Park, K.W.; Cho, S.H.; Kwon, O.D.; et al. Weed Flora Diversity and Composition on Upland Field of Korea. Weed Turf. Sci. 2015, 4, 159–175, (In Korean with English Abstract). [Google Scholar] [CrossRef]
- Ochwo-Ssemakula, M.; Sengooba, T.; Hakiza, J.J.; Adipala, E.; Edema, R.; Redinbaugh, M.G.; Aritua, V.; Winter, S. Characterization and Distribution of a Potyvirus Associated with Passion Fruit Woodiness Disease in Uganda. Plant Dis. 2012, 96, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Kim, G.S.; Kwak, H.R.; Kim, J.E.; Seo, J.K.; Hong, S.J.; Lee, G.J.; Kim, J.H.; Choi, M.K.; Kim, B.R.; et al. Occurrence and eradication of Plum pox virus on Ornamentals in Korea, 2016–2017. Res. Plant Dis. 2019, 25, 8–15, (In Korean with English Abstract). [Google Scholar] [CrossRef]
- Choi, M.K. Evaluation of the Weeds around Capsicum annuum (CA) Cultivation Fields as Potential Habitats of CA-Infecting Viruses. Plant Pathol. J. 2023, 39, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Kil, E.J.; Seo, H.; Byun, H.S.; Suh, S.S.; Lee, T.K.; Lee, K.Y.; Lee, J.H.; Kim, J.K.; Ko, S.J.; Choi, H.S.; et al. First Report of Euphorbia leaf curl virus in Passion Fruits in South Korea and its Natural Occurrence in Papaya. Plant Dis. 2016, 100, 865. [Google Scholar] [CrossRef]
- Pleše, N.; Wrischer, M. A mixed infection of Passiflora caerulea L. with two viruses. Acta Bot. Croat. 1984, 43, 1–6. Available online: https://hrcak.srce.hr/158905 (accessed on 2 February 2025).
- Tomlinson, J.A.; Carter, A.L.; Dale, W.; Simpson, C.J. Weed plants as sources of cucumber mosaic virus. Ann. Appl. Biol. 1970, 66, 11–16. [Google Scholar] [CrossRef]
- Ko, S.J.; Kang, B.R.; Choi, D.S.; Kim, D.I.; Lee, G.S.; Kim, C.S.; Choi, H.S. Pattern of the occurrence of Tomato spotted wilt virus in Jeonnam Province. Res. Plant Dis. 2013, 19, 273–280, (In Korean with English Abstract). [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Boezen, D.; Zwart, M.P. Metagenomic Studies of Viruses in Weeds and Wild Plants: A Powerful Approach to Characterise Variable Virus Communities. Viruses 2021, 13, 1939. [Google Scholar] [CrossRef]
| Virus | Primer | Primer Sequences (5′→3′) | Size (bp) | Accession No. Referred |
|---|---|---|---|---|
| EuLCV | EuLCV-F | AGTGGTCCCCCCTCCACTAAC | 339 | AJ811911.1 |
| EuLCV-R | CAGCCTCCGTCGAACCTTCG | |||
| PaLCuGdV | PaLCuGdV-m-2F | CTGTCTTACGTGCAAGGA | 605 | MZ130299.1 |
| PaLCuGdV-m-2R | GCTTGCATATTGACCACCAG | |||
| TYLCV | TYLCV 1F | GTC AAC CAA TCA AAT TGC ATC CTC AA | 712 | AM691759.1 |
| TYLCV 1R | GTC CAA AAT CCA TTG GGC | |||
| CMV | CMV 1-5 u | CTTGTGCGTTTRATGGCTACGAAGGC | 473 | M57602.1 |
| CMV 1-5 d | CACGGACCGAAGTCCTTCCGAAGAAA | |||
| EAPV | EAPV-IB-1F | CATTGATAATGGCACCTCACC | 231 | MH922997.1 |
| EAPV-IB-1R | AGCCAAACTCAAGTCCCTCA |
| Year | PaLCuGdV | EuLCV | EAPV | CMV | TYLCV | Uninfected | No. of Samples Tested |
|---|---|---|---|---|---|---|---|
| 2017 | 28 (87.5) | 28 (87.5) | 24 (75.0) | 1 (3.1) | 0 (0.0) | 1 | 32 (100.0) |
| 2018 | 33 (58.9) | 30 (53.6) | 22 (39.3) | 6 (10.7) | 0 (0.0) | 12 | 56 (100.0) |
| 2019 | 62 (73.8) | 37 (44.0) | 26 (31.0) | 19 (22.6) | 0 (0.0) | 11 | 84 (100.0) |
| 2020 | 67 (78.8) | 64 (75.3) | 37 (43.5) | 25 (29.4) | 0 (0.0) | 11 | 85 (100.0) |
| 2021 | 21 (80.8) | 18 (69.2) | 5 (19.2) | 13 (50.0) | 0 (0.0) | 3 | 26 (100.0) |
| Total | 211 (74.6) | 177 (62.5) | 114 (40.3) | 64 (22.6) | 0 (0.0) | 38 | 283 (100.0) |
| Year | Infection Type | No. of Samples Tested | ||
|---|---|---|---|---|
| Single | Mixed | Total | ||
| 2017 | 1 (3.1) | 30 (93.8) | 31 (96.9) | 32 (100.0) |
| 2018 | 14 (25.0) | 30 (53.6) | 44 (78.6) | 56 (100.0) |
| 2019 | 32 (38.1) | 41 (48.8) | 73 (86.9) | 84 (100.0) |
| 2020 | 11 (12.9) | 63 (74.1) | 74 (87.1) | 85 (100.0) |
| 2021 | 2 (7.7) | 21 (80.8) | 23 (88.5) | 26 (100.0) |
| Total | 60 (21.2) | 185 (65.4) | 245 (86.6) | 283 (100.0) |
| Year | PaLCuGdV | EuLCV | EAPV | CMV | TYLCV | Uninfected | No. of Samples Tested |
|---|---|---|---|---|---|---|---|
| 2018 | 12 (48.0) | 10 (40.0) | 1 (4.0) | 0 (0.0) | 0 (0.0) | 12 | 25 (100.0) |
| 2019 | 6 (9.2) | 2 (3.1) | 0 (0.0) | 5 (7.7) | 0 (0.0) | 54 | 65 (100.0) |
| 2020 | 30 (22.7) | 29 (22.0) | 0 (0.0) | 11 (8.3) | 0 (0.0) | 85 | 132 (100.0) |
| 2021 | 9 (3.8) | 11 (4.6) | 0 (0.0) | 22 (9.2) | 0 (0.0) | 209 | 239 (100.0) |
| Total | 57 (12.4) | 52 (11.3) | 1 (0.2) | 38 (8.2) | 0 (0.0) | 360 | 461 (100.0) |
| Year | Infection Type | No. of Samples Tested | ||
|---|---|---|---|---|
| Single | Mixed | Total | ||
| 2018 | 4 (16.0) | 9 (36.0) | 13 (52.0) | 25 (100.0) |
| 2019 | 10 (15.4) | 1 (1.5) | 11 (16.9) | 65 (100.0) |
| 2020 | 26 (19.7) | 21 (15.9) | 47 (35.6) | 132 (100.0) |
| 2021 | 19 (7.9) | 11 (4.6) | 30 (12.6) | 239 (100.0) |
| Total | 59 (12.8) | 42 (9.1) | 101 (21.9) | 461 (100.0) |
| Family | Species (Tested Plants) | No. of Viral Infections | ||||
|---|---|---|---|---|---|---|
| PaLCuGdV | EuLCV | EAPV | CMV | Total | ||
| Asteraceae (14 species) | Erigeron annuus (14) | 3 | 3 | 0 | 0 | 6 |
| Sonchus asper (8) | 2 | 1 | 0 | 2 | 5 | |
| Erigeron canadensis (19) | 3 | 0 | 0 | 0 | 3 | |
| Taraxacum mongolicum (12) | 2 | 0 | 0 | 1 | 3 | |
| Bidens frondosa (5) | 1 | 1 | 0 | 0 | 2 | |
| Youngia japonica (1) | 1 | 1 | 0 | 0 | 2 | |
| Cirsium japonicum var. maackii (1) | 1 | 1 | 0 | 0 | 2 | |
| Eclipta thermalis (17) | 1 | 1 | 0 | 0 | 2 | |
| Artemisia annua (1) | 1 | 0 | 0 | 1 | 2 | |
| Sonchus oleraceus (9) | 0 | 1 | 0 | 0 | 1 | |
| Artemisia indica (3) | 0 | 1 | 0 | 0 | 1 | |
| Centipeda minima (14) | 0 | 1 | 0 | 0 | 1 | |
| Erigeron sumatrensis (1) | 0 | 0 | 0 | 1 | 1 | |
| Symphyotrichum expansum (2) | 0 | 0 | 0 | 1 | 1 | |
| Subtotal (107) | 15 | 11 | 0 | 6 | 32 | |
| (46.9) | (34.4) | (0.0) | (18.8) | (100.0) | ||
| Amaranthaceae (4 species) | Amaranthus blitum (4) | 2 | 1 | 0 | 0 | 3 |
| Amaranthus tricolor (2) | 1 | 1 | 0 | 0 | 2 | |
| Achyranthes bidentata var. japonica (10) | 1 | 0 | 0 | 1 | 2 | |
| Chenopodium album var. centrorubrum (14) | 2 | 0 | 0 | 1 | 3 | |
| Subtotal (30) | 6 | 2 | 0 | 2 | 10 | |
| (60.0) | (20.0) | (0.0) | (20.0) | (100.0) | ||
| Poaceae (3 species) | Digitaria ciliaris (16) | 3 | 3 | 0 | 3 | 9 |
| Setaria viridis (1) | 0 | 0 | 0 | 1 | 1 | |
| Poa annua (7) | 0 | 0 | 0 | 1 | 1 | |
| Subtotal (24) | 3 | 3 | 0 | 5 | 11 | |
| (27.3) | (27.3) | (0.0) | (45.5) | (100.0) | ||
| Lamiaceae (3 species) | Lamium amplexicaule (3) | 1 | 1 | 0 | 1 | 3 |
| Salvia rosmarinus (1) | 1 | 1 | 0 | 0 | 2 | |
| Lavandula angustifolia (5) | 0 | 0 | 0 | 1 | 1 | |
| Subtotal (9) | 2 | 2 | 0 | 2 | 6 | |
| (33.3) | (33.3) | (0.0) | (33.3) | (100.0) | ||
| Fabaceae (3 species) | Trifolium repens (3) | 1 | 1 | 0 | 0 | 2 |
| Glycine max subsp. soja (3) | 1 | 0 | 0 | 0 | 1 | |
| Amorpha fruticosa (1) | 0 | 0 | 0 | 1 | 1 | |
| Subtotal (7) | 2 | 1 | 0 | 1 | 4 | |
| (50.0) | (25.0) | (0.0) | (25.0) | (100.0) | ||
| Solanaceae (2 species) | Solanum nigrum (19) | 4 | 5 | 0 | 2 | 11 |
| Capsicum annuum (4) | 1 | 3 | 0 | 2 | 6 | |
| Subtotal (23) | 5 | 8 | 0 | 4 | 17 | |
| (29.4) | (47.1) | (0.0) | (23.5) | (100.0) | ||
| Liliaceae (2 species) | Chlorophytum comosum var. variegatum (3) | 1 | 2 | 0 | 1 | 4 |
| Liriope muscari (4) | 1 | 1 | 0 | 0 | 2 | |
| Subtotal (7) | 2 | 3 | 0 | 1 | 6 | |
| Caryophyllaceae (2 species) | Stellaria aquatica (4) | 2 | 1 | 0 | 0 | 3 |
| Stellaria media (3) | 1 | 0 | 0 | 1 | 2 | |
| Subtotal (7) | 3 | 1 | 0 | 1 | 5 | |
| Ranunculaceae (2 species) | Aquilegia buergeriana var. oxysepala (1) | 1 | 1 | 0 | 0 | 2 |
| Ranunculus sceleratus (1) | 1 | 0 | 0 | 0 | 1 | |
| Subtotal (2) | 2 | 1 | 0 | 0 | 3 | |
| Boraginaceae (2 species) | Trigonotis peduncularis (1) | 1 | 1 | 0 | 0 | 2 |
| Bothriospermum zeylanicum (1) | 0 | 0 | 0 | 1 | 1 | |
| Subtotal (2) | 1 | 1 | 0 | 1 | 3 | |
| Brassicaceae (2 species) | Cardamine flexuosa (3) | 0 | 1 | 0 | 1 | 2 |
| Turritis glabra (1) | 0 | 1 | 0 | 0 | 1 | |
| Subtotal (4) | 0 | 2 | 0 | 1 | 3 | |
| Euphorbiaceae (2 species) | Acalypha australis (12) | 1 | 0 | 0 | 0 | 1 |
| Euphorbia maculata (1) | 0 | 0 | 0 | 1 | 1 | |
| Subtotal (13) | 1 | 0 | 0 | 1 | 2 | |
| Oxalidaceae (1 species) | Oxalis corniculate (49) | 7 (41.2) | 5 (29.4) | 0 (0.0) | 5 (29.4) | 17 (100.0) |
| Plantaginaceae (1 species) | Plantago asiatica (3) | 1 | 3 | 0 | 0 | 4 |
| Rosaceae (1 species) | Prunus serrulate f. spontanea (1) | 1 | 1 | 1 | 0 | 3 |
| Apiaceae (1 species) | Peucedanum japonicum (4) | 1 | 1 | 0 | 1 | 3 |
| Cyperaceae (1 species) | Cyperus amuricus (1) | 1 | 1 | 0 | 0 | 2 |
| Cannabaceae (1 species) | Humulus scandens (2) | 1 | 1 | 0 | 0 | 2 |
| Brassicaceae (1 species) | Cardamine fallax (4) | 1 | 1 | 0 | 0 | 2 |
| Phytolaccaceae (1 species) | Phytolacca americana (2) | 0 | 2 | 0 | 0 | 2 |
| Moraceae (1 species) | Morus alba (4) | 0 | 1 | 0 | 1 | 2 |
| Portulacaceae (1 species) | Portulaca oleracea (25) | 0 | 0 | 0 | 2 | 2 |
| Violaceae (1 species) | Viola mandshurica (3) | 1 | 0 | 0 | 0 | 1 |
| Commelinaceae (1 species) | Commelina communis (7) | 0 | 0 | 0 | 1 | 1 |
| Convolvulaceae (1 species) | Calystegia pubescens (1) | 0 | 0 | 0 | 1 | 1 |
| Equisetaceae (1 species) | Equisetum arvense (6) | 0 | 0 | 0 | 1 | 1 |
| Araceae (1 species) | Pinellia ternata (19) | 0 | 0 | 0 | 1 | 1 |
| 27 families | 56 species (366) | 56 (38.4) | 51 (34.9) | 1 (0.7) | 38 (26.0) | 146 (100.0) |
| Unidentified (1 species) | Unidentified (5) | 1 | 1 | 0 | 0 | 2 |
| Total (371) | 57 (38.5) | 52 (35.1) | 1 (0.7) | 38 (25.7) | 148 (100.0) | |
| Classification z | P_CMV | P_PaLCuGdV | P_EuLCV | P_EAPV | W_CMV | W_PaLCuGdV | W_EuLCV |
|---|---|---|---|---|---|---|---|
| P_PALCuGdV | 0.23 ** | ||||||
| P_EuLCV | 0.23 ** | 0.57 ** | |||||
| P_EAPV | 0.07 | 0.12 | 0.33 ** | ||||
| W_CMV | −0.00 | 0.13 * | 0.12 | −0.04 | |||
| W_PaLCuGdV | −0.00 | −0.00 | 0.07 | 0.11 | 0.06 | ||
| W_EuLCV | −0.00 | 0.06 | 0.09 | 0.16 * | 0.14 * | 0.61 ** | |
| W_EAPV | −0.00 | 0.44 | 0.06 | −0.00 | −0.00 | 0.13 | 0.15 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, M.K. Integrating Viral Infection and Correlation Analysis in Passiflora edulis and Surrounding Weeds to Enhance Sustainable Agriculture in Republic of Korea. Viruses 2025, 17, 383. https://doi.org/10.3390/v17030383
Choi MK. Integrating Viral Infection and Correlation Analysis in Passiflora edulis and Surrounding Weeds to Enhance Sustainable Agriculture in Republic of Korea. Viruses. 2025; 17(3):383. https://doi.org/10.3390/v17030383
Chicago/Turabian StyleChoi, Min Kyung. 2025. "Integrating Viral Infection and Correlation Analysis in Passiflora edulis and Surrounding Weeds to Enhance Sustainable Agriculture in Republic of Korea" Viruses 17, no. 3: 383. https://doi.org/10.3390/v17030383
APA StyleChoi, M. K. (2025). Integrating Viral Infection and Correlation Analysis in Passiflora edulis and Surrounding Weeds to Enhance Sustainable Agriculture in Republic of Korea. Viruses, 17(3), 383. https://doi.org/10.3390/v17030383





