AliMarko: A Pipeline for Virus Identification Using an Expert-Guided Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Read Filtration
2.2. Read Mapping and Analysis
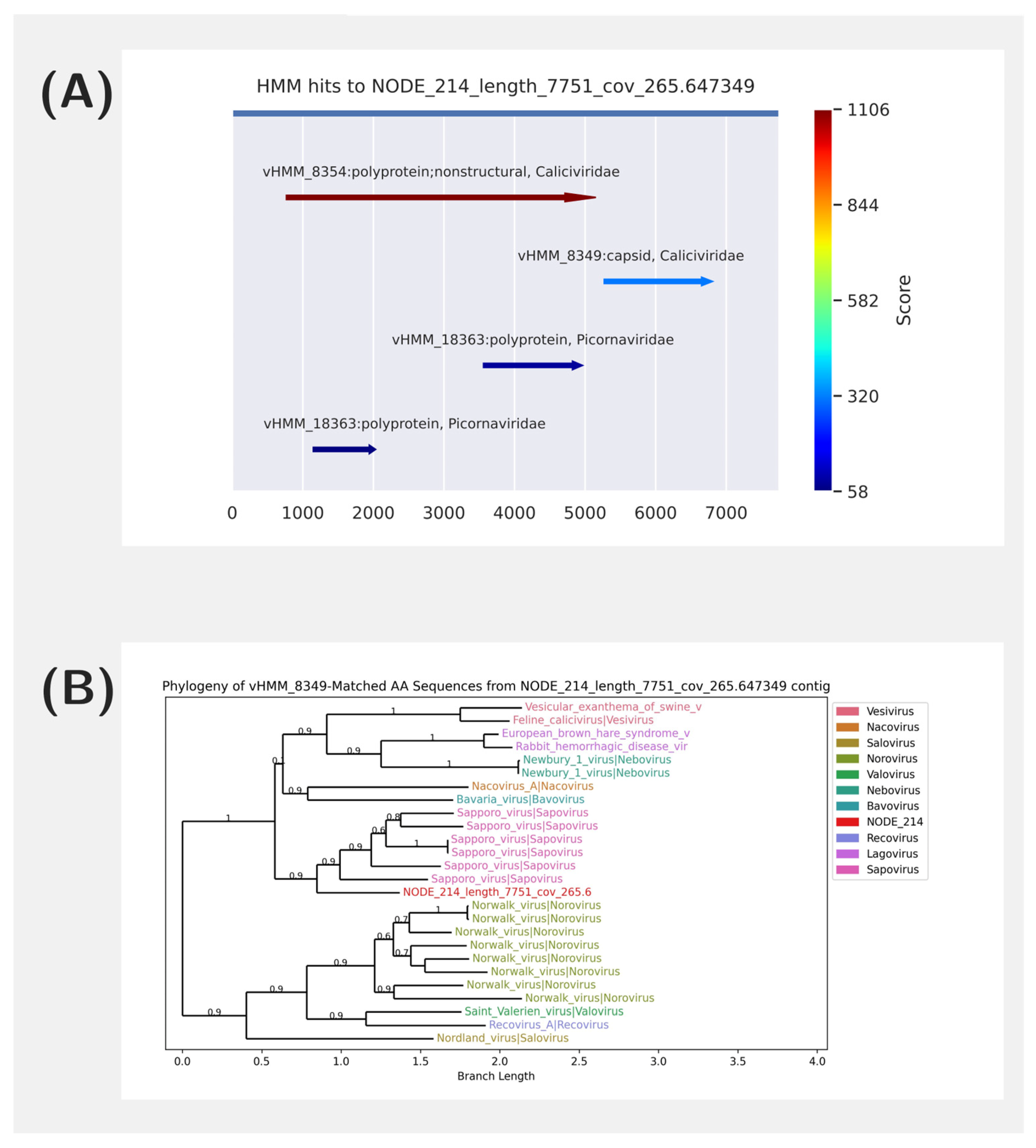
2.3. HMM Analysis
2.4. Phylogenetic Analysis
2.5. Task Management
2.6. Materials
3. Results
3.1. Pipeline Overview
3.2. AliMarko Reports
3.2.1. One Sample Report
3.2.2. Multisample Report
3.3. Applying the Pipeline to RNA Metagenomic Data
3.4. Comparison of HMM vs. Mapping for Viral Detection
3.5. Applying the Pipeline to Diverse Set of Metagenomic Data
3.6. Validation and Performance Evaluation of AliMarko on Simulated Viral Metagenomes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Simmonds, P.; Adriaenssens, E.M.; Zerbini, F.M.; Abrescia, N.G.A.; Aiewsakun, P.; Alfenas-Zerbini, P.; Bao, Y.; Barylski, J.; Drosten, C.; Duffy, S.; et al. Four principles to establish a universal virus taxonomy. PLoS Biol. 2023, 21, e3001922. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Dolja, V.V.; Krupovic, M.; Varsani, A.; Wolf, Y.I.; Yutin, N.; Zerbini, F.M.; Kuhn, J.H. Global Organization and Proposed Megataxonomy of the Virus World. Microbiol. Mol. Biol. Rev. 2020, 84, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Dolja, V.V.; Koonin, E.V. Origin of viruses: Primordial replicators recruiting capsids from hosts. Nat. Rev. Microbiol. 2019, 17, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P.; Adams, M.J.; Benkő, M.; Breitbart, M.; Brister, J.R.; Carstens, E.B.; Davison, A.J.; Delwart, E.; Gorbalenya, A.E.; Harrach, B.; et al. Consensus statement: Virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 2017, 15, 161–168. [Google Scholar] [CrossRef]
- Jurasz, H.; Pawłowski, T.; Perlejewski, K. Contamination Issue in Viral Metagenomics: Problems, Solutions, and Clinical Perspectives. Front. Microbiol. 2021, 12, 745076. [Google Scholar] [CrossRef]
- Yolken, R.H.; Jones-Brando, L.; Dunigan, D.D.; Kannan, G.; Dickerson, F.; Severance, E.; Sabunciyan, S.; Talbot, C.C., Jr.; Prandovszky, E.; Gurnon, J.R.; et al. Chlorovirus ATCV-1 is part of the human oropharyngeal virome and is associated with changes in cognitive functions in humans and mice. Proc. Natl. Acad. Sci. USA 2014, 111, 16106–16111. [Google Scholar] [CrossRef]
- Lombardi, V.C.; Ruscetti, F.W.; Gupta, J.D.; Pfost, M.A.; Hagen, K.S.; Peterson, D.L.; Ruscetti, S.K.; Bagni, R.K.; Petrow-Sadowski, C.; Gold, B.; et al. Detection of an Infectious Retrovirus, XMRV, in Blood Cells of Patients with Chronic Fatigue Syndrome. Science 2009, 326, 585–589. [Google Scholar] [CrossRef]
- Kjartansdóttir, K.R.; Friis-Nielsen, J.; Asplund, M.; Mollerup, S.; Mourier, T.; Jensen, R.H.; Hansen, T.A.; Rey-Iglesia, A.; Richter, S.R.; Alquezar-Planas, D.E.; et al. Traces of ATCV-1 associated with laboratory component contamination. Proc. Natl. Acad. Sci. USA 2015, 112, E925–E926. [Google Scholar] [CrossRef]
- Delviks-Frankenberry, K.; Cingöz, O.; Coffin, J.M.; Pathak, V.K. Recombinant origin, contamination, and de-discovery of XMRV. Curr. Opin. Virol. 2012, 2, 499–507. [Google Scholar] [CrossRef]
- Asplund, M.; Kjartansdóttir, K.R.; Mollerup, S.; Vinner, L.; Fridholm, H.; Herrera, J.A.R.; Friis-Nielsen, J.; Hansen, T.; Jensen, R.; Nielsen, I.; et al. Contaminating viral sequences in high-throughput sequencing viromics: A linkage study of 700 sequencing libraries. Clin. Microbiol. Infect. 2019, 25, 1277–1285. [Google Scholar] [CrossRef]
- Duan, J.; Keeler, E.; McFarland, A.; Scott, P.; Collman, R.G.; Bushman, F.D. The virome of the kitome: Small circular virus-like genomes in laboratory reagents. Microbiol. Resour. Announc. 2024, 13, e0126123. [Google Scholar] [CrossRef] [PubMed]
- Privitera, G.F.; Alaimo, S.; Ferro, A.; Pulvirenti, A. Virus finding tools: Current solutions and limitations. Briefings Bioinform. 2022, 23, bbac235. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.; Gish, W.; Miller, W.; Myers, E.; Lipman, D. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Wu, G.; Lim, E.S.; Droit, L.; Krishnamurthy, S.; Barouch, D.H.; Virgin, H.W.; Wang, D. VirusSeeker, a computational pipeline for virus discovery and virome composition analysis. Virology 2017, 503, 21–30. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef]
- Babaian, A.; Edgar, R. Ribovirus classification by a polymerase barcode sequence. PeerJ 2022, 10, e14055. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Guo, J.; Bolduc, B.; Zayed, A.A.; Varsani, A.; Dominguez-Huerta, G.; Delmont, T.O.; Pratama, A.A.; Gazitúa, M.C.; Vik, D.; Sullivan, M.B.; et al. VirSorter2: A multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome 2021, 9, 37. [Google Scholar] [CrossRef]
- Ren, J.; Ahlgren, N.A.; Lu, Y.Y.; Fuhrman, J.A.; Sun, F. VirFinder: A novel k-mer based tool for identifying viral sequences from assembled metagenomic data. Microbiome 2017, 5, 69. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Shan, Y.; Liu, X.; Chen, G.; Pan, Y.; Gou, Q.; Zou, J.; Chang, Z.; Zeng, Q.; Yang, C.; et al. VirID: Beyond virus discovery—An integrated platform for comprehensive RNA virus characterization. bioRxiv 2024. Available online: https://www.biorxiv.org/content/early/2024/07/09/2024.07.05.602175 (accessed on 7 February 2024).
- Gwak, H.-J.; Rho, M. ViBE: A hierarchical BERT model to identify eukaryotic viruses using metagenome sequencing data. Brief. Bioinform. 2022, 23, bbac204. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Song, K.; Deng, C.; Ahlgren, N.A.; Fuhrman, J.A.; Li, Y.; Xie, X.; Poplin, R.; Sun, F. Identifying viruses from metagenomic data using deep learning. Quant. Biol. 2020, 8, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Kohl, C.; Kurth, A. European Bats as Carriers of Viruses with Zoonotic Potential. Viruses 2014, 6, 3110–3128. [Google Scholar] [CrossRef]
- Letko, M.; Seifert, S.N.; Olival, K.J.; Plowright, R.K.; Munster, V.J. Bat-borne virus diversity, spillover and emergence. Nat. Rev. Microbiol. 2020, 18, 461–471. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Kwon, M.; Lee, S.; Berselli, M.; Chu, C.; Park, P.J. BamSnap: A lightweight viewer for sequencing reads in BAM files. Bioinformatics 2021, 37, 263–264. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed]
- Larralde, M.; Zeller, G. PyHMMER: A Python library binding to HMMER for efficient sequence analysis. Bioinformatics 2023, 39, btad214. [Google Scholar] [CrossRef] [PubMed]
- Viral Minion DB: A Database of Viral Profile HMMs. Icb.usp.br. 2020. Available online: http://www.bioinfovir.icb.usp.br/minion_db/ (accessed on 11 September 2024).
- Oliveira, L.S.; Gruber, A. Rational Design of Profile Hidden Markov Models for Viral Classification and Discovery; Exon Publications eBooks: Brisbane, Australia, 2021; pp. 151–170. [Google Scholar]
- Boeckmann, B.; Bairoch, A.; Apweiler, R.; Blatter, M.C.; Estreicher, A.; Gasteiger, E.; Martin, M.J.; Michoud, K.; O’Donovan, C.; Phan, I.; et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 365. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Speranskaya, A.S.; Artiushin, I.V.; Samoilov, A.E.; Korneenko, E.V.; Khabudaev, K.V.; Ilina, E.N.; Yusefovich, A.P.; Safonova, M.V.; Dolgova, A.S.; Gladkikh, A.S.; et al. Identification and Genetic Characterization of MERS-Related Coronavirus Isolated from Nathusius’ Pipistrelle (Pipistrellus nathusii) near Zvenigorod (Moscow Region, Russia). Int. J. Environ. Res. Public Health 2023, 20, 3702. [Google Scholar] [CrossRef]
- Huang, W.; Li, L.; Myers, J.R.; Marth, G.T. ART: A next-generation sequencing read simulator. Bioinformatics 2011, 28, 593–594. [Google Scholar] [CrossRef]
- Gourlé, H.; Karlsson-Lindsjö, O.; Hayer, J.; Bongcam-Rudloff, E. Simulating Illumina metagenomic data with InSilicoSeq. Bioinformatics 2019, 35, 521–522. [Google Scholar] [CrossRef]
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2017, 46, D708–D717. [Google Scholar] [CrossRef]
- Vinjé, J.; Estes, M.K.; Esteves, P.; Green, K.Y.; Katayama, K.; Knowles, N.J.; L’homme, Y.; Martella, V.; Vennema, H.; White, P.A.; et al. ICTV Virus Taxonomy Profile: Caliciviridae. J. Gen. Virol. 2019, 100, 1469–1470. [Google Scholar] [CrossRef]
- Barry, A.F.; Durães-Carvalho, R.; Oliveira-Filho, E.F.; Alfieri, A.A.; Van der Poel, W.H.M. High-resolution phylogeny providing insights towards the epidemiology, zoonotic aspects and taxonomy of sapoviruses. Infect. Genet. Evol. 2017, 56, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Pereira-Marques, J.; Hout, A.; Ferreira, R.M.; Weber, M.; Pinto-Ribeiro, I.; van Doorn, L.J.; Knetsch, C.W.; Figueiredo, C. Impact of Host DNA and Sequencing Depth on the Taxonomic Resolution of Whole Metagenome Sequencing for Microbiome Analysis. Front. Microbiol. 2019, 10, 1277. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Cassi, X.; Kohn, T. Comparison of Three Viral Nucleic Acid Preamplification Pipelines for Sewage Viral Metagenomics. Food Environ. Virol. 2024, 16, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Cobbin, J.C.; Charon, J.; Harvey, E.; Holmes, E.C.; Mahar, J.E. Current challenges to virus discovery by meta-transcriptomics. Curr. Opin. Virol. 2021, 51, 48–55. [Google Scholar] [CrossRef]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Popov, N.; Sonets, I.; Evdokimova, A.; Molchanova, M.; Panova, V.; Korneenko, E.; Manolov, A.; Ilina, E. AliMarko: A Pipeline for Virus Identification Using an Expert-Guided Approach. Viruses 2025, 17, 355. https://doi.org/10.3390/v17030355
Popov N, Sonets I, Evdokimova A, Molchanova M, Panova V, Korneenko E, Manolov A, Ilina E. AliMarko: A Pipeline for Virus Identification Using an Expert-Guided Approach. Viruses. 2025; 17(3):355. https://doi.org/10.3390/v17030355
Chicago/Turabian StylePopov, Nikolay, Ignat Sonets, Anastasia Evdokimova, Maria Molchanova, Vera Panova, Elena Korneenko, Alexander Manolov, and Elena Ilina. 2025. "AliMarko: A Pipeline for Virus Identification Using an Expert-Guided Approach" Viruses 17, no. 3: 355. https://doi.org/10.3390/v17030355
APA StylePopov, N., Sonets, I., Evdokimova, A., Molchanova, M., Panova, V., Korneenko, E., Manolov, A., & Ilina, E. (2025). AliMarko: A Pipeline for Virus Identification Using an Expert-Guided Approach. Viruses, 17(3), 355. https://doi.org/10.3390/v17030355






