Reduction of Influenza A Virus Prevalence in Pigs at Weaning After Using Custom-Made Influenza Vaccines in the Breeding Herds of an Integrated Swine Farm System
Abstract
1. Introduction
2. Materials and Methods
2.1. Farm Selection
2.2. Sample Collection and Processing
2.3. Diagnostic Tests
2.3.1. Influenza A Virus rRT-PCR
2.3.2. Cell Culture for Influenza A Virus Isolation
2.3.3. Viral Genetic Sequencing
2.4. Custom Vaccine Strain Selection and Sequence Analysis
- For H3 strains: the 7 key amino acids at positions 144, 155, 156, 158, 159, 189, and 193 [22];
- For H1 strains of swine 1A lineage (alpha, beta, gamma, and pandemic), the 51 amino acids are listed in quotation marks at these antigenic sites: Sa “124,125,153,154,155, 156, 157, 159, 160, 161,162, 163, and 164”; Sb “184, 185, 186, 187, 188, 189, 190, 191, 192, 193, 194, 195, and 196”; Ca1 “166, 67, 168, 169, 202, 203, 204, 205, 235, 236, and 237”; Ca2 “137,138, 139, 140, 141, 142, 143, 221, and 222”; and Cb “70, 71, 72, 73, 74, and 75” [21];
- For H1 strains of human 1B lineage (delta 1a, delta 1b, and delta 2), the 23 putative key amino acids, specifically those at sites 69, 119, 121, 127, 129, 133, 140, 152, 167, 174, 185, 188, 208, 214, 215, 221, 255, 258, 269, 272, 288, 307, and 309 [19].
2.5. Vaccine Manufacturing and Administration
2.6. Data Analysis
3. Results
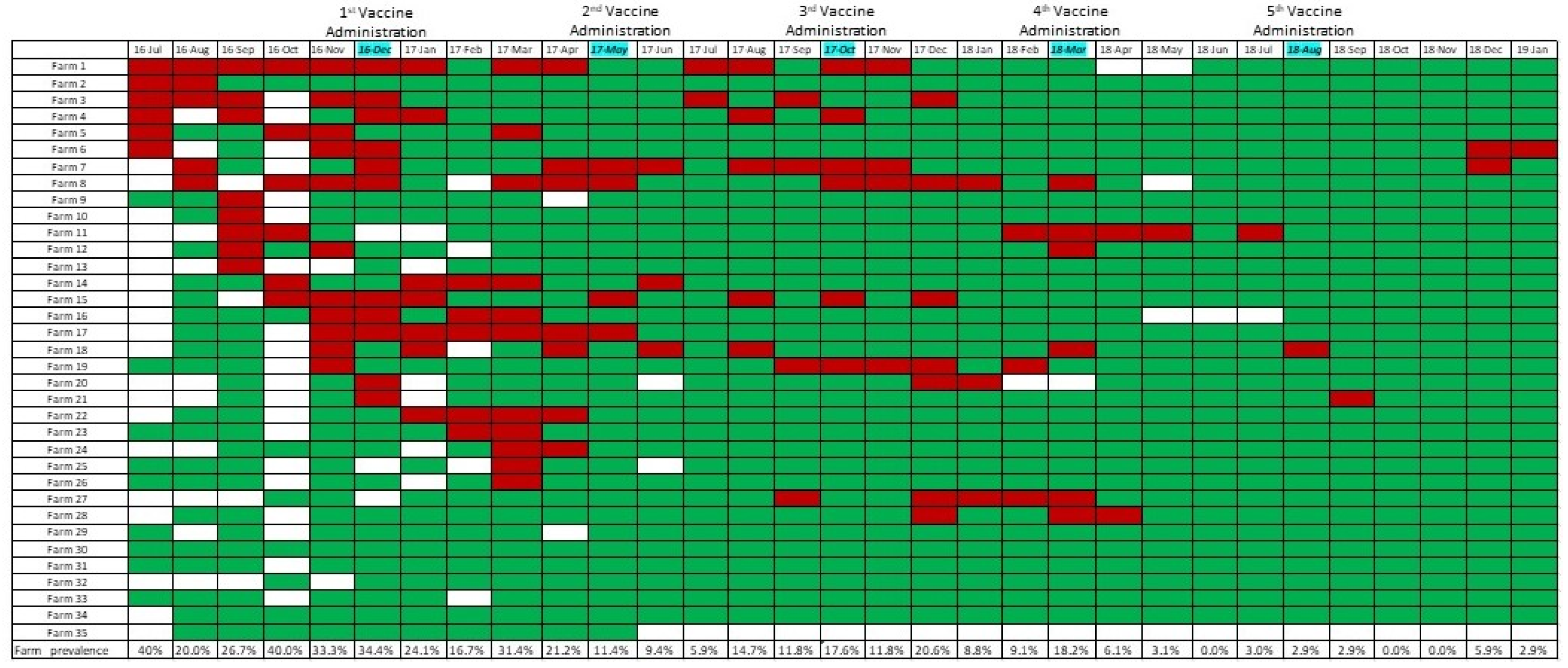
3.1. Summary of IAV rRT-PCR Results and Prevalence Calculations
3.2. Custom-Made Vaccine Composition and Updates
3.3. Effect of Vaccination on IAV Occurrence
3.4. Analyses of Hemagglutinin Protein Amino Acid Sequences and Amino Acids at Antigenic Sites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CPE | Cytopathic effect |
| Ct | Cycle threshold |
| GISRS | Global Influenza Surveillance and Response System |
| GLMM | General linear mixed model |
| HA | Hemagglutinin |
| HI | Hemagglutination inhibition |
| IAV | Influenza A virus |
| IRD | Influenza Research Database |
| MDCK | Madin–Darby canine kidney cells |
| NOVACC | Non-vaccinated |
| PBS | Phosphate-buffered saline |
| PRRSV | Porcine reproductive and respiratory syndrome virus |
| rRT-PCR | reverse transcription, real-time polymerase chain reaction |
| USD | United States dollars |
| WHO | World Health Organization |
References
- Vincent, A.; Van Reek, K. Influenza Viruses. In Diseases of Swine, 11th ed.; John Wiley & Sons: Hoboken, NJ, USA, 2019; pp. 576–593. [Google Scholar]
- Nelson, M.I.; Vincent, A.L.; Kitikoon, P.; Holmes, E.C.; Gramer, M.R. Evolution of novel reassortant A/H3N2 influenza viruses in North American swine and humans, 2009–2011. J. Virol. 2012, 86, 8872–8878. [Google Scholar] [CrossRef]
- Dykhuis Haden, C.; Painter, T.; Fangman, T. Assessing production parameters and economic impact of swine influenza, PRRS and Mycoplasma hyopneumoniae on finishing pigs in a large production system. In Proceedings of the American Association of Swine Veterinarians Annual Meeting, Denver, CO, USA, 2 March 2012; pp. 75–76. [Google Scholar]
- Beaudoin, A.; Johnson, S.; Davies, P.; Bender, J.; Gramer, M. Characterization of Influenza A Outbreaks in Minnesota Swine Herds and Measures Taken to Reduce the Risk of Zoonotic Transmission. Zoonoses Public Health 2012, 59, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Bush, E. Swine 2012 Part III: Changes in the U.S. Swine Industry, 1995−2012; United States Department of Agriculture: Washington, DC, USA, 2015. Available online: https://www.aphis.usda.gov/sites/default/files/swine2012-dr-trends.pdf (accessed on 3 November 2019).
- Cador, C.; Hervé, S.; Andraud, M.; Gorin, S.; Paboeuf, F.; Barbier, N.; Quéguiner, S.; Deblanc, C.; Simon, G.; Rose, N. Maternally-derived antibodies do not prevent transmission of swine influenza A virus between pigs. Vet. Res. 2016, 47, 86. [Google Scholar] [CrossRef] [PubMed]
- Sandbulte, M.R.; Spickler, A.R.; Zaabel, P.K.; Roth, J.A. Optimal Use of Vaccines for Control of Influenza A Virus in Swine. Vaccines 2015, 3, 22–73. [Google Scholar] [CrossRef] [PubMed]
- Electronic Code of Federal Regulations (e-CFR). Title 9, Animals and Animal Products CHAPTER I—Animal and Plant Health Inspection Service, Department of Agriculture, Subchapter E—Viruses, Serums, Toxins, and Analogous Products; Organisms and Vectors. Part 113—Standard Requirements; Government Publishing Office: Washington, DC, USA, 2025.
- Lewis, N.S.; Russell, C.A.; Langat, P.; Anderson, T.K.; Berger, K.; Bielejec, F.; Burke, D.F.; Dudas, G.; Fonville, J.M.; Fouchier, R.A.; et al. The global antigenic diversity of swine influenza A viruses. eLife 2016, 5, e12217. [Google Scholar] [CrossRef] [PubMed]
- de Jong, J.C.; Smith, D.J.; Lapedes, A.S.; Donatelli, I.; Campitelli, L.; Barigazzi, G.; Van Reeth, K.; Jones, T.C.; Rimmelzwaan, G.F.; Osterhaus, A.D.M.E.; et al. Antigenic and genetic evolution of swine influenza A (H3N2) viruses in Europe. J. Virol. 2007, 81, 4315–4322. [Google Scholar] [CrossRef] [PubMed]
- Rajão, D.S.; Pérez, D.R. Universal Vaccines and Vaccine Platforms to Protect against Influenza Viruses in Humans and Agriculture. Front. Microbiol. 2018, 9, 123. [Google Scholar] [CrossRef] [PubMed]
- Corzo, C.A.; Gramer, M.; Kuhn, M.; Mohr, M.; Morrison, R. Observations regarding influenza A virus shedding in a swine breeding farm after mass vaccination. J. Swine Health Prod. 2012, 20, 7. [Google Scholar] [CrossRef] [PubMed]
- Romagosa, A.; Allerson, M.; Gramer, M.; Joo, H.S.; Deen, J.; Detmer, S.; Torremorell, M. Vaccination of influenza a virus decreases transmission rates in pigs. Vet. Res. 2011, 42, 120. [Google Scholar] [CrossRef] [PubMed]
- Chamba Pardo, F.O.; Wayne, S.; Culhane, M.R.; Perez, A.; Allerson, M.; Torremorell, M. Effect of strain-specific maternally-derived antibodies on influenza A virus infection dynamics in nursery pigs. PLoS ONE 2019, 14, e0210700. [Google Scholar] [CrossRef] [PubMed]
- Thompson, R.; Coleman, L.; Culhane, M.; Basynger, A. Elimination of two consecutive swine influenza subtypes in a large breed to wean herd. In Proceedings of the American Association of Swine Veterinarians Annual Meeting, Dallas, TX, USA, 2 March 2014; pp. 221–223. [Google Scholar]
- Tricco, A.C.; Chit, A.; Soobiah, C.; Hallett, D.; Meier, G.; Chen, M.H.; Tashkandi, M.; Bauch, C.T.; Loeb, M. Comparing influenza vaccine efficacy against mismatched and matched strains: A systematic review and meta-analysis. BMC Med. 2013, 11, 153. [Google Scholar] [CrossRef] [PubMed]
- Diaz, A.; Perez, A.; Sreevatsan, S.; Davies, P.; Culhane, M.; Torremorell, M. Association between Influenza A Virus Infection and Pigs Subpopulations in Endemically Infected Breeding Herds. PLoS ONE 2015, 10, e0129213. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, N.M.; Galvani, A.P.; Bush, R.M. Ecological and immunological determinants of influenza evolution. Nature 2003, 422, 428–433. [Google Scholar] [CrossRef]
- WHO Writing Group. Improving influenza vaccine virus selection: Report of a WHO informal consultation held at WHO headquarters, Geneva, Switzerland, 14–16 June 2010. Influenza Other Respir. Viruses 2013, 7 (Suppl. 2), 52–53. [Google Scholar] [CrossRef]
- Rajao, D.S.; Anderson, T.K.; Kitikoon, P.; Stratton, J.; Lewis, N.S.; Vincent, A.L. Antigenic and genetic evolution of contemporary swine H1 influenza viruses in the United States. Virology 2018, 518, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Caton, A.J.; Brownlee, G.G.; Yewdell, J.W.; Gerhard, W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 1982, 31 Pt 1, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Abente, E.J.; Santos, J.; Lewis, N.S.; Gauger, P.C.; Stratton, J.; Skepner, E.; Anderson, T.K.; Rajao, D.S.; Perez, D.R.; Vincent, A.L. The Molecular Determinants of Antibody Recognition and Antigenic Drift in the H3 Hemagglutinin of Swine Influenza A Virus. J. Virol. 2016, 90, 8266–8280. [Google Scholar] [CrossRef]
- Xie, H.; Wan, X.F.; Ye, Z.; Plant, E.P.; Zhao, Y.; Xu, Y.; Li, X.; Finch, C.; Zhao, N.; Kawano, T.; et al. H3N2 Mismatch of 2014-15 Northern Hemisphere Influenza Vaccines and Head-to-head Comparison between Human and Ferret Antisera derived Antigenic Maps. Sci. Rep. 2015, 5, 15279. [Google Scholar] [CrossRef] [PubMed]
- Sow and Gilt Management Manual. Pig Improvement Company—PIC. 2015. Available online: http://na.picgenus.com/sites/genuspic_com/Uploads/sowgilt_manual.pdf (accessed on 22 September 2024).
- Holtkamp, D.J.; Polson, D.D.; Torremorell, M.; Morrison, B.; Classen, D.M.; Becton, L.; Henry, S.; Rodibaugh, M.T.; Rowland, R.R.; Snelson, H.; et al. Terminologie zur Klassifizierung des PRRSV-Status von Schweineherden. Tierarztl Prax Ausg G Grosstiere Nutztiere 2011, 39, 101–112. [Google Scholar] [CrossRef]
- Lee, C.; Allerson, M.; Torremorell, M. The impact of pooling nasal swabs on the diagnostic sensitivity of influrnza A virus RRT-PCR. In Proceedings of the American Association of Swine Veterinarians Annual Meeting, Dallas, TX, USA, 2 March 2014; 299p. [Google Scholar]
- Garrido-Mantilla, J.; Alvarez, J.; Culhane, M.; Nirmala, J.; Cano, J.P.; Torremorell, M. Comparison of individual, group and environmental sampling strategies to conduct influenza surveillance in pigs. BMC Vet. Res. 2019, 15, 61. [Google Scholar] [CrossRef] [PubMed]
- Spackman, E.; Senne, D.A.; Bulaga, L.L.; Myers, T.J.; Perdue, M.L.; Garber, L.P.; Lohman, K.; Daum, L.T.; Suarez, D.L. Development of Real-Time RT-PCR for the Detection of Avian Influenza Virus. Avian Dis. 2003, 47, 1079–1082. [Google Scholar] [CrossRef] [PubMed]
- Animal and Plant Health Inspection Service, United States Department of Agriculture, 2007. pp. 721–724. Available online: https://www.govinfo.gov/content/pkg/CFR-2012-title9-vol1/pdf/CFR-2012-title9-vol1-sec113-113.pdf (accessed on 8 October 2019).
- Gall, A.; Hoffmann, B.; Harder, T.; Grund, C.; Beer, M. Universal Primer Set for Amplification and Sequencing of HA0 Cleavage Sites of All Influenza A Viruses. J. Clin. Microbiol. 2008, 46, 2561–2567. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinforma Oxf. Engl. 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Geneious-SBGrid Consortium—Supported Software. Available online: https://sbgrid.org/software/titles/geneious (accessed on 2 September 2019).
- Zhang, Y.; Aevermann, B.D.; Anderson, T.K.; Burke, D.F.; Dauphin, G.; Gu, Z.; He, S.; Kumar, S.; Larsen, C.N.; Lee, A.J.; et al. Influenza Research Database: An integrated bioinformatics resource for influenza virus research. Nucleic Acids Res. 2017, 45, D466–D474. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Anderson, T.K.; Zeller, M.A.; Gauger, P.C.; Vincent, A.L. octoFLU: Automated Classification for the Evolutionary Origin of Influenza A Virus Gene Sequences Detected in U.S. Swine. Microbiol. Resour. Announc. 2019, 8, e00673-19. [Google Scholar] [CrossRef]
- Zeller, M.A.; Gauger, P.C.; Arendsee, Z.W.; Souza, C.K.; Vincent, A.L.; Anderson, T.K. Machine Learning Prediction and Experimental Validation of Antigenic Drift in H3 Influenza A Viruses in Swine. mSphere 2021, 6, e00920-20. [Google Scholar] [CrossRef] [PubMed]
- Christ, A. Mixed Effects Models and Extensions in Ecology with R. J. Stat. Softw. 2009, 32, 1–3. [Google Scholar] [CrossRef]
- Loeffen, W.L.A.; Heinen, P.P.; Bianchi, A.T.J.; Hunneman, W.A.; Verheijden, J.H.M. Effect of maternally derived antibodies on the clinical signs and immune response in pigs after primary and secondary infection with an influenza H1N1 virus. Vet. Immunol. Immunopathol. 2003, 92, 23–35. [Google Scholar] [CrossRef]
- Osterholm, M.T.; Kelley, N.S.; Sommer, A.; Belongia, E.A. Efficacy and effectiveness of influenza vaccines: A systematic review and meta-analysis. Lancet Infect Dis. 2012, 12, 36–44. [Google Scholar] [CrossRef]
- USDA, 2007. Swine 2006, Part II: Reference of Swine Health and Health Management Practices in the United States, 2006 USDA:APHIS:VS, CEAH. Fort Collins, CO #N479.1207. Available online: https://www.aphis.usda.gov/sites/default/files/swine2006-dr-partii.pdf (accessed on 29 September 2019).
- White, L.A.; Torremorell, M.; Craft, M.E. Influenza A virus in swine breeding herds: Combination of vaccination and biosecurity practices can reduce likelihood of endemic piglet reservoir. Prev. Vet. Med. 2017, 138, 55–69. [Google Scholar] [CrossRef]
- Kitikoon, P.; Vincent, A.; Jones, K.; Nilubol, D.; Yu, S.; Janke, B.; Thacker, B.; Thacker, E. Vaccine efficacy and immune response to swine influenza virus challenge in pigs infected with porcine reproductive and respiratory syndrome virus at the time of SIV vaccination. Vet. Microbiol. 2009, 139, 235–244. [Google Scholar] [CrossRef]
- Chamba Pardo, F.O.; Alba-Casals, A.; Nerem, J.; Morrison, R.B.; Puig, P.; Torremorell, M. Influenza Herd-Level Prevalence and Seasonality in Breed-to-Wean Pig Farms in the Midwestern United States. Front. Vet. Sci. 2017, 4, 167. [Google Scholar] [CrossRef]
- Tamerius, J.; Nelson, M.I.; Zhou, S.Z.; Viboud, C.; Miller, M.A.; Alonso, W.J. Global Influenza Seasonality: Reconciling Patterns across Temperate and Tropical Regions. Environ. Health Perspect. 2011, 119, 439–445. [Google Scholar] [CrossRef]
- Lipsitch, M.; Viboud, C. Influenza seasonality: Lifting the fog. Proc. Natl. Acad. Sci. USA 2009, 106, 3645–3646. [Google Scholar] [CrossRef] [PubMed]
- Monto, A.S.; Malosh, R.E.; Petrie, J.G.; Martin, E.T. The Doctrine of Original Antigenic Sin: Separating Good from Evil. J. Infect Dis. 2017, 215, 1782–1788. [Google Scholar] [CrossRef]
- Sautto, G.A.; Kirchenbaum, G.A.; Ross, T.M. Towards a universal influenza vaccine: Different approaches for one goal. Virol. J. 2018, 15, 17. [Google Scholar] [CrossRef]
- Webster, R.G.; Laver, W.G.; Air, G.M.; Schild, G.C. Molecular mechanisms of variation in influenza viruses. Nature 1982, 296, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Bolton, M.J.; Abente, E.J.; Venkatesh, D.; Stratton, J.A.; Zeller, M.; Anderson, T.K.; Lewis, N.S.; Vincent, A.L. Antigenic evolution of H3N2 influenza A viruses in swine in the United States from 2012 to 2016. Influenza Other Respir Viruses 2019, 13, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Soundararajan, V.; Tharakaraman, K.; Raman, R.; Raguram, S.; Shriver, Z.; Sasisekharan, V.; Sasisekharan, R. Extrapolating from sequence--the 2009 H1N1 ‘swine’ influenza virus. Nat. Biotechnol. 2009, 27, 510–513. [Google Scholar] [CrossRef]

| Vaccine Name | Vaccine Code | Strain Code | Strain Collection Date | Subtype, US Clade, Global Clade (and Amino Acid Motif of H3) | Vaccine Administration/ Season |
|---|---|---|---|---|---|
| A | 1317 | 1A | 22-March-2016 | H1N1, gamma, 1A.3.3.3 | Fall 2016 (n = 30) * Spring 2017 (n = 32) Spring 2018 (n = 2) |
| 2A | 20-January-2016 | H1N2, delta2, 1B.2.1 | |||
| 3A | 21-April-2016 | H3N2, IVA, 3.1990.4.1 (NYKNYSS) | |||
| B | 1350 NEW | 1B | 10-January-2017 | H1N2 delta2 1B.2.1 | Fall 2017 (n = 31) Spring 2018 (n = 9) |
| 2B | 17-November-2016 | H3N2 IVA 3.1990.4.1 (KYNNYKY) | |||
| C | 1379 | 1C | 02-May-2016 | H1N1gamma 1A3.3.3 | Spring 2018 (n = 21) Fall 2018 (n = 32) |
| 1B | 10-January-2017 | H1N2 delta 2 1B.2.1 | |||
| 2B | 17-November-2016 | H3N2 IVA 3.1990.4.1 (KYNNYKY) |
| Vaccine Administration | Vaccination Date | Vaccine Used | No. Pos/No. Tot—Before (%) | No. Pos/No. Tot—After (%) | p Value |
|---|---|---|---|---|---|
| First | 16-December | A | 170/1050 (16.2) | 125/965 (13) | 0.381 |
| Second | 17-May | A | 114/978 (11.7) | 26/975 (2.7) | 0.003 |
| Third | 17-October | B | 30/975 (3.1) | 67/1004 (6.7) | 0.099 |
| Fourth | 18-March | A, B, C | 30/975 (5.1) | 9/407 (2.2) | 0.008 |
| Fifth | 18-August | C | 3/366 (0.8) | 8/421 (1.9) | 0.824 |
| Vaccine Administration | Vaccination Year-Month | N Herds | Vaccine Used (Code) | Time Period Relative to Vaccination | Estimated Proportions of IAV- Positive Samples | 95% CI | p Value |
|---|---|---|---|---|---|---|---|
| First | 2016-December | 6 | NO VACC | Before | 4.41% | 0.80–21.11 | <0.0001 |
| After | 16.24% | 3.32–52.31 | |||||
| 29 | A | Before | 7.15% | 2.96–16.27 | <0.0001 | ||
| After | 2.30% | 0.89–5.78 | |||||
| Second | 2017-May | 4 | NO VACC | Before | 1.78% | 0.10–28.52 | 0.34 |
| After | 0.70% | 0.02–13.59 | |||||
| 31 | A | Before | 1.99% | 0.53–7.10 | <0.0001 | ||
| After | 0.30% | 0.07–1.13 | |||||
| Third | 2017-October | 2 | NO VACC | Before | 3.10% | 0.05–63.66 | 0.99 |
| After | 4.85% | 0.09–72.55 | |||||
| 31 | B | Before | 0.01% | 0.02–0.75 | <0.0001 | ||
| After | 0.05% | 0.08–2.75 | |||||
| Fourth | 2018-March | 5 | NO VACC | Before | NA * | NA | NA |
| After | NA | NA | |||||
| 2 | A | Before | 2.68 | 0.03–68.90 | 0.80 | ||
| After | 1.15 | 0.01–49.91 | |||||
| 9 | B | Before | NA | NA | NA | ||
| After | NA | NA | |||||
| 18 | C | Before | NA | NA | NA | ||
| After | NA | NA | |||||
| Fifth | 2018-August | 1 | NO VACC | Before | NA | NA | NA |
| After | NA | NA | |||||
| 34 | C | Before | NA | NA | |||
| After | NA | NA |
| Vaccine Name | Vaccine Strain (Vaccine Letter Code) | Herd-Strain Subtype | % Nucleotide Identity | % Amino Acid Identity | ||
|---|---|---|---|---|---|---|
| Minimum | Maximum | Minimum | Maximum | |||
| A | H3N2 IVA (3A) | H3 | 94.35% | 99.94% | 95.05% | 100.00% |
| B | H3N2 IVA (2B) | H3 | 86.28% | NA * | 80.07% | 93.98% |
| C | H1N1 gamma 1A3.3.3 (1C) | H1 | 88.54% | 100.00% | 90.64% | 100.00% |
| A | H1N1 gamma 1A3.3.3(1A) | H1 | 90.24% | 99.29% | 91.70% | 99.12% |
| C | H1N2 delta 2 1B.2.1 (1B) | H1 | 94.80% | 99.86% | 93.44% | 100.00% |
| A | H1N2 delta 2 1B.2.1 (2A) | H1 | 94.35% | 99.85% | 93.98% | 100.00% |
| B | H1N2 delta2 1B.2.1 (1B) | H1 | 96.09% | 96.35% | 95.93% | 96.28% |
| Vaccine Name | Vaccine Number Code | Vaccine Strain/Antigenic Site | Minimum Number of a.a. Differences/ Number of Key a.a. Sites Evaluated (%) | Maximum Number of a.a. Differences/ Number of Key a.a. Sites Evaluated (%) | |
|---|---|---|---|---|---|
| A | 1317 | H3 IVA_3.1990.4.1 | 1/7 (14%) | 5/7 (71%) | |
| B | 1350 NEW | H3 IVA_3.1990.4.1 | 4/7 (57%) | ||
| C | 1379 | H1 gamma_1A.3.3.3 | Sa antigenic site | 2/13 (15%) | 4/13 (31%) |
| Sb antigenic site | 2/13 (15%) | 4/13 (31%) | |||
| Cb antigenic site | 2/6 (33%) | ||||
| Ca1 antigenic site | 2/11 (18%) | ||||
| Ca2 antigenic site | 0/9 (0%) | 3/9 (33%) | |||
| A | 1317 | H1 gamma_1A.3.3.3 | Sa antigenic site | 2/13 (15%) | 4/13 (31%) |
| Sb antigenic site | 2/13 (15%) | 4/13 (31%) | |||
| Cb antigenic site | 1/6 (17%) | 2/6 (33%) | |||
| Ca1 antigenic site | 0/11 (0%) | 4/11 (36%) | |||
| Ca2 antigenic site | 0/9 (0%) | 2/9 (22%) | |||
| A | 1317 | H1 delta2_1B.2.1 | 1/23 (4%) | 13/23 (57%) | |
| B | 1350 NEW | H1 delta2_1B.2.1 | 1/23 (4%) | 7/23 (30%) | |
| Strain/Clade | Amino Acid Differences | Number of Sequences/Total Sequences * | |
|---|---|---|---|
| H3 | N145K | 8/10 | |
| N156K | 3/10 | ||
| K189S | 5/10 | ||
| Y193S | 3/10 | ||
| H1 gamma_1A.3.3.3 | Sa antigenic site | E155G | 4/27 |
| Q163K | 5/27 | ||
| Sb antigenic site | T185S | 5/27 | |
| Cb antigenic site | S71Y | 2/27 | |
| Ca1 antigenic site | D168N | 5/27 | |
| T203S | 5/27 | ||
| Ca2 antigenic site | A141T | 5/27 | |
| K142N | 12/27 | ||
| H1 delta2_1B.2.1 | G121D | 10/27 | |
| V152E | 24/27 | ||
| A214V | 13/27 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garrido-Mantilla, J.; Sanhueza, J.; Alvarez, J.; Pittman, J.S.; Davies, P.; Torremorell, M.; Culhane, M.R. Reduction of Influenza A Virus Prevalence in Pigs at Weaning After Using Custom-Made Influenza Vaccines in the Breeding Herds of an Integrated Swine Farm System. Viruses 2025, 17, 240. https://doi.org/10.3390/v17020240
Garrido-Mantilla J, Sanhueza J, Alvarez J, Pittman JS, Davies P, Torremorell M, Culhane MR. Reduction of Influenza A Virus Prevalence in Pigs at Weaning After Using Custom-Made Influenza Vaccines in the Breeding Herds of an Integrated Swine Farm System. Viruses. 2025; 17(2):240. https://doi.org/10.3390/v17020240
Chicago/Turabian StyleGarrido-Mantilla, Jorge, Juan Sanhueza, Julio Alvarez, Jeremy S. Pittman, Peter Davies, Montserrat Torremorell, and Marie R. Culhane. 2025. "Reduction of Influenza A Virus Prevalence in Pigs at Weaning After Using Custom-Made Influenza Vaccines in the Breeding Herds of an Integrated Swine Farm System" Viruses 17, no. 2: 240. https://doi.org/10.3390/v17020240
APA StyleGarrido-Mantilla, J., Sanhueza, J., Alvarez, J., Pittman, J. S., Davies, P., Torremorell, M., & Culhane, M. R. (2025). Reduction of Influenza A Virus Prevalence in Pigs at Weaning After Using Custom-Made Influenza Vaccines in the Breeding Herds of an Integrated Swine Farm System. Viruses, 17(2), 240. https://doi.org/10.3390/v17020240








