ZPR1 Is Dispensable for HPV R-Loop Resolution but Regulates Host R-Loop Dynamics
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmids and Antibodies
2.2. Cell Culture
2.3. DNA-RNA Co-Immunoprecipitation (DRIP)
2.4. Viral Transcription Analysis
2.5. Chromatin Immunoprecipitation (ChIP) Assay
2.6. Co-Immunoprecipitation (Co-IP)
2.7. Exonuclease V Digestion
2.8. SETX and ZPR1 Expression Analysis
2.9. Statistical Analysis
2.10. Data Availability
3. Results
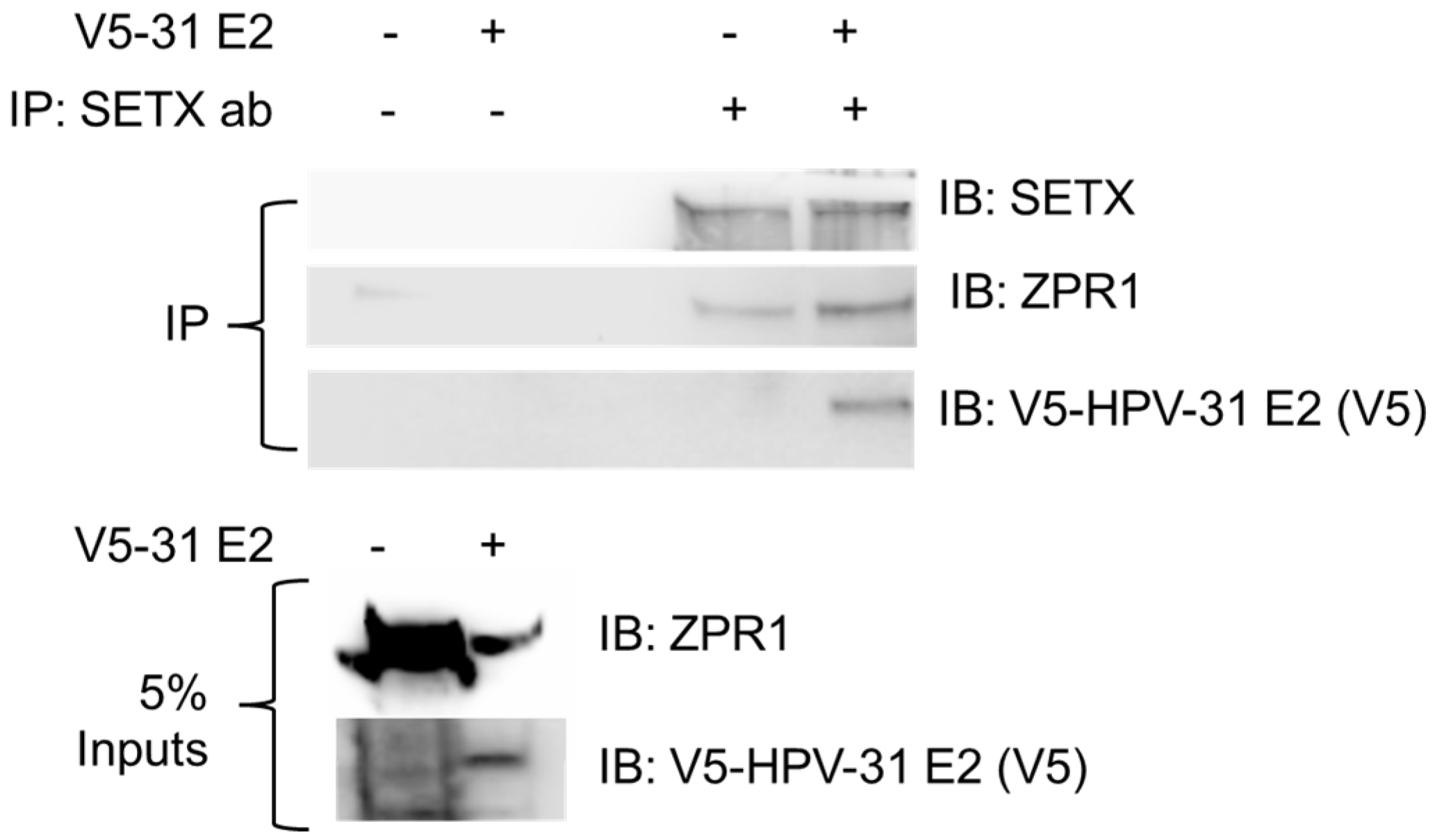
3.1. ZPR1 Is in Complex with SETX-E2
3.2. ZPR1 Depletion Decreases HPV R-Loops and Increases SETX Loading to the Viral Promoter
3.3. E2 Associates with RNA-DNA Hybrid-Containing Chromatin
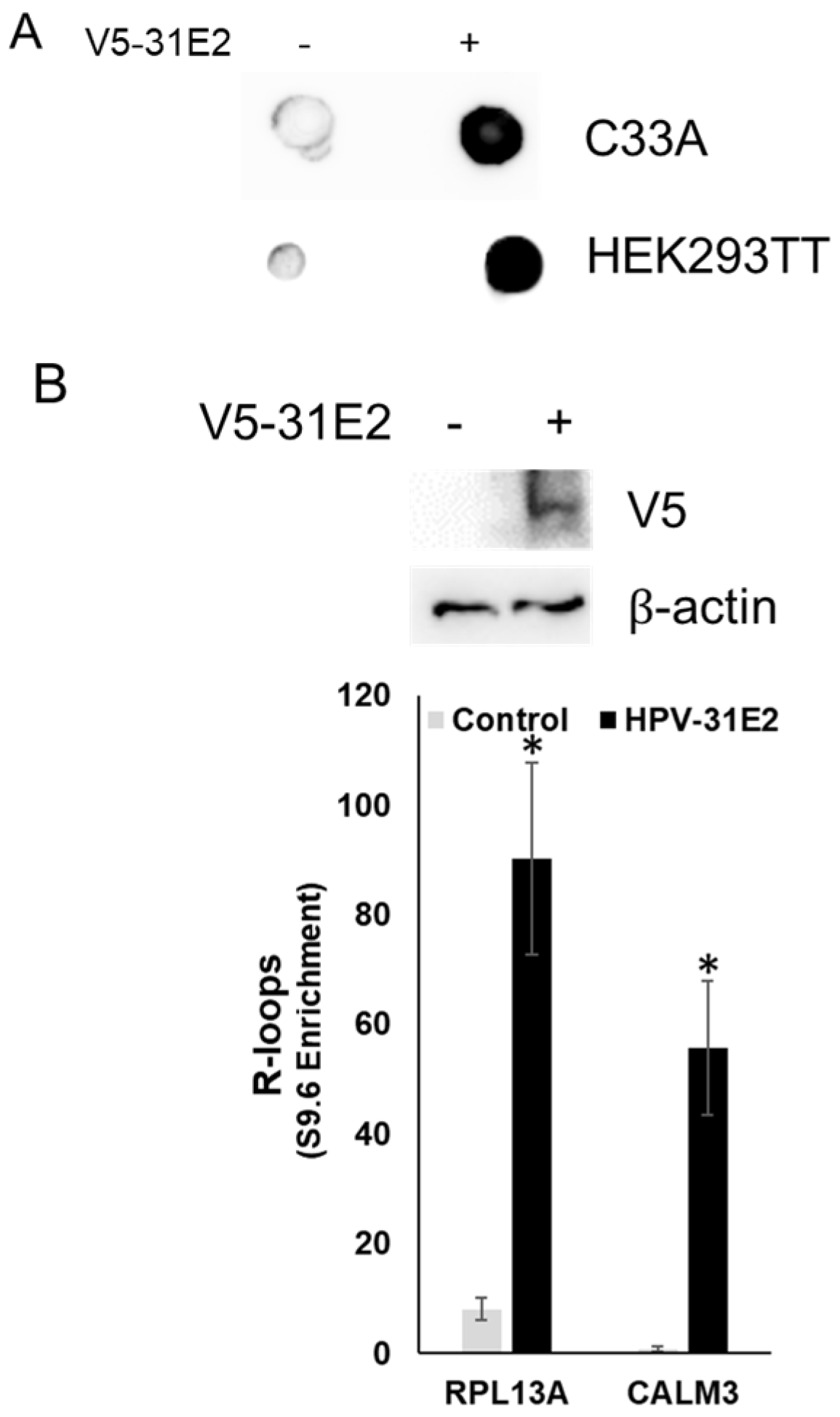
3.4. E2 Overexpression Increases Host R-Loop Formation
3.5. ZPR1, but Not SETX Levels Are Lower in HPV Positive Cervical and Head And Neck Cancers
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Allison, D.F.; Wang, G.G. R-loops: Formation, function, and relevance to cell stress. Cell Stress 2019, 3, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Niehrs, C.; Luke, B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol. 2020, 21, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Jose, L.; Smith, K.; Crowner, A.; Androphy, E.J.; DeSmet, M. Senataxin mediates R-loop resolution on HPV episomes. J. Virol. 2024, 98, e0100324. [Google Scholar] [CrossRef]
- Templeton, C.W.; Laimins, L.A. p53-dependent R-loop formation and HPV pathogenesis. Proc. Natl. Acad. Sci. USA 2023, 120, e2305907120. [Google Scholar] [CrossRef]
- Templeton, C.W.; Laimins, L.A. HPV induced R-loop formation represses innate immune gene expression while activating DNA damage repair pathways. PLoS Pathog. 2024, 20, e1012454. [Google Scholar] [CrossRef]
- Crowner, A.; Smith, K.; DeSmet, M. Regulation of R-Loops in DNA Tumor Viruses. Pathogens 2024, 13, 863. [Google Scholar] [CrossRef]
- Wongsurawat, T.; Gupta, A.; Jenjaroenpun, P.; Owens, S.; Forrest, J.C.; Nookaew, I. R-loop-forming Sequences Analysis in Thousands of Viral Genomes Identify A New Common Element in Herpesviruses. Sci. Rep. 2020, 10, 6389. [Google Scholar] [CrossRef]
- Rennekamp, A.J.; Lieberman, P.M. Initiation of Epstein-Barr virus lytic replication requires transcription and the formation of a stable RNA-DNA hybrid molecule at OriLyt. J. Virol. 2011, 85, 2837–2850. [Google Scholar] [CrossRef]
- Yiu, S.P.T.; Guo, R.; Zerbe, C.; Weekes, M.P.; Gewurz, B.E. Epstein-Barr virus BNRF1 destabilizes SMC5/6 cohesin complexes to evade its restriction of replication compartments. Cell Rep. 2022, 38, 110411. [Google Scholar] [CrossRef]
- Skourti-Stathaki, K.; Proudfoot, N.J.; Gromak, N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell 2011, 42, 794–805. [Google Scholar] [CrossRef]
- Mischo, H.E.; Gómez-González, B.; Grzechnik, P.; Rondón, A.G.; Wei, W.; Steinmetz, L.; Aguilera, A.; Proudfoot, N.J. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol. Cell 2011, 41, 21–32. [Google Scholar] [CrossRef]
- Alzu, A.; Bermejo, R.; Begnis, M.; Lucca, C.; Piccini, D.; Carotenuto, W.; Saponaro, M.; Brambati, A.; Cocito, A.; Foiani, M.; et al. Senataxin Associates with Replication Forks to Protect Fork Integrity across RNA-Polymerase-II-Transcribed Genes. Cell 2012, 151, 835–846. [Google Scholar] [CrossRef] [PubMed]
- Brambati, A.; Zardoni, L.; Achar, Y.J.; Piccini, D.; Galanti, L.; Colosio, A.; Foiani, M.; Liberi, G. Dormant origins and fork protection mechanisms rescue sister forks arrested by transcription. Nucleic Acids Res. 2018, 46, 1227–1239. [Google Scholar] [CrossRef]
- Gatti, V.; De Domenico, S.; Melino, G.; Peschiaroli, A. Senataxin and R-loops homeostasis: Multifaced implications in carcinogenesis. Cell Death Discov. 2023, 9, 145. [Google Scholar] [CrossRef] [PubMed]
- Morgan, I.M. The functions of papillomavirus E2 proteins. Virology 2025, 603, 110387. [Google Scholar] [CrossRef] [PubMed]
- Kannan, A.; Jiang, X.; He, L.; Ahmad, S.; Gangwani, L. ZPR1 prevents R-loop accumulation, upregulates SMN2 expression and rescues spinal muscular atrophy. Brain 2020, 143, 69–93. [Google Scholar] [CrossRef]
- Kannan, A.; Gangadharan Leela, S.; Branzei, D.; Gangwani, L. Role of senataxin in R-loop-mediated neurodegeneration. Brain Commun. 2024, 6, fcae239. [Google Scholar] [CrossRef]
- Kannan, A.; Cuartas, J.; Gangwani, P.; Branzei, D.; Gangwani, L. Mutation in senataxin alters the mechanism of R-loop resolution in amyotrophic lateral sclerosis 4. Brain 2022, 145, 3072–3094. [Google Scholar] [CrossRef]
- Perego, M.G.L.; Taiana, M.; Bresolin, N.; Comi, G.P.; Corti, S. R-Loops in Motor Neuron Diseases. Mol. Neurobiol. 2019, 56, 2579–2589. [Google Scholar] [CrossRef]
- Cuartas, J.; Gangwani, L. R-loop Mediated DNA Damage and Impaired DNA Repair in Spinal Muscular Atrophy. Front. Cell. Neurosci. 2022, 16, 826608. [Google Scholar] [CrossRef]
- Simon, C.M.; Dai, Y.; Van Alstyne, M.; Koutsioumpa, C.; Pagiazitis, J.G.; Chalif, J.I.; Wang, X.; Rabinowitz, J.E.; Henderson, C.E.; Pellizzoni, L.; et al. Converging Mechanisms of p53 Activation Drive Motor Neuron Degeneration in Spinal Muscular Atrophy. Cell Rep. 2017, 21, 3767–3780. [Google Scholar] [CrossRef]
- Kannan, A.; Bhatia, K.; Branzei, D.; Gangwani, L. Combined deficiency of Senataxin and DNA-PKcs causes DNA damage accumulation and neurodegeneration in spinal muscular atrophy. Nucleic Acids Res. 2018, 46, 8326–8346. [Google Scholar] [CrossRef] [PubMed]
- Jose, L.; Androphy, E.J.; DeSmet, M. SETD6 Regulates E2-Dependent Human Papillomavirus Transcription. J. Virol. 2022, 96, e0129522. [Google Scholar] [CrossRef] [PubMed]
- Evans, A.M.; Salnikov, M.; Tessier, T.M.; Mymryk, J.S. Reduced MHC Class I and II Expression in HPV-Negative vs. HPV-Positive Cervical Cancers. Cells 2022, 11, 3911. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Gopaulakrishnan, S.; Pollack, S.; Stubbs, B.J.; Pagès, H.; Readey, J.; Davis, S.; Waldron, L.; Morgan, M.; Carey, V. restfulSE: A semantically rich interface for cloud-scale genomics with Bioconductor. F1000Research 2019, 8, 21. [Google Scholar] [CrossRef]
- Broatch, J.E.; Dietrich, S.; Goelman, D. Introducing Data Science Techniques by Connecting Database Concepts and dplyr. J. Stat. Educ. 2019, 27, 147–153. [Google Scholar] [CrossRef]
- Skidmore, Z.L.; Wagner, A.H.; Lesurf, R.; Campbell, K.M.; Kunisaki, J.; Griffith, O.L.; Griffith, M. GenVisR: Genomic Visualizations in R. Bioinformatics 2016, 32, 3012–3014. [Google Scholar] [CrossRef]
- Li, F.; Zafar, A.; Luo, L.; Denning, A.M.; Gu, J.; Bennett, A.; Yuan, F.; Zhang, Y. R-Loops in Genome Instability and Cancer. Cancers 2023, 15, 4986. [Google Scholar] [CrossRef]
- Massimi, P.; Pim, D.; Bertoli, C.; Bouvard, V.; Banks, L. Interaction between the HPV-16 E2 transcriptional activator and p53. Oncogene 1999, 18, 7748–7754. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moffitt, R.; Brooks, S.; Androphy, E.J.; DeSmet, M. ZPR1 Is Dispensable for HPV R-Loop Resolution but Regulates Host R-Loop Dynamics. Viruses 2025, 17, 1502. https://doi.org/10.3390/v17111502
Moffitt R, Brooks S, Androphy EJ, DeSmet M. ZPR1 Is Dispensable for HPV R-Loop Resolution but Regulates Host R-Loop Dynamics. Viruses. 2025; 17(11):1502. https://doi.org/10.3390/v17111502
Chicago/Turabian StyleMoffitt, Rylann, Steven Brooks, Elliot J. Androphy, and Marsha DeSmet. 2025. "ZPR1 Is Dispensable for HPV R-Loop Resolution but Regulates Host R-Loop Dynamics" Viruses 17, no. 11: 1502. https://doi.org/10.3390/v17111502
APA StyleMoffitt, R., Brooks, S., Androphy, E. J., & DeSmet, M. (2025). ZPR1 Is Dispensable for HPV R-Loop Resolution but Regulates Host R-Loop Dynamics. Viruses, 17(11), 1502. https://doi.org/10.3390/v17111502





