Comparative Analysis Reveals Host Species-Dependent Diversity Among 16 Virulent Bacteriophages Isolated Against Soybean Bradyrhizobium spp.
Abstract
1. Introduction
2. Materials and Methods
2.1. Lytic Phage Isolation
2.2. Lysate Preparation and Titering
2.3. Electron Microscopy of Phages
2.4. Host Range Determination
2.5. DNA Extraction and Sequencing
2.6. Assembly and Annotation of Lytic Phage Genomes
2.7. Taxonomic Classification
2.8. Average Nucleotide Identity and Coverage
2.9. Genome Maps and Alignments
2.10. Geographical Analysis
3. Results
3.1. Lytic Phage Isolation
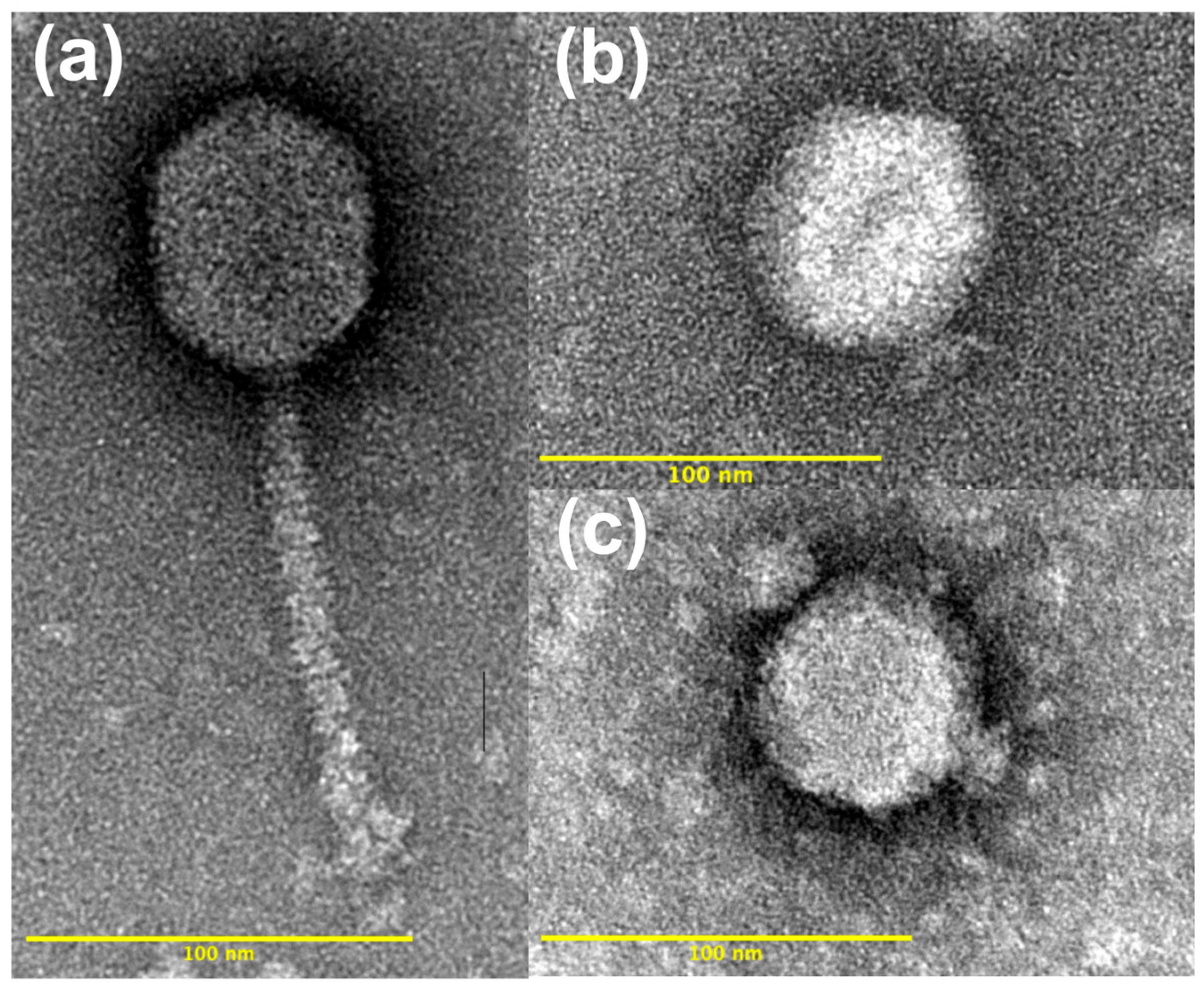
3.2. Transmission Electron Microscopy and Phage Morphology
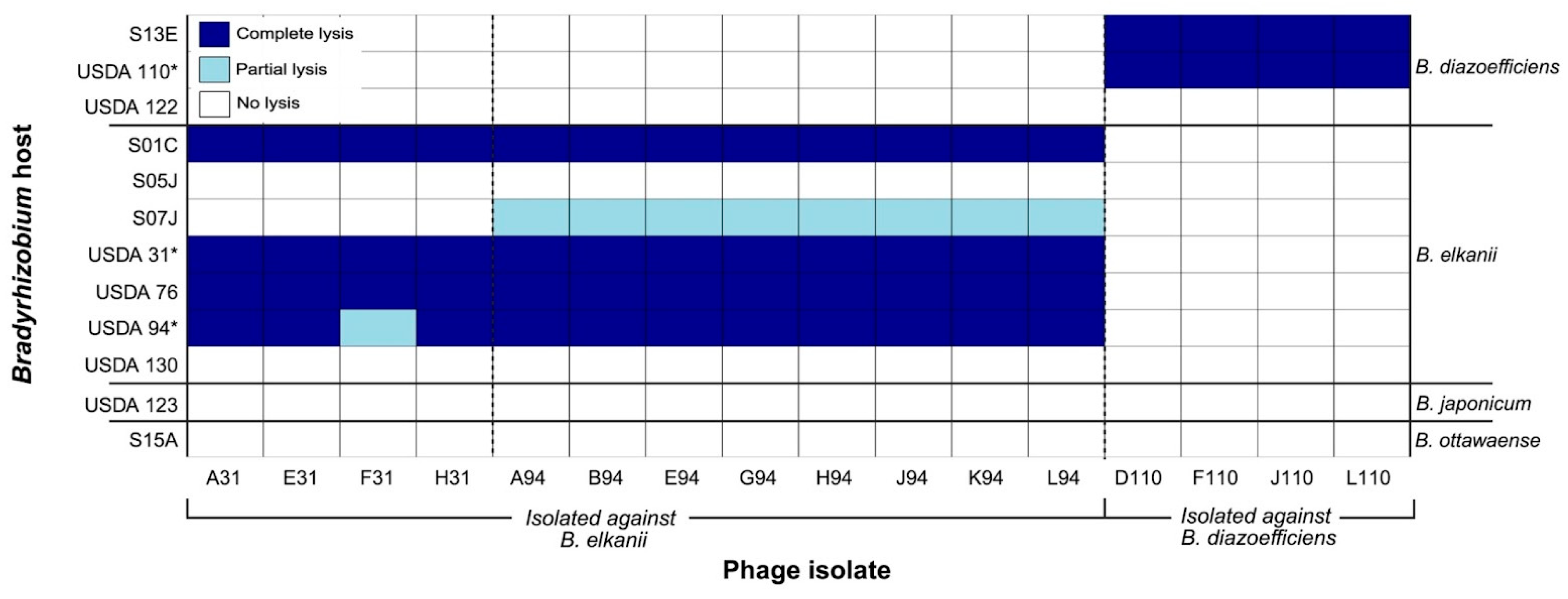
3.3. Host Range
3.4. Genome Sequencing
3.5. Taxonomic Classification
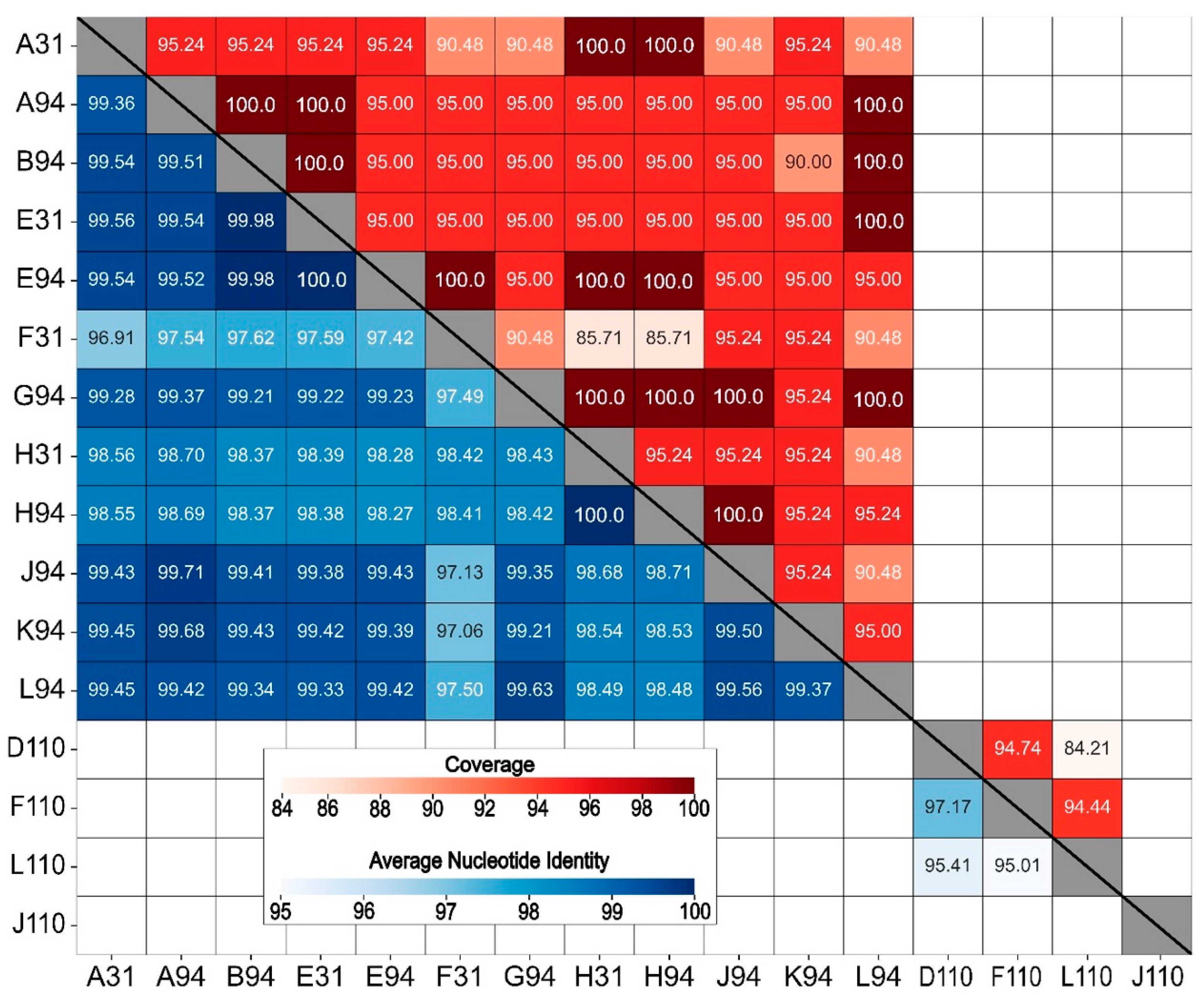
3.6. Average Nucleotide Identity
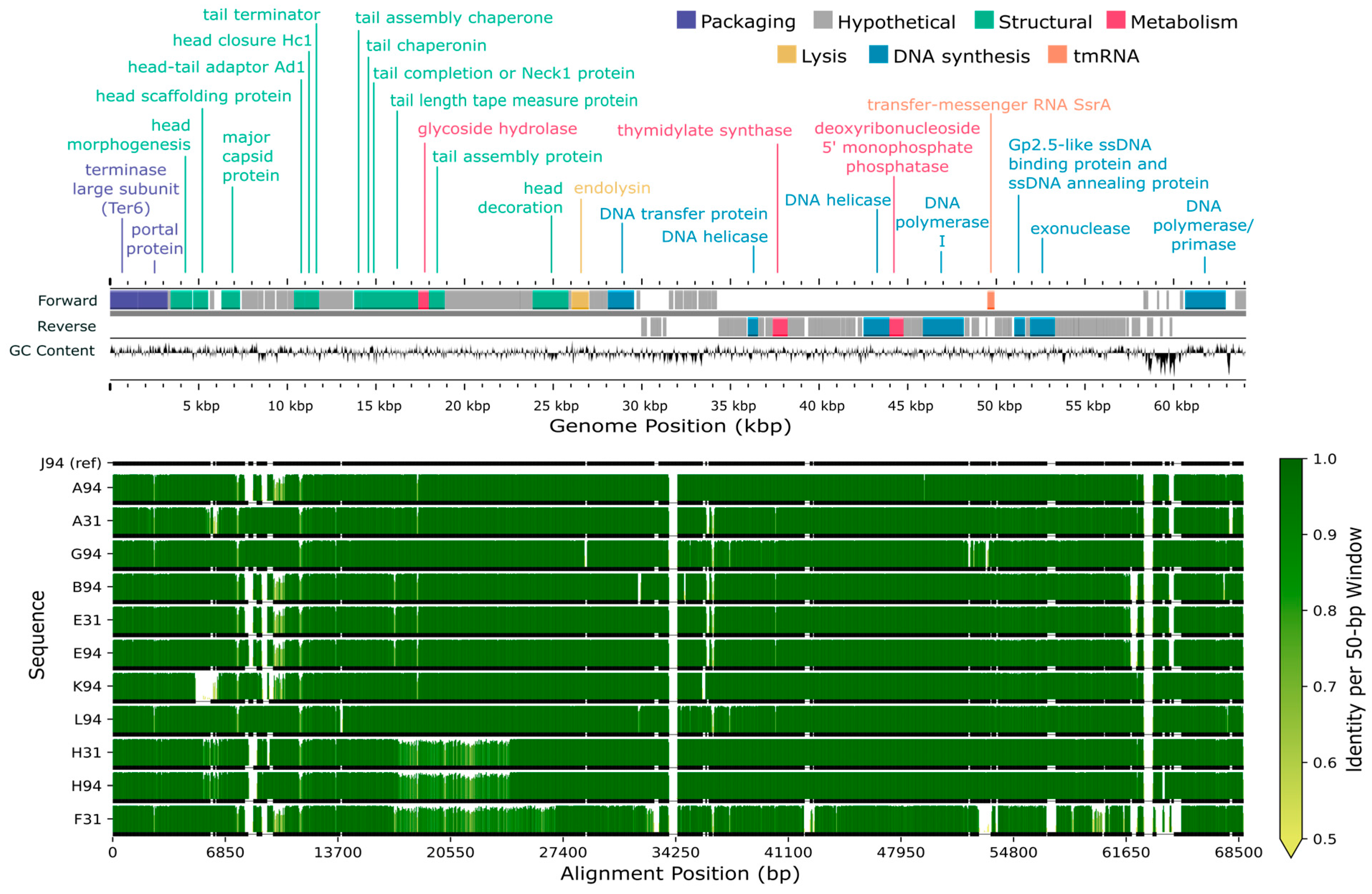
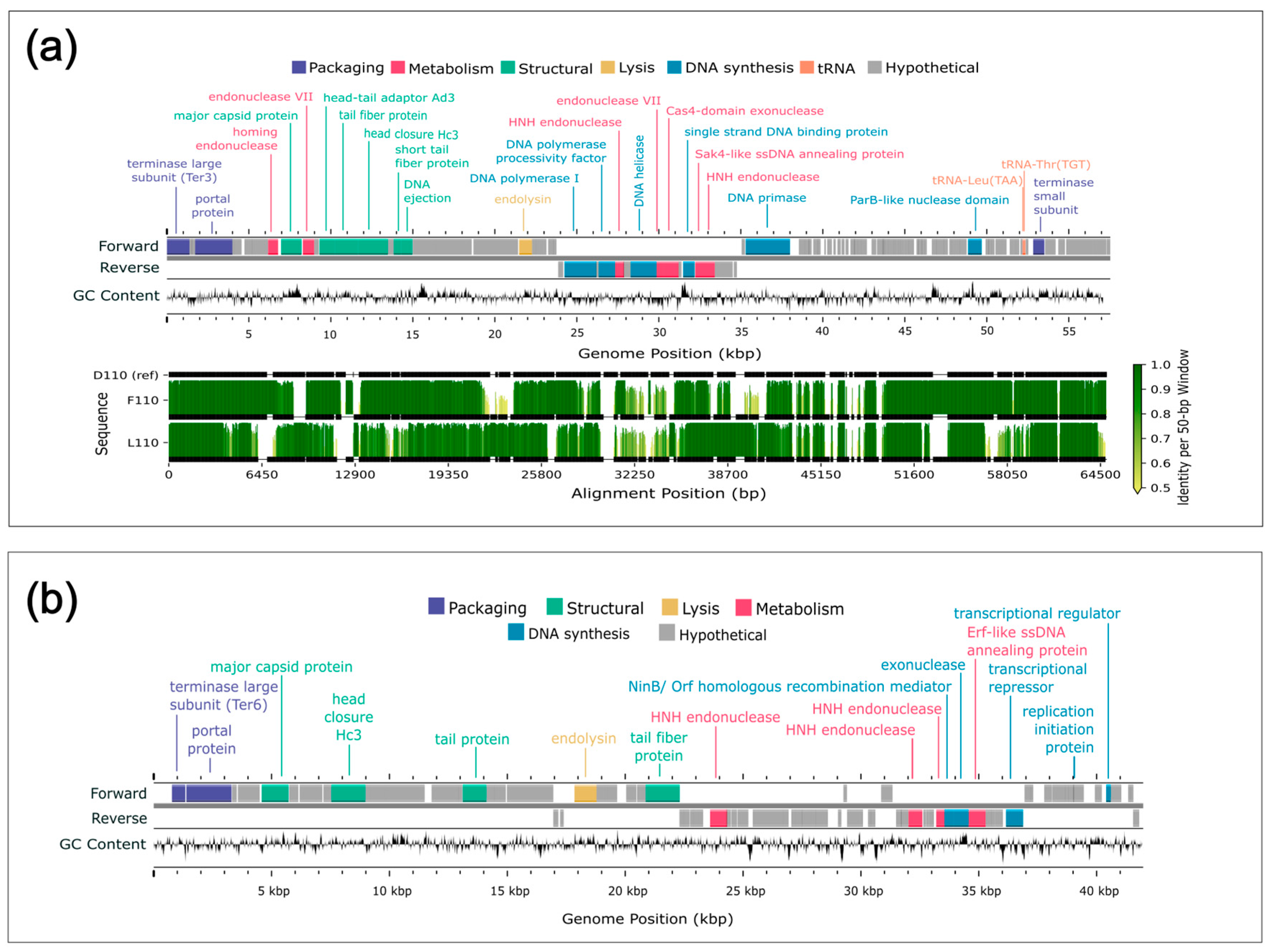
3.7. Gene Prediction and Annotation
3.8. Comparative Genome Alignments
3.9. Geographical Analysis
4. Discussion
4.1. Phage Isolation and Host Range
4.2. Morphology
4.3. Genome Composition
4.4. Comparative Genomics
4.5. Conserved Genes and Genetic Diversity
4.6. Functional Insights into terL and polA Genes
4.7. Geographic Influence on Phage Diversity
4.8. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Soybeans|USDA Foreign Agricultural Service. Available online: https://www.fas.usda.gov/data/production/commodity/2222000 (accessed on 30 May 2025).
- Sugiyama, A.; Ueda, Y.; Takase, H.; Yazaki, K. Do Soybeans Select Specific Species of Bradyrhizobium during Growth? Commun. Integr. Biol. 2015, 8, e992734. [Google Scholar] [CrossRef]
- Messina, M. Perspective: Soybeans Can Help Address the Caloric and Protein Needs of a Growing Global Population. Front. Nutr. 2022, 9, 909464. [Google Scholar] [CrossRef]
- Shober, A.L.; Taylor, R. Nitrogen Management for Soybeans|Cooperative Extension|University of Delaware. Available online: https://www.udel.edu/academics/colleges/canr/cooperative-extension/fact-sheets/nitrogen-management-soybeans/ (accessed on 30 May 2025).
- Tamagno, S.; Sadras, V.O.; Haegele, J.W.; Armstrong, P.R.; Ciampitti, I.A. Interplay between Nitrogen Fertilizer and Biological Nitrogen Fixation in Soybean: Implications on Seed Yield and Biomass Allocation. Sci. Rep. 2018, 8, 17502. [Google Scholar] [CrossRef]
- Menegat, S.; Ledo, A.; Tirado, R. Greenhouse Gas Emissions from Global Production and Use of Nitrogen Synthetic Fertilisers in Agriculture. Sci. Rep. 2022, 12, 14490. [Google Scholar] [CrossRef] [PubMed]
- Norton, J.; Ouyang, Y. Controls and Adaptive Management of Nitrification in Agricultural Soils. Front. Microbiol. 2019, 10, 449199. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Beusen, A.H.W.; Middelburg, J.J. Surface-Water Nitrate Exposure to World Populations Has Expanded and Intensified during 1970–2010. Environ. Sci. Technol. 2023, 57, 19395–19406. [Google Scholar] [CrossRef] [PubMed]
- Paerl, H.W. Coastal Eutrophication and Harmful Algal Blooms: Importance of Atmospheric Deposition and Groundwater as “New” Nitrogen and Other Nutrient Sources. Limnol. Oceanogr. 1997, 42, 1154–1165. [Google Scholar] [CrossRef]
- Tian, H.; Xu, R.; Canadell, J.G.; Thompson, R.L.; Winiwarter, W.; Suntharalingam, P.; Davidson, E.A.; Ciais, P.; Jackson, R.B.; Janssens-Maenhout, G.; et al. A Comprehensive Quantification of Global Nitrous Oxide Sources and Sinks. Nature 2020, 586, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Fuhrmann, J.J.; Vasilas, B.L. Field Response of the Glycine-bradyrhizobium Symbiosis to Modified Early-Nodule Occupancy. Soil Biol. Biochem. 1993, 25, 1203–1209. [Google Scholar] [CrossRef]
- Zimmer, S.; Messmer, M.; Haase, T.; Piepho, H.-P.; Mindermann, A.; Schulz, H.; Habekuß, A.; Ordon, F.; Wilbois, K.-P.; Heß, J. Effects of Soybean Variety and Bradyrhizobium Strains on Yield, Protein Content and Biological Nitrogen Fixation under Cool Growing Conditions in Germany. Europ. J. Agron. 2016, 72, 38–46. [Google Scholar] [CrossRef]
- Mushegian, A.R. Are There 1031 virus Particles on Earth, or More, or Fewer? J. Bacteriol. 2020, 202. [Google Scholar] [CrossRef]
- Hendrix, R.W.; Smith, M.C.M.; Burns, R.N.; Ford, M.E.; Hatfull, G.F. Evolutionary Relationships among Diverse Bacteriophages and Prophages: All the World’s a Phage. Proc. Natl. Acad. Sci. USA 1999, 96, 2192–2197. [Google Scholar] [CrossRef]
- Domingo, E. Quasispecies Dynamics in Disease Prevention and Control. Virus Popul. 2016, 263–297. [Google Scholar] [CrossRef]
- Williamson, K.E.; Fuhrmann, J.J.; Wommack, K.E.; Radosevich, M. Viruses in Soil Ecosystems: An Unknown Quantity within an Unexplored Territory. Annu. Rev. Virol. 2017, 4, 201–219. [Google Scholar] [CrossRef]
- Ali, F.S.; Loynachan, T.E.; Hammad, A.M.M.; Aharchi, Y. Polyvirulent Rhizobiophage from a Soybean Rhizosphere Soil. Soil Biol. Biochem. 1998, 30, 2171–2175. [Google Scholar] [CrossRef]
- Appunu, C.; Dhar, B. Morphology and General Characteristics of Lytic Phages Infective on Strains of Bradyrhizobium japonicum. Curr. Microbiol. 2008, 56, 21–27. [Google Scholar] [CrossRef]
- Appunu, C.; Dhar, B. Isolation and Symbiotic Characteristics of Two Tn5-Derived Phage-Resistant Bradyrhizobium japonicum Strains That Nodulate Soybean. Curr. Microbiol. 2008, 57, 212–217. [Google Scholar] [CrossRef]
- Hashem, F.M.; Angle, J.S. Rhizobiophage Effects on Bradyrhizobium japonicum, Nodulation and Soybean Growth. Soil Biol. Biochem. 1988, 20, 69–73. [Google Scholar] [CrossRef]
- Hashem, F.M.; Angle, J.S.; Ristiano, P.A. Isolation and Characterization of Rhizobiophages Specific for Bradyrhizobium japonicum USDA 117. Can. J. Microbiol. 1986, 32, 326–329. [Google Scholar] [CrossRef]
- Msimbira, L.A.; Jaiswal, S.K.; Dakora, F.D. Identification and Characterization of Phages Parasitic on Bradyrhizobia Nodulating Groundnut (Arachis hypogaea L.) in South Africa. Appl. Soil Ecol. 2016, 108, 334–340. [Google Scholar] [CrossRef]
- Shahaby, A.F.; Alharthi, A.A.; El-Tarras, A.E. Characterization of Rhizobiophages Specific for Rhizobium sp. Sinorhizobum sp. and Bradyrhizobium sp. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 155–171. [Google Scholar]
- Kowalski, M.; Ham, G.E.; Frederick, L.R.; Anderson, I.C. Relationship between Strains of Rhizobium japonicum and Their Bacteriophages from Soil and Nodules of Field-Grown Soybeans. Soil Sci. 1974, 118, 221–228. [Google Scholar] [CrossRef]
- Joglekar, P.; Ferrell, B.D.; Jarvis, T.; Haramoto, K.; Place, N.; Dums, J.T.; Polson, S.W.; Wommack, K.E.; Fuhrmann, J.J. Spontaneously Produced Lysogenic Phages Are an Important Component of the Soybean Bradyrhizobium Mobilome. mBio 2023, 14, e0029523. [Google Scholar] [CrossRef] [PubMed]
- Richards, V.A.; Ferrell, B.D.; Polson, S.W.; Wommack, K.E.; Fuhrmann, J.J. Soybean Bradyrhizobium spp. Spontaneously Produce Abundant and Diverse Temperate Phages in Culture. Viruses 2024, 16, 1750. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.; Domingues, R.; Evans, B.; Sutton, J.M.; Adriaenssens, E.M.; Turner, D. Genomic Diversity of Bacteriophages Infecting the Genus Acinetobacter. Viruses 2022, 14, 181. [Google Scholar] [CrossRef]
- Adriaenssens, E.M.; Brister, J.R. How to Name and Classify Your Phage: An Informal Guide. Viruses 2017, 9, 70. [Google Scholar] [CrossRef]
- Turner, D.; Kropinski, A.M.; Adriaenssens, E.M. A Roadmap for Genome-Based Phage Taxonomy. Viruses 2021, 13, 506. [Google Scholar] [CrossRef]
- Ackermann, H.W. Bacteriophage Taxonomy. Microbiol. Aust. 2011, 32, 90. [Google Scholar] [CrossRef]
- Aldeguer-Riquelme, B.; Conrad, R.E.; Antón, J.; Rossello-Mora, R.; Konstantinidis, K.T. A Natural ANI Gap That Can Define Intra-Species Units of Bacteriophages and Other Viruses. mBio 2024, 15, e01536-24. [Google Scholar] [CrossRef]
- Chaudhari, H.V.; Inamdar, M.M.; Kondabagil, K. Scaling Relation between Genome Length and Particle Size of Viruses Provides Insights into Viral Life History. iScience 2021, 24, 102452. [Google Scholar] [CrossRef]
- Dion, M.B.; Oechslin, F.; Moineau, S. Phage Diversity, Genomics and Phylogeny. Nat. Rev. Microbiol. 2020, 18, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Valencia-Toxqui, G.; Ramsey, J. How to Introduce a New Bacteriophage on the Block: A Short Guide to Phage Classification. J. Virol. 2024, 98, e01821-23. [Google Scholar] [CrossRef]
- Joglekar, P.; Mesa, C.P.; Richards, V.A.; Polson, S.W.; Wommack, K.E.; Fuhrmann, J.J. Polyphasic Analysis Reveals Correlation between Phenotypic and Genotypic Analysis in Soybean Bradyrhizobia (Bradyrhizobium spp.). Syst. Appl. Microbiol. 2020, 43, 126073. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Cui, J.; Schlub, T.E.; Holmes, E.C. An Allometric Relationship between the Genome Length and Virion Volume of Viruses. J. Virol. 2014, 88, 6403–6410. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2; Use R! Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-24275-0. [Google Scholar]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Bouras, G.; Nepal, R.; Houtak, G.; Psaltis, A.J.; Wormald, P.J.; Vreugde, S. Pharokka: A Fast Scalable Bacteriophage Annotation Tool. Bioinformatics 2023, 39, btac776. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Merrill, B.D.; Ward, A.T.; Grose, J.H.; Hope, S. Software-Based Analysis of Bacteriophage Genomes, Physical Ends, and Packaging Strategies. BMC Genom. 2016, 17, 679. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The Protein Families Database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
- Duffy, C.; Feiss, M. The Large Subunit of Bacteriophage λ’s Terminase Plays a Role in DNA Translocation and Packaging Termination. J. Mol. Biol. 2002, 316, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Casjens, S. Prophages and Bacterial Genomics: What Have We Learned so Far? Mol. Microbiol. 2003, 49, 277–300. [Google Scholar] [CrossRef]
- Keown, R.A.; Dums, J.T.; Brumm, P.J.; MacDonald, J.; Mead, D.A.; Ferrell, B.D.; Moore, R.M.; Harrison, A.O.; Polson, S.W.; Wommack, K.E. Novel Viral DNA Polymerases from Metagenomes Suggest Genomic Sources of Strand-Displacing Biochemical Phenotypes. Front. Microbiol. 2022, 13, 858366. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- van Kempen, M.; Kim, S.S.; Tumescheit, C.; Mirdita, M.; Lee, J.; Gilchrist, C.L.M.; Söding, J.; Steinegger, M. Fast and Accurate Protein Structure Search with Foldseek. Nat. Biotechnol. 2023, 42, 243–246. [Google Scholar] [CrossRef] [PubMed]
- Millard, A.; Denise, R.; Lestido, M.; Thomas, M.T.; Webster, D.; Turner, D.; Sicheritz-Pontén, T. TaxMyPhage: Automated Taxonomy of dsDNA Phage Genomes at the Genus and Species Level. Phage 2025, 6, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P.; Adriaenssens, E.M.; Lefkowitz, E.J.; Oksanen, H.M.; Siddell, S.G.; Zerbini, F.M.; Alfenas-Zerbini, P.; Aylward, F.O.; Dempsey, D.M.; Dutilh, B.E.; et al. Changes to Virus Taxonomy and the ICTV Statutes Ratified by the International Committee on Taxonomy of Viruses. Arch. Virol. 2024, 169, 236. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High Throughput ANI Analysis of 90K Prokaryotic Genomes Reveals Clear Species Boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-Depth Characterization and Visualization of Bacterial Genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Gilchrist, C.L.M.; Chooi, Y.H. clinker & clustermap.js: Automatic Generation of Gene Cluster Comparison Figures. Bioinformatics 2021, 37, 2473–2475. [Google Scholar] [CrossRef]
- Delmont, T.O.; Eren, E.M. Linking Pangenomes and Metagenomes: The Prochlorococcus Metapangenome. PeerJ 2018, 6, e4320. [Google Scholar] [CrossRef]
- Eren, A.M.; Kiefl, E.; Shaiber, A.; Veseli, I.; Miller, S.E.; Schechter, M.S.; Fink, I.; Pan, J.N.; Yousef, M.; Fogarty, E.C.; et al. Community-Led, Integrated, Reproducible Multi-Omics with Anvi’o. Nat. Microbiol. 2021, 6, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Mantel, N. The Detection of Disease Clustering and a Generalized Regression Approach. Cancer Res. 1967, 27, 209–220. [Google Scholar] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Lopez Gonzalez-Nieto, P.; Gomez Flechoso, M.; Arribas Mocoroa, M.E.; Muñoz Martin, A.; Garcia Lorenzo, M.L.; Cabrera Gomez, G.; Alvarez Gomez, J.A.; Caso Fraile, A.; Orosco Dagan, J.M.; Merinero Palomares, R.; et al. Design and Development of a Virtual Laboratory in Python for the Teaching of Data Analysis and Mathematics in Geology: GeoPy. INTED2020 Proc. 2020, 1, 2236–2242. [Google Scholar] [CrossRef]
- Choi, K.H. Viral Polymerases. Adv. Exp. Med. Biol. 2012, 726, 267–304. [Google Scholar] [CrossRef]
- Rost, B. Twilight Zone of protein sequence alignments. Protein Eng. Des. Sel. 1999, 12, 85–94. [Google Scholar] [CrossRef]
- Nobrega, F.L.; Vlot, M.; de Jonge, P.A.; Dreesens, L.L.; Beaumont, H.J.E.; Lavigne, R.; Dutilh, B.E.; Brouns, S.J.J. Targeting Mechanisms of Tailed Bacteriophages. Nat. Rev. Microbiol. 2018, 16, 760–773. [Google Scholar] [CrossRef]
- Vincent, A.T. Bacterial Hypothetical Proteins May Be of Functional Interest. Front. Bacteriol. 2024, 3, 1334712. [Google Scholar] [CrossRef]
- Gómez, P.; Bennie, J.; Gaston, K.J.; Buckling, A. The Impact of Resource Availability on Bacterial Resistance to Phages in Soil. PLoS ONE 2015, 10, e0123752. [Google Scholar] [CrossRef]
- Fuhrmann, J. Symbiotic Effectiveness of Indigenous Soybean Bradyrhizobia as Related to Serological, Morphological, Rhizobitoxine, and Hydrogenase Phenotypes. Appl. Environ. Microbiol. 1990, 56, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Williamson, K.E.; Wommack, K.E.; Radosevich, M. Sampling Natural Viral Communities from Soil for Culture-Independent Analyses. Appl. Environ. Microbiol. 2003, 69, 6628–6633. [Google Scholar] [CrossRef] [PubMed]
- Williamson, K.E.; Radosevich, M.; Wommack, K.E. Abundance and Diversity of Viruses in Six Delaware Soils. Appl. Environ. Microbiol. 2005, 71, 3119–3125. [Google Scholar] [CrossRef] [PubMed]
- Fuhrmann, J. Serological Distribution of Bradyrhizobium japonicum as Influenced by Soybean Cultivar and Sampling Location. Soil Biol. Biochem. 1989, 21, 1079–1081. [Google Scholar] [CrossRef]
- Minamisawa, K.; Onodera, S.; Tanimura, Y.; Kobayashi, N.; Yuhashi, K.I.; Kubota, M. Preferential Nodulation of Glycine max, Glycine soja and Macroptilium atropurpureum by Two Bradyrhizobium Species japonicum and elkanii. FEMS Microbiol. Ecol. 1997, 24, 49–56. [Google Scholar] [CrossRef]
- Korytowski, D.A.; Smith, H. Permanence and Stability of a Kill the Winner Model in Marine Ecology. Bull. Math. Biol. 2017, 79, 995–1004. [Google Scholar] [CrossRef]
- Winter, C.; Bouvier, T.; Weinbauer, M.G.; Thingstad, T.F. Trade-offs between competition and defense specialists among unicellular planktonic organisms: The “killing the winner” hypothesis revisited. Microbiol. Mol. Biol. Rev. MMBR 2010, 74, 42–57. [Google Scholar] [CrossRef]
- Holtappels, D.; Alfenas-Zerbini, P.; Koskella, B. Drivers and Consequences of Bacteriophage Host Range. FEMS Microbiol. Rev. 2023, 47, fuad038. [Google Scholar] [CrossRef]
- Hyman, P.; Abedon, S.T. Bacteriophage Host Range and Bacterial Resistance. Adv. Appl. Microbiol. 2010, 70, 217–248. [Google Scholar] [CrossRef]
- Almpanis, A.; Swain, M.; Gatherer, D.; McEwan, N. Correlation between Bacterial G+C Content, Genome Size and the G+C Content of Associated Plasmids and Bacteriophages. Microb. Genom. 2018, 4, e000168. [Google Scholar] [CrossRef]
- Das, R.; Rahlff, J. Phage Genome Architecture and GC Content: Structural Genes and Where to Find Them. bioRxiv 2024. [Google Scholar] [CrossRef]
- Bailly-Bechet, M.; Vergassola, M.; Rocha, E. Causes for the Intriguing Presence of tRNAs in Phages. Genome Res. 2007, 17, 1486. [Google Scholar] [CrossRef]
- Rocha, P.C.E.; Danchin, A. Base Composition Bias Might Result from Competition for Metabolic Resources. TRENDS Genet. 2002, 18, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, N.; Lindell, M.; Pavlov, M.; Schiavone, L.H.; Wagner, E.G.H.; Ehrenberg, M. Structure Probing of tmRNA in Distinct Stages of Trans-Translation. RNA 2007, 13, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Hatfull, G.F.; Pedulla, M.L.; Jacobs-Sera, D.; Cichon, P.M.; Foley, A.; Ford, M.E.; Gonda, R.M.; Houtz, J.M.; Hryckowian, A.J.; Kelchner, V.A.; et al. Exploring the Mycobacteriophage Metaproteome: Phage Genomics as an Educational Platform. PLoS Genet. 2006, 2, e92. [Google Scholar] [CrossRef]
- Felden, B.; Gillet, R.; Metzinger, L. Protein Tagging and Ribosome Rescue in Bacteria Requires the Recognition of Transfer-Messenger RNA by an Aminoacyl-tRNA Synthetase. In Madame Curie Bioscience Database [Internet]; Landes Bioscience: Austin, TX, USA, 2013. Available online: https://www.ncbi.nlm.nih.gov/books/NBK6065/ (accessed on 20 October 2025).
- Withey, J.; Friedman, D. Analysis of the Role of Trans-Translation in the Requirement of tmRNA for LambdaimmP22 Growth in Escherichia coli. J. Bacteriol. 1999, 181, 2148–2157. [Google Scholar] [CrossRef]
- Retallack, D.M.; Johnson, L.L.; Friedman, D.I. Role for 10Sa RNA in the Growth of Lambda-P22 Hybrid Phage. J. Bacteriol. 1994, 176, 2082–2089. [Google Scholar] [CrossRef]
- Ranquet, C.; Geiselmann, J.; Toussaint, A. The tRNA Function of SsrA Contributes to Controlling Repression of Bacteriophage Mu Prophage. Proc. Natl. Acad. Sci. USA 2001, 98, 10220–10225. [Google Scholar] [CrossRef]
- Taslem Mourosi, J.; Awe, A.; Guo, W.; Batra, H.; Ganesh, H.; Wu, X.; Zhu, J. Understanding Bacteriophage Tail Fiber Interaction with Host Surface Receptor: The Key “Blueprint” for Reprogramming Phage Host Range. Int. J. Mol. Sci. 2022, 23, 12146. [Google Scholar] [CrossRef]
- Stern, A.; Sorek, R. The Phage-Host Arms-Race: Shaping the Evolution of Microbes. Bioessays 2011, 33, 43. [Google Scholar] [CrossRef]
- Ryu, S. Grand Challenges in Phage Biology. Front. Microbiol. 2021, 12, 715039. [Google Scholar] [CrossRef]
- Grose, J.H.; Casjens, S.R. Understanding the Enormous Diversity of Bacteriophages: The Tailed Phages That Infect the Bacterial Family Enterobacteriaceae. Virology 2014, 468–470, 421–443. [Google Scholar] [CrossRef]
- Wangchuk, J.; Chatterjee, A.; Patil, S.; Madugula, S.K.; Kondabagil, K. The Coevolution of Large and Small Terminases of Bacteriophages Is a Result of Purifying Selection Leading to Phenotypic Stabilization. Virology 2021, 564, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Casjens, S.; Weigele, P. DNA Packaging by Bacteriophage P22. In Madame Curie Bioscience Database [Internet]; Landes Bioscience: Austin, TX, USA, 2013; NBK6430. Available online: https://www.ncbi.nlm.nih.gov/books/NBK6430/ (accessed on 29 October 2025).
- Wommack, K.E.; Nasko, D.J.; Chopyk, J.; Sakowski, E.G. Counts and Sequences, Observations That Continue to Change Our Understanding of Viruses in Nature. J. Microbiol. 2015, 53, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Astatke, M.; Ng, K.; Grindley, N.D.F.; Joyce, C.M. A Single Side Chain Prevents Escherichia coli DNA Polymerase I (Klenow Fragment) from Incorporating Ribonucleotides. Biochemistry 1998, 95, 3402–3407. [Google Scholar] [CrossRef]
- Suzuki, M.; Yoshida, S.; Adman, E.T.; Blank, A.; Loeb, L.A.; Gottstein, J. Thermus Aquaticus DNA Polymerase I Mutants with Altered Fidelity. Interacting Mutations in the O-Helix. J. Biol. Chem. 2000, 275, 32728–32735. [Google Scholar] [CrossRef]
- Tabor, S.; Richardson, C.C. A Single Residue in DNA Polymerases of the Escherichia coli DNA Polymerase I Family Is Critical for Distinguishing between Deoxy- and Dideoxyribonucleotides. Proc. Natl. Acad. Sci. USA 1995, 92, 6339–6343. [Google Scholar] [CrossRef]
- Tabor, S.; Richardson, C.C. DNA Sequence Analysis with a Modified Bacteriophage T7 DNA Polymerase (DNA Polymerase I/Reverse Transcriptase/Chain-Terminating Inhibitors/2′-Deoxyinosine 5′-Triphosphate/Processivity). Biochemistry 1987, 84, 4767–4771. [Google Scholar]
- Nasko, D.J.; Chopyk, J.; Sakowski, E.G.; Ferrell, B.D.; Polson, S.W.; Wommack, K.E. Family A DNA Polymerase Phylogeny Uncovers Diversity and Replication Gene Organization in the Virioplankton. Front. Microbiol. 2018, 9, 35053. [Google Scholar] [CrossRef]
- Schmidt, H.F.; Sakowski, E.G.; Williamson, S.J.; Polson, S.W.; Wommack, K.E. Shotgun Metagenomics Indicates Novel Family A DNA Polymerases Predominate within Marine Virioplankton. ISME J. 2014, 8, 103–114. [Google Scholar] [CrossRef]
- Tuttle, A.R.; Trahan, N.D.; Son, M.S. Growth and Maintenance of Escherichia coli Laboratory Strains. Curr. Protoc. 2021, 1, e20. [Google Scholar] [CrossRef] [PubMed]
- Howard-Varona, C.; Hargreaves, K.R.; Abedon, S.T.; Sullivan, M.B. Lysogeny in nature: Mechanisms, impact and ecology of temperate phages. ISME J. 2017, 11, 1511–1520. [Google Scholar] [CrossRef] [PubMed]
- Altamirano, F.L.G.; Barr, J.J. Screening for Lysogen Activity in Therapeutically Relevant Bacteriophages. Bio-Protocol 2021, 11, e3997. [Google Scholar] [CrossRef] [PubMed]
- Vargas Gil, S.; Meriles, J.; Conforto, C.; Basanta, M.; Radl, V.; Hagn, A.; Schloter, M.; March, G.J. Response of Soil Microbial Communities to Different Management Practices in Surface Soils of a Soybean Agroecosystem in Argentina. Eur. J. Soil Biol. 2011, 47, 55–60. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morgese, E.A.; Ferrell, B.D.; Toth, S.C.; Polson, S.W.; Wommack, K.E.; Fuhrmann, J.J. Comparative Analysis Reveals Host Species-Dependent Diversity Among 16 Virulent Bacteriophages Isolated Against Soybean Bradyrhizobium spp. Viruses 2025, 17, 1474. https://doi.org/10.3390/v17111474
Morgese EA, Ferrell BD, Toth SC, Polson SW, Wommack KE, Fuhrmann JJ. Comparative Analysis Reveals Host Species-Dependent Diversity Among 16 Virulent Bacteriophages Isolated Against Soybean Bradyrhizobium spp. Viruses. 2025; 17(11):1474. https://doi.org/10.3390/v17111474
Chicago/Turabian StyleMorgese, Emily A., Barbra D. Ferrell, Spencer C. Toth, Shawn W. Polson, K. Eric Wommack, and Jeffry J. Fuhrmann. 2025. "Comparative Analysis Reveals Host Species-Dependent Diversity Among 16 Virulent Bacteriophages Isolated Against Soybean Bradyrhizobium spp." Viruses 17, no. 11: 1474. https://doi.org/10.3390/v17111474
APA StyleMorgese, E. A., Ferrell, B. D., Toth, S. C., Polson, S. W., Wommack, K. E., & Fuhrmann, J. J. (2025). Comparative Analysis Reveals Host Species-Dependent Diversity Among 16 Virulent Bacteriophages Isolated Against Soybean Bradyrhizobium spp. Viruses, 17(11), 1474. https://doi.org/10.3390/v17111474







