Within-Subtype HIV-1 Polymorphisms and Their Impacts on Intact Proviral DNA Assay (IPDA) for Viral Reservoir Quantification
Abstract
1. Introduction
2. Methods
2.1. Phylogenetic Analysis
2.2. In Silico Evaluation of the Compatibility of IPDA Primers/Probes per Geographical Region
3. Results
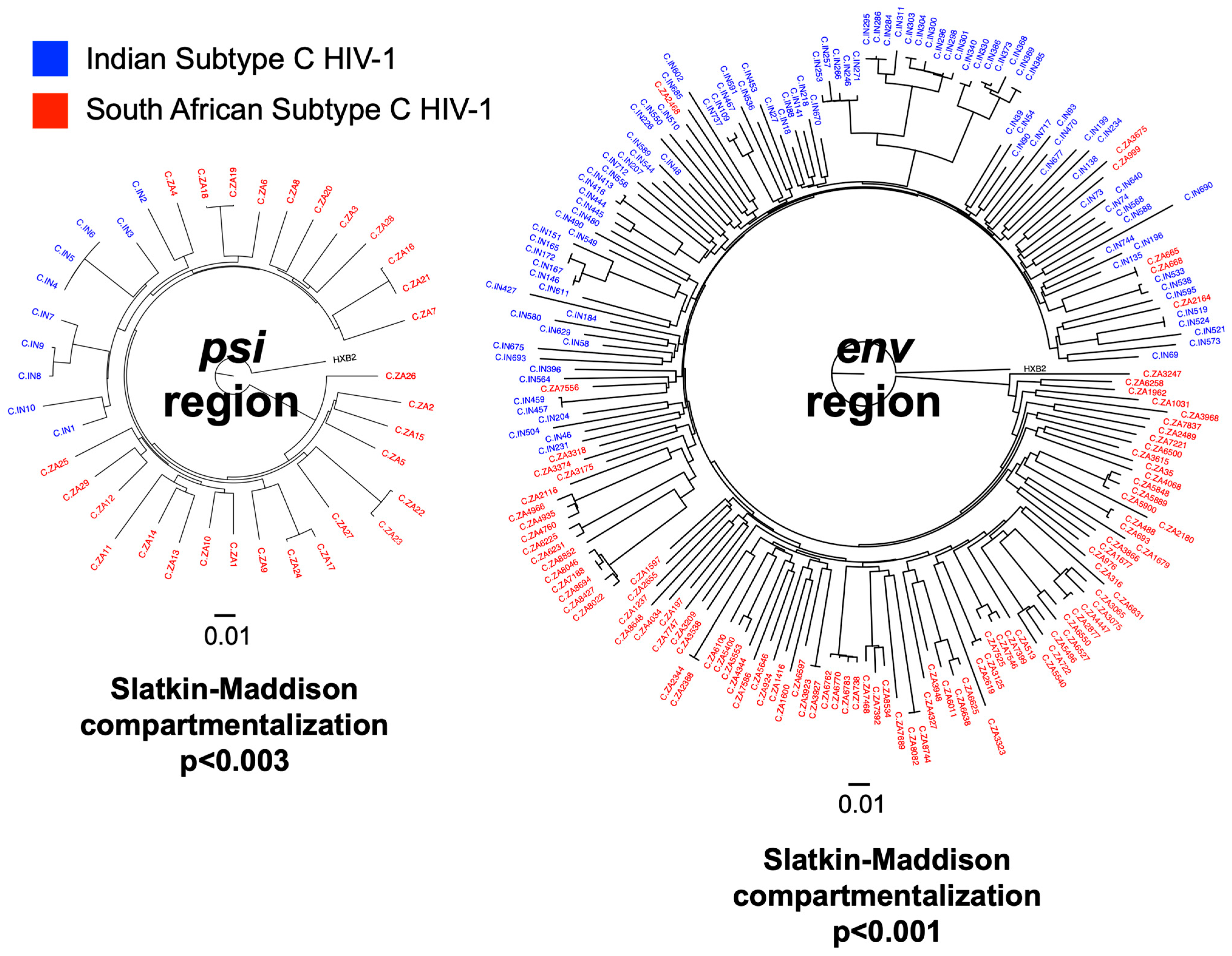
3.1. Subtype C HIV-1 Isolated from India (IN) and South Africa (ZA) Were Genetically Distinct
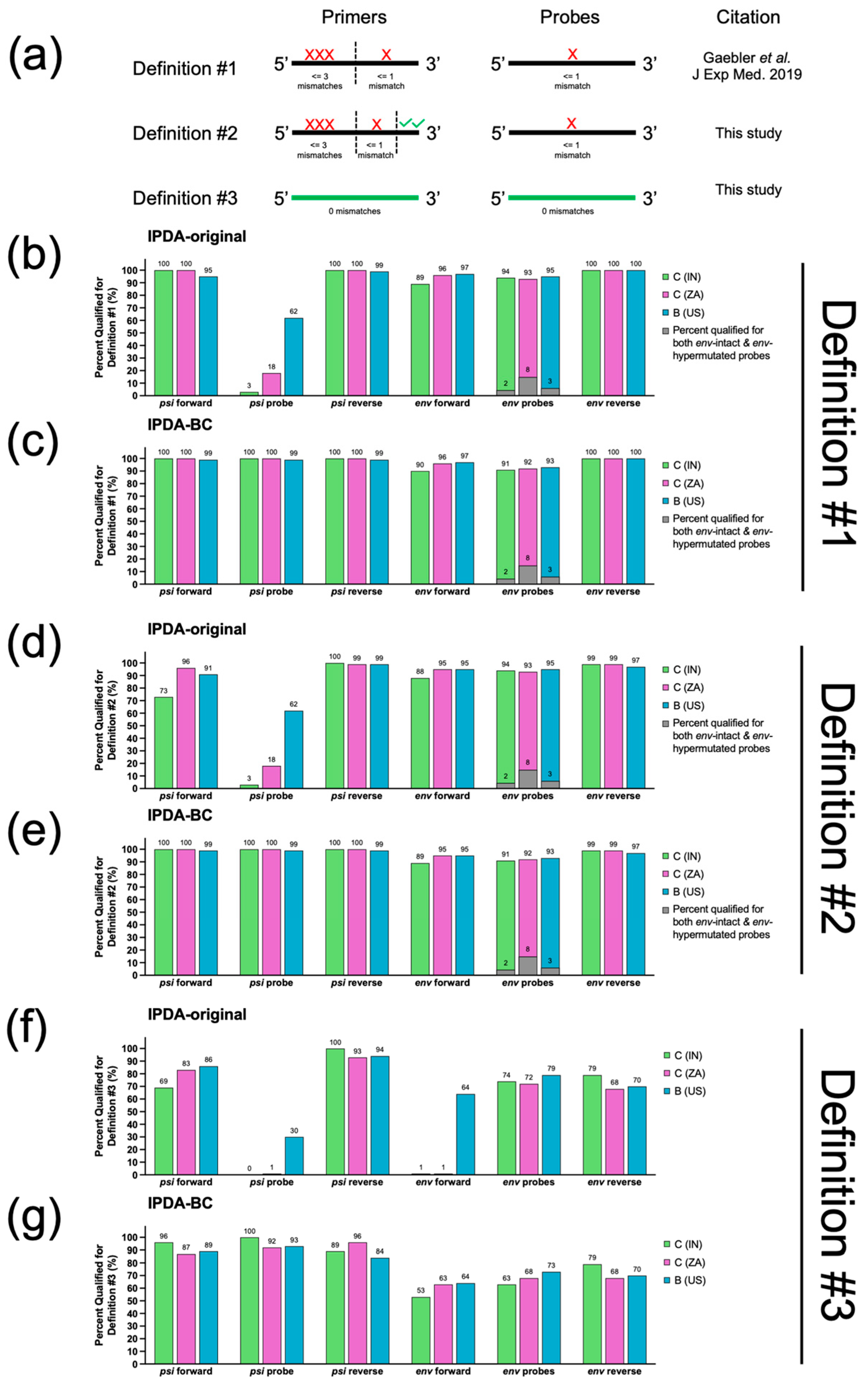
3.2. In Silico Analysis Predicted That IPDA-BC Primers/Probes Were 6–10% More Likely to Fail in IN than ZA Samples
3.3. Ability to Distinguish Between Hypermutated and Non-Hypermutated Genomes
3.4. Performance Evaluation of the Three Mismatch Definitions Used in This Study
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bruner, K.M.; Wang, Z.; Simonetti, F.R.; Bender, A.M.; Kwon, K.J.; Sengupta, S.; Fray, E.J.; Beg, S.A.; Antar, A.A.R.R.; Jenike, K.M.; et al. A Quantitative Approach for Measuring the Reservoir of Latent HIV-1 Proviruses. Nature 2019, 566, 120–125. [Google Scholar] [CrossRef]
- Ho, Y.C.; Shan, L.; Hosmane, N.N.; Wang, J.; Laskey, S.B.; Rosenbloom, D.I.S.; Lai, J.; Blankson, J.N.; Siliciano, J.D.; Siliciano, R.F. Replication-Competent Noninduced Proviruses in the Latent Reservoir Increase Barrier to HIV-1 Cure. Cell 2013, 155, 540. [Google Scholar] [CrossRef]
- Lee, G.Q. Chemistry and Bioinformatics Considerations in Using Next-Generation Sequencing Technologies to Inferring HIV Proviral DNA Genome-Intactness. Viruses 2021, 13, 1874. [Google Scholar] [CrossRef]
- Kinloch, N.N.; Ren, Y.; Conce Alberto, W.D.; Dong, W.; Khadka, P.; Huang, S.H.; Mota, T.M.; Wilson, A.; Shahid, A.; Kirkby, D.; et al. HIV-1 Diversity Considerations in the Application of the Intact Proviral DNA Assay (IPDA). Nat. Commun. 2021, 12, 165. [Google Scholar] [CrossRef]
- Gaebler, C.; Falcinelli, S.D.; Stoffel, E.; Read, J.; Murtagh, R.; Oliveira, T.Y.; Ramos, V.; Lorenzi, J.C.C.; Kirchherr, J.; James, K.S.; et al. Sequence Evaluation and Comparative Analysis of Novel Assays for Intact Proviral HIV-1 DNA. J. Virol. 2021, 95, e01986-20. [Google Scholar] [CrossRef]
- Williams, A.; Menon, S.; Crowe, M.; Agarwal, N.; Biccler, J.; Bbosa, N.; Ssemwanga, D.; Adungo, F.; Moecklinghoff, C.; Macartney, M.; et al. Geographic and Population Distributions of Human Immunodeficiency Virus (HIV)-1 and HIV-2 Circulating Subtypes: A Systematic Literature Review and Meta-Analysis (2010–2021). J. Infect. Dis. 2023, 228, 1583–1591. [Google Scholar] [CrossRef]
- Gunst, J.D.; Højen, J.F.; Pahus, M.H.; Rosás-Umbert, M.; Stiksrud, B.; McMahon, J.H.; Denton, P.W.; Nielsen, H.; Johansen, I.S.; Benfield, T.; et al. Impact of a TLR9 Agonist and Broadly Neutralizing Antibodies on HIV-1 Persistence: The Randomized Phase 2a TITAN Trial. Nat. Med. 2023, 29, 2547–2558. [Google Scholar] [CrossRef]
- Lee, G.Q.; Khadka, P.; Gowanlock, S.N.; Copertino, D.C.; Duncan, M.C.; Omondi, F.H.; Kinloch, N.N.; Kasule, J.; Kityamuweesi, T.; Buule, P.; et al. HIV-1 Subtype A1, D, and Recombinant Proviral Genome Landscapes during Long-Term Suppressive Therapy. Nat. Commun. 2024, 15, 5480. [Google Scholar] [CrossRef]
- Cassidy, N.A.J.; Fish, C.S.; Levy, C.N.; Roychoudhury, P.; Reeves, D.B.; Hughes, S.M.; Schiffer, J.T.; Benki-Nugent, S.; John-Stewart, G.; Wamalwa, D.; et al. HIV Reservoir Quantification Using Cross-Subtype Multiplex DdPCR. iScience 2021, 25, 103615. [Google Scholar] [CrossRef]
- Buchholtz, N.V.E.J.; Nühn, M.M.; de Jong, T.C.M.; Stienstra, T.A.T.; Reddy, K.; Ndung’u, T.; Ndhlovu, Z.M.; Fisher, K.; Palmer, S.; Wensing, A.M.J.; et al. Development of a Highly Sensitive and Specific Intact Proviral DNA Assay for HIV-1 Subtype B and C. Virol. J. 2024, 21, 36. [Google Scholar] [CrossRef]
- Hiener, B.; Horsburgh, B.A.; Eden, J.-S.S.; Barton, K.; Schlub, T.E.; Lee, E.; von Stockenstrom, S.; Odevall, L.; Milush, J.M.; Liegler, T.; et al. Identification of Genetically Intact HIV-1 Proviruses in Specific CD4+T Cells from Effectively Treated Participants. Cell Rep. 2017, 21, 813–822. [Google Scholar] [CrossRef]
- Lee, G.Q.; Orlova-Fink, N.; Einkauf, K.; Chowdhury, F.Z.; Sun, X.; Harrington, S.; Kuo, H.-H.; Hua, S.; Chen, H.-R.; Ouyang, Z.; et al. Clonal Expansion of Genome-Intact HIV-1 in Functionally-Polarized Th1 CD4 T Cells. J. Clin. Investig. 2017, 127, 2689–2696. [Google Scholar] [CrossRef]
- Reddy, K.; Lee, G.Q.; Reddy, N.; Chikowore, T.J.; Baisley, K.; Dong, K.L.; Walker, B.D.; Yu, X.G.; Lichterfeld, M.; Ndung’u, T. Differences in HIV-1 Reservoir Size, Landscape Characteristics, and Decay Dynamics in Acute and Chronic Treated HIV-1 Clade C Infection. Elife 2025, 13, RP96617. [Google Scholar] [CrossRef]
- Los Alamos National Laboratory. Los Alamos HIV Sequence Database. Available online: http://www.hiv.lanl.gov/ (accessed on 20 August 2024).
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Paradis, E.; Schliep, K. Ape 5.0: An Environment for Modern Phylogenetics and Evolutionary Analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Kosakovsky Pond, S.L.; Poon, A.F.Y.; Velazquez, R.; Weaver, S.; Hepler, N.L.; Murrell, B.; Shank, S.D.; Magalis, B.R.; Bouvier, D.; Nekrutenko, A.; et al. HyPhy 2.5—A Customizable Platform for Evolutionary Hypothesis Testing Using Phylogenies. Mol. Biol. Evol. 2019, 37, 295. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Gaebler, C.; Lorenzi, J.; Oliveira, T.; Nogueira, L.; Ramos, V.; Lu, C.; Pai, J.; Mendoza, P.; Jankovic, M.; Caskey, M.; et al. Combination of Quadruplex QPCR and Next-Generation Sequencing for Qualitative and Quantitative Analysis of the HIV-1 Latent Reservoir. J. Exp. Med. 2019, 216, 2253–2264. [Google Scholar] [CrossRef]
- Simsek, M.; Adnan, H. Effect of Single Mismatches at 3′–End of Primers on Polymerase Chain Reaction. J. Sci. Res. Med. Sci. 2000, 2, 11. [Google Scholar]
- Gunst, J.D.; Pahus, M.H.; Rosás-Umbert, M.; Lu, I.N.; Benfield, T.; Nielsen, H.; Johansen, I.S.; Mohey, R.; Østergaard, L.; Klastrup, V.; et al. Early Intervention with 3BNC117 and Romidepsin at Antiretroviral Treatment Initiation in People with HIV-1: A Phase 1b/2a, Randomized Trial. Nat. Med. 2022, 28, 2424. [Google Scholar] [CrossRef]
- Neogi, U.; Bontell, I.; Shet, A.; de Costa, A.; Gupta, S.; Diwan, V.; Laishram, R.S.; Wanchu, A.; Ranga, U.; Banerjea, A.C.; et al. Molecular Epidemiology of HIV-1 Subtypes in India: Origin and Evolutionary History of the Predominant Subtype C. PLoS ONE 2012, 7, e39819. [Google Scholar] [CrossRef]
- Souto, B.; Triunfante, V.; Santos-Pereira, A.; Martins, J.; Araújo, P.M.M.; Osório, N.S. Evolutionary Dynamics of HIV-1 Subtype C in Brazil. Sci. Rep. 2021, 11, 23060. [Google Scholar] [CrossRef]
- Yukl, S.A.; Kaiser, P.; Kim, P.; Telwatte, S.; Joshi, S.K.; Vu, M.; Lampiris, H.; Wong, J.K. HIV Latency in Isolated Patient CD4+ T Cells May Be Due to Blocks in HIV Transcriptional Elongation, Completion, and Splicing. Sci. Transl. Med. 2018, 10, eaap9927. [Google Scholar] [CrossRef]
- Stevenson, E.M.; Terry, S.; Copertino, D.; Leyre, L.; Danesh, A.; Weiler, J.; Ward, A.R.; Khadka, P.; McNeil, E.; Bernard, K.; et al. SARS CoV-2 MRNA Vaccination Exposes Latent HIV to Nef-Specific CD8 + T-Cells. Nat. Commun. 2022, 13, 4888. [Google Scholar] [CrossRef]
- Lee, G.Q.; Reddy, K.; Einkauf, K.B.; Gounder, K.; Chevalier, J.M.; Dong, K.L.; Walker, B.D.; Yu, X.G.; Ndung’u, T.; Lichterfeld, M. HIV-1 DNA Sequence Diversity and Evolution during Acute Subtype C Infection. Nat. Commun. 2019, 10, 2737. [Google Scholar] [CrossRef]
- Palma, P.; Zangari, P.; Alteri, C.; Tchidjou, H.K.; Manno, E.C.; Liuzzi, G.; Perno, C.F.; Rossi, P.; Bertoli, A.; Bernardi, S. Early Antiretroviral Treatment (EART) Limits Viral Diversity over Time in a Long-Term HIV Viral Suppressed Perinatally Infected Child. BMC Infect. Dis. 2016, 16, 742. [Google Scholar] [CrossRef]
- Lee, G.Q. A Daisy Chain of Inferences: The Role of Single-Cell and Single-Genome Proviral Sequencing in Characterizing HIV-1 Reservoirs. Curr. Opin. HIV AIDS 2025, 20, 512–517. [Google Scholar] [CrossRef]
- Kinloch, N.N.; Shahid, A.; Dong, W.; Kirkby, D.; Jones, B.R.; Beelen, C.J.; MacMillan, D.; Lee, G.Q.; Mota, T.M.; Sudderuddin, H.; et al. HIV Reservoirs Are Dominated by Genetically Younger and Clonally Enriched Proviruses. MBio 2023, 14, e02417-23. [Google Scholar] [CrossRef]
- Bustin, S.A.; Mueller, R.; Nolan, T. Parameters for Successful PCR Primer Design. Methods Mol. Biol. 2020, 2065, 5–22. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arikatla, M.R.; Mathad, J.S.; Reddy, K.; Reddy, N.; Ndung’u, T.; Dupnik, K.M.; Lee, G.Q. Within-Subtype HIV-1 Polymorphisms and Their Impacts on Intact Proviral DNA Assay (IPDA) for Viral Reservoir Quantification. Viruses 2025, 17, 1453. https://doi.org/10.3390/v17111453
Arikatla MR, Mathad JS, Reddy K, Reddy N, Ndung’u T, Dupnik KM, Lee GQ. Within-Subtype HIV-1 Polymorphisms and Their Impacts on Intact Proviral DNA Assay (IPDA) for Viral Reservoir Quantification. Viruses. 2025; 17(11):1453. https://doi.org/10.3390/v17111453
Chicago/Turabian StyleArikatla, Mohith Reddy, Jyoti S. Mathad, Kavidha Reddy, Nicole Reddy, Thumbi Ndung’u, Kathryn M. Dupnik, and Guinevere Q. Lee. 2025. "Within-Subtype HIV-1 Polymorphisms and Their Impacts on Intact Proviral DNA Assay (IPDA) for Viral Reservoir Quantification" Viruses 17, no. 11: 1453. https://doi.org/10.3390/v17111453
APA StyleArikatla, M. R., Mathad, J. S., Reddy, K., Reddy, N., Ndung’u, T., Dupnik, K. M., & Lee, G. Q. (2025). Within-Subtype HIV-1 Polymorphisms and Their Impacts on Intact Proviral DNA Assay (IPDA) for Viral Reservoir Quantification. Viruses, 17(11), 1453. https://doi.org/10.3390/v17111453






