Charting the Proteins of Oropouche Virus
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Sequence of Oropouche Virus
2.2. Protein Modeling
3. Results
3.1. Identification of Conserved Domains in Oropouche Virus Proteins
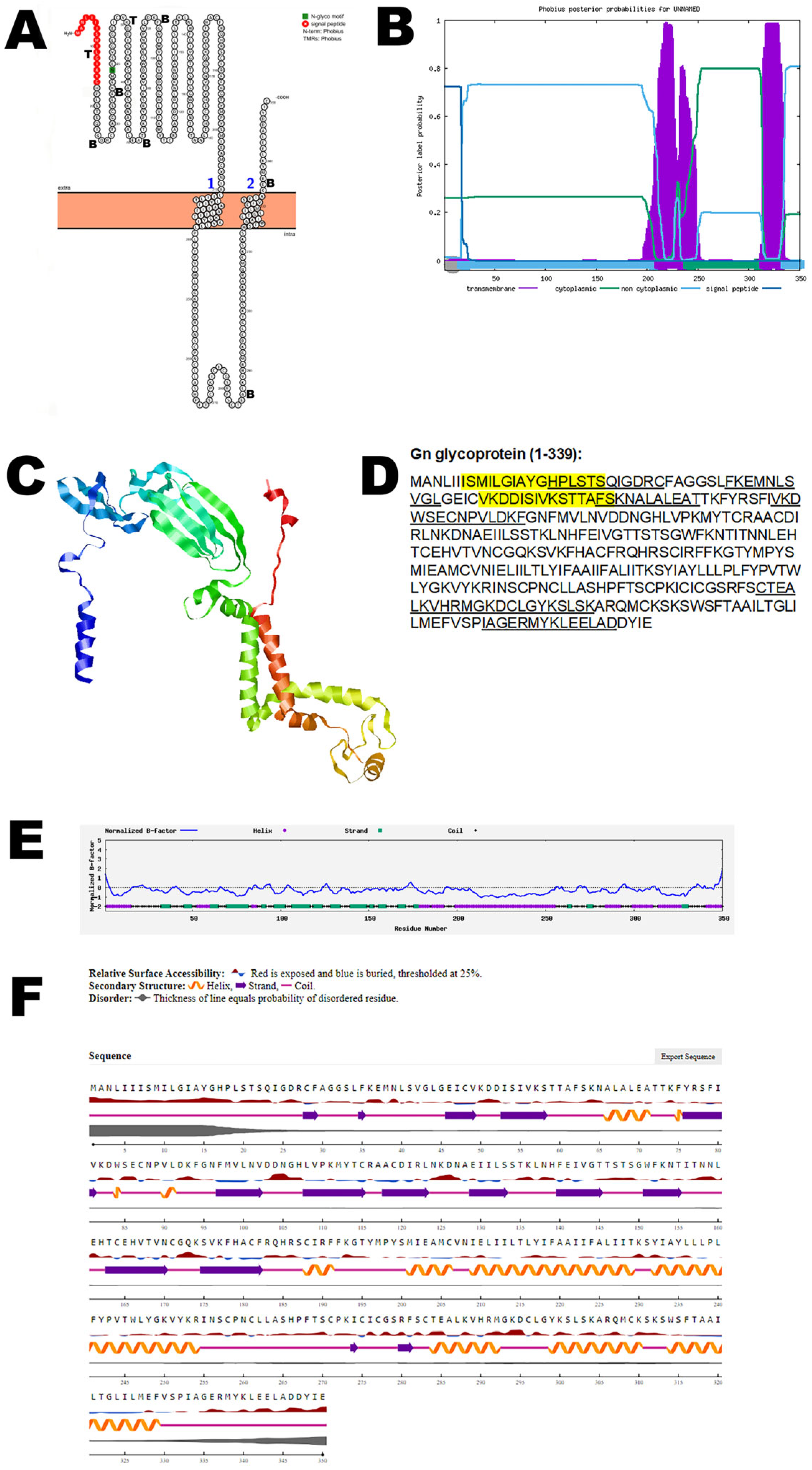
3.2. Gn Glycoprotein
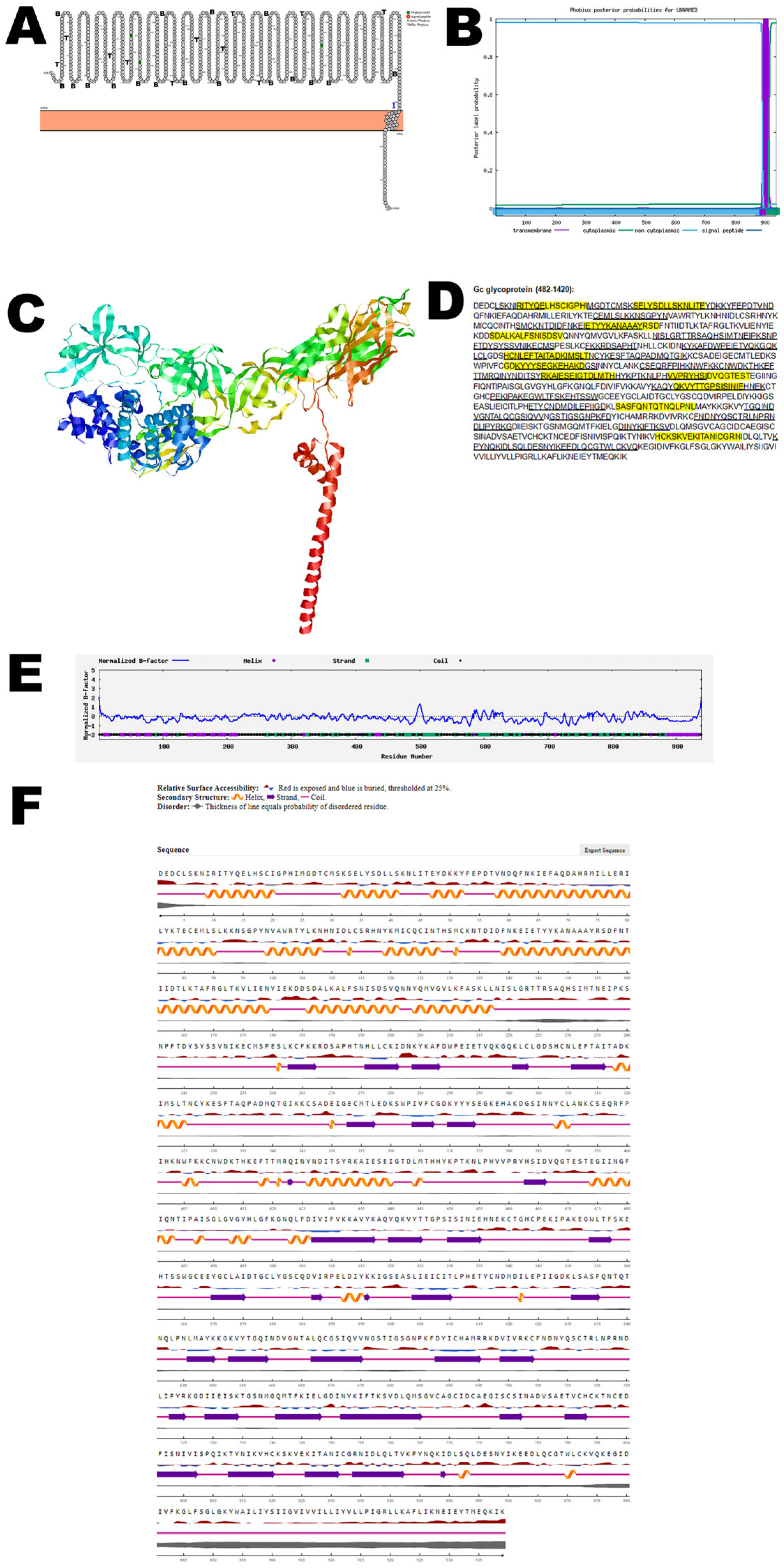
3.3. Gc Glycoprotein
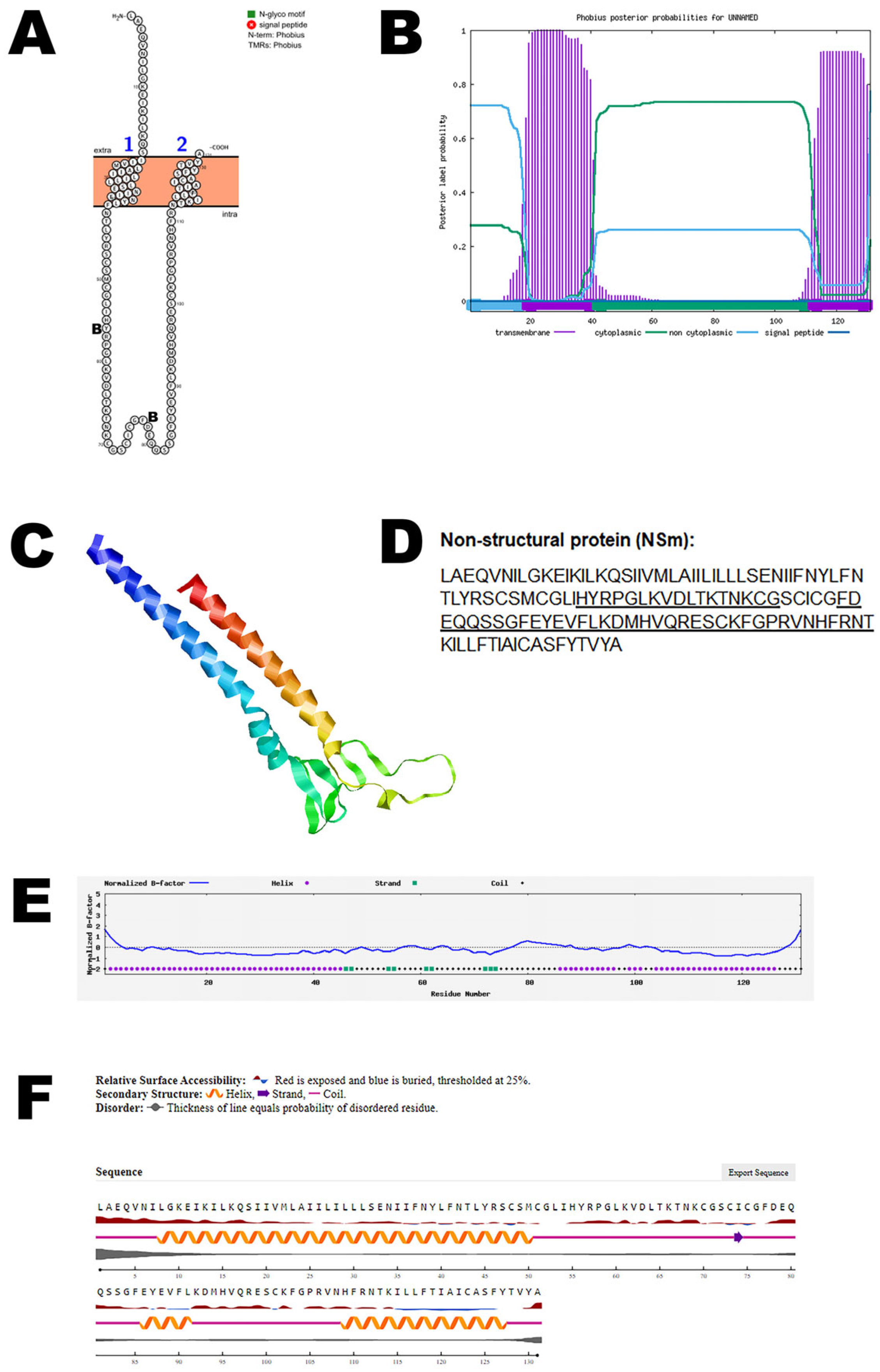
3.4. NSm Protein
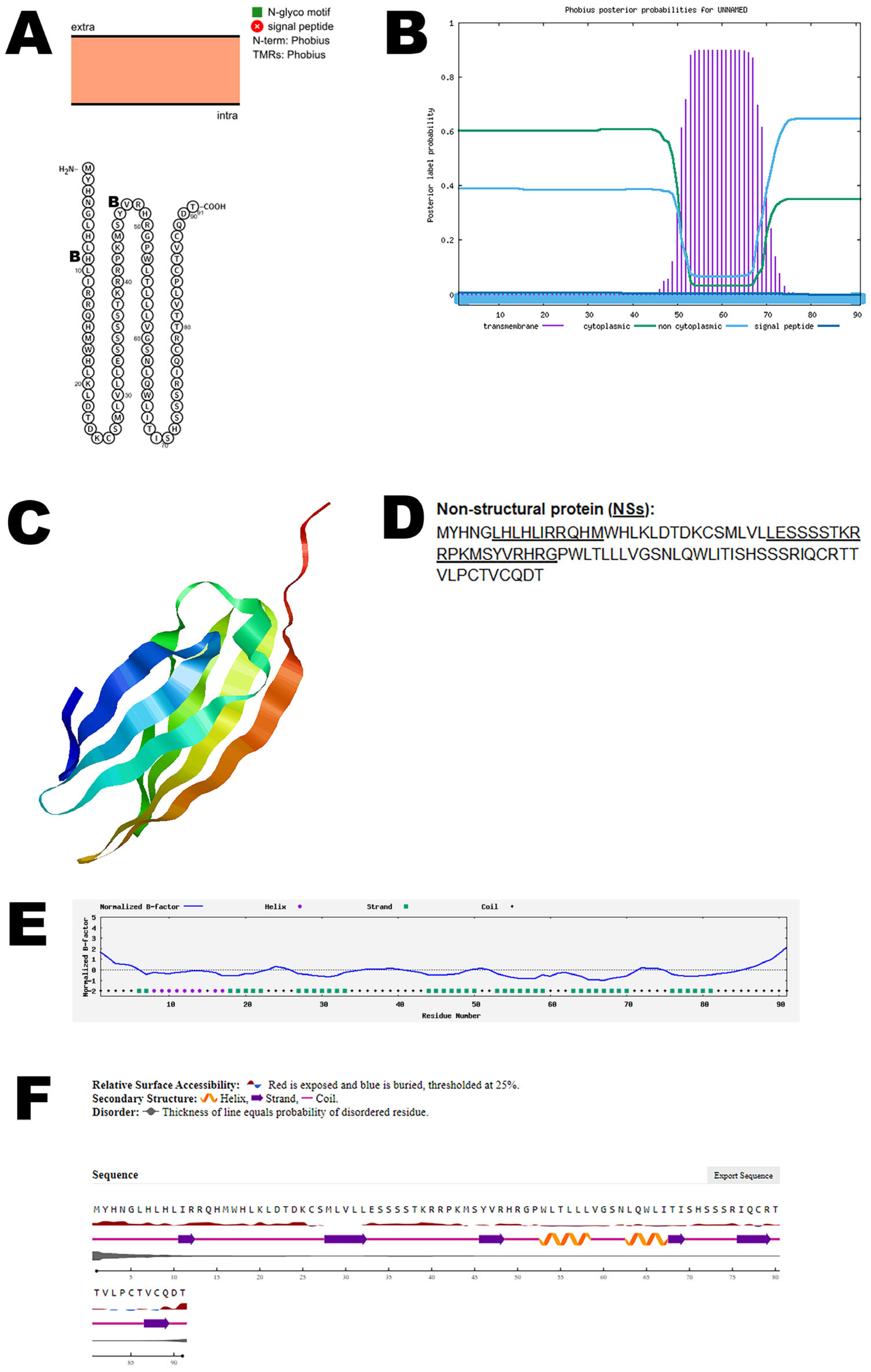
3.5. NSs Protein
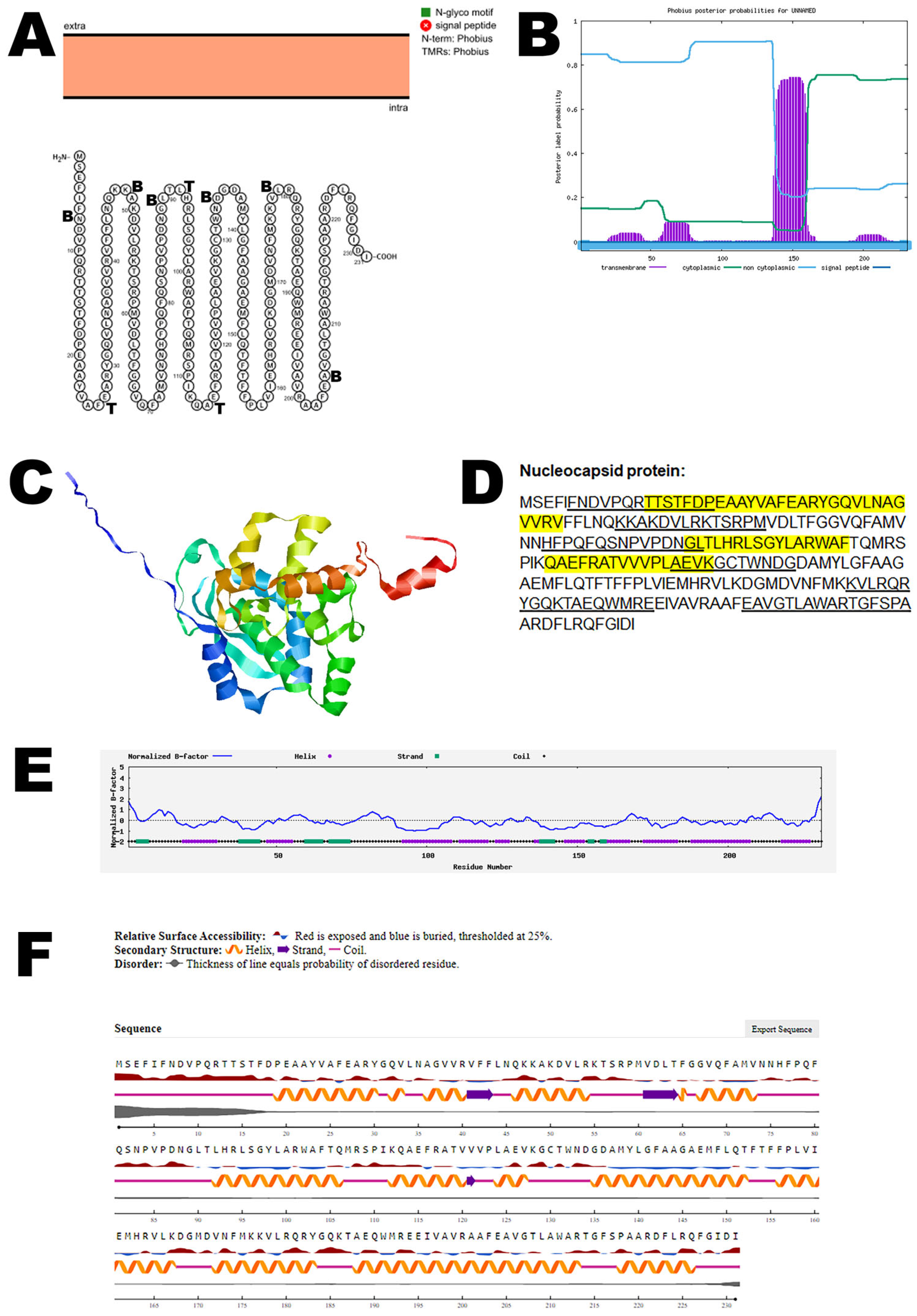
3.6. Nucleocapsid Protein
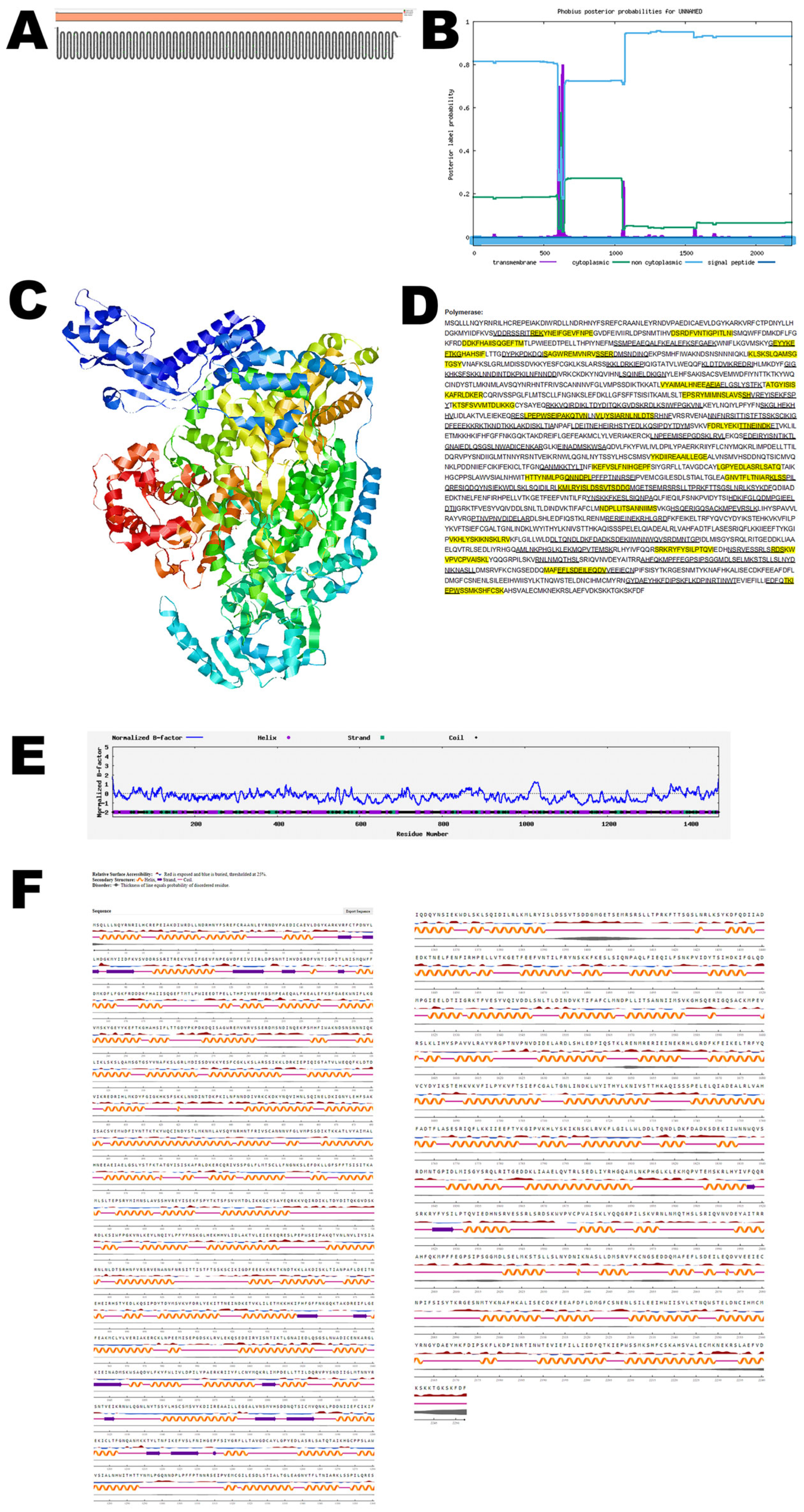
3.7. RNA Polymerase
3.8. Comparative Analysis
4. Discussion
Supplementary Materials
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anderson, C.R.; Spence, L.; Downs, W.G.; Aitken, T.H. Oropouche virus: A new human disease agent from Trinidad, West Indies. Am. J. Trop. Med. Hyg. 1961, 10, 574–578. [Google Scholar] [CrossRef]
- Files, M.A.; Hansen, C.A.; Herrera, V.C.; Schindewolf, C.; Barrett, A.D.; Beasley, D.W.; Bourne, N.; Milligan, G.N. Baseline mapping of Oropouche virology, epidemiology, therapeutics, and vaccine research and development. NPJ Vaccines 2022, 7, 38. [Google Scholar] [CrossRef]
- Pinheiro, F.P.; Travassos da Rosa, A.P.; Travassos da Rosa, J.F.; Bensabath, G. An outbreak of Oropouche virus disease in the vicinity of Santarem, Para, Brazil. Tropenmed. Parasitol. 1976, 27, 213–223. [Google Scholar]
- Desai, A.N.; Otter, A.; Koopmans, M.; Granata, G.; Grobusch, M.P.; Tunali, V.; Astorri, R.; Jokelainen, P.; Greub, G.; Ergönül, Ö.; et al. Oropouche virus: A re-emerging arbovirus of clinical significance. IJID Reg. 2024, 13, 100456. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention/CDC. Countries and Territories with Recent or Previous Oropouche Virus Transmission. 2025. Available online: https://www.cdc.gov/oropouche/data-maps/countries-and-territories-at-risk-for-oropouche.html (accessed on 20 August 2025).
- Morrison, A.; White, J.L.; Hughes, H.R.; Guagliardo, S.A.J.; Velez, J.O.; Fitzpatrick, K.A.; Davis, E.H.; Stanek, D.; Kopp, E.; Dumoulin, P.; et al. Oropouche Virus Disease Among U.S. Travelers—United States. MMWR Morb. Mortal. Wkly. Rep. 2024, 73, 769–773. [Google Scholar] [CrossRef]
- Pan American Health Organization/PAHO. PAHO Publishes New Update on Oropouche Fever in the Americas. 2025. Available online: https://www.paho.org/en/news/14-8-2025-paho-publishes-new-update-oropouche-fever-americas (accessed on 24 August 2025).
- Vasconcelos, H.B.; Azevedo, R.S.; Casseb, S.M.; Nunes-Neto, J.P.; Chiang, J.O.; Cantuária, P.C.; Segura, M.N.; Martins, L.C.; Monteiro, H.A.; Rodrigues, S.G.; et al. Oropouche fever epidemic in Northern Brazil: Epidemiology and molecular characterization of isolates. J. Clin. Virol. 2009, 44, 129–133. [Google Scholar] [CrossRef]
- Vernal, S.; Martini, C.C.R.; da Fonseca, B.A.L. Oropouche virus-associated aseptic meningoencephalitis, Southeastern Brazil. Emerg. Infect. Dis. 2019, 25, 380–382. [Google Scholar] [CrossRef]
- Chiang, J.O.; Azevedo, R.S.; Justino, M.C.A.; Matos, H.J.; Cabeça, H.L.S.; Silva, S.P.; Henriques, D.F.; Silva, E.V.P.; Andrade, G.S.S.; Vasconcelos, P.F.; et al. Neurological disease caused by Oropouche virus in northern Brazil: Should it be included in the scope of clinical neurological diseases? J. Neurovirol. 2021, 27, 626–630. [Google Scholar] [CrossRef] [PubMed]
- Cain, M.; Ly, H. Oropouche virus: Understanding “sloth fever” disease dynamics and novel intervention strategies against this emerging neglected tropical disease. Virulence 2024, 15, 2439521. [Google Scholar] [CrossRef] [PubMed]
- Tilston-Lunel, N.L.; Acrani, G.O.; Randall, R.E.; Elliott, R.M. Generation of Recombinant Oropouche Viruses Lacking the Nonstructural Protein NSm or NSs. J. Virol. 2015, 90, 2616–2627. [Google Scholar] [CrossRef] [PubMed]
- Sakkas, H.; Bozidis, P.; Franks, A.; Papadopoulou, C. Oropouche fever: A review. Viruses 2018, 10, 175. [Google Scholar] [CrossRef]
- Romero-Alvarez, D.; Escobar, L.E. Oropouche fever, an emergent disease from the Americas. Microbes Infect. 2018, 20, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Salvato, R.S. Re-emergence of Oropouche virus as a novel global threat. Curr. Res. Microb. Sci. 2025, 8, 100406. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, F.P.; Hoch, A.L.; Gomes, M.L.; Roberts, D.R. Oropouche virus. IV. Laboratory transmission by Culicoides paraensis. Am. J. Trop. Med. Hyg. 1981, 30, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, F.P.; Travassos da Rosa, A.P.; Gomes, M.L.; LeDuc, J.W.; Hoch, A.L. Transmission of Oropouche virus from man to hamster by the midge Culicoides paraensis. Science 1982, 215, 1251–1253. [Google Scholar] [CrossRef]
- de Mendonça, S.F.; Rocha, M.N.; Ferreira, F.V.; Leite, T.H.J.F.; Amadou, S.C.G.; Sucupira, P.H.F.; Marques, J.T.; Ferreira, A.G.A.; Moreira, L.A. Evaluation of Aedes aegypti, Aedes albopictus, and Culex quinquefasciatus mosquitoes competence to Oropouche virus infection. Viruses 2021, 13, 755. [Google Scholar] [CrossRef]
- Gutierrez, B.; Wise, E.L.; Pullan, S.T.; Logue, C.H.; Bowden, T.A.; Escalera-Zamudio, M.; Trueba, G.; Nunes, M.R.T.; Faria, N.R.; Pybus, O.G. Evolutionary Dynamics of Oropouche Virus in South America. J. Virol. 2020, 94, e01127-19. [Google Scholar] [CrossRef]
- Schwarz, M.M.; Price, D.A.; Ganaie, S.S.; Feng, A.; Mishra, N.; Hoehl, R.M.; Fatma, F.; Stubbs, S.H.; Whelan, S.P.J.; Cui, X.; et al. Oropouche orthobunyavirus infection is mediated by the cellular host factor Lrp1. Proc. Natl. Acad. Sci. USA 2022, 119, e2204706119. [Google Scholar] [CrossRef]
- National Center for Biological Information. Available online: https://www.ncbi.nlm.nih.gov/protein/ (accessed on 8 August 2024).
- National Center for Biological Information. Conserved Domains. Available online: https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi (accessed on 12 August 2024).
- PTMGPT2: Post-Translational Modification Prediction via Prompt-Based Fine-Tuning of a GPT-2 Model. Available online: https://nsclbio.jbnu.ac.kr/tools/ptmgpt2 (accessed on 7 October 2025).
- Shrestha, P.; Kandel, J.; Tayara, H.; Chong, K.T. Post-translational modification prediction via prompt-based fine-tuning of a GPT-2 model. Nat. Commun. 2024, 15, 6699. [Google Scholar] [CrossRef]
- Skrabanek, L.; Campagne, F.; Weinstein, H. Building protein diagrams on the web with the residue-based diagram editor RbDe. Nucleic Acids Res. 2003, 31, 3856–3858. [Google Scholar] [CrossRef]
- Protter. Interactive Protein Feature Visualization. Available online: https://wlab.ethz.ch/protter/start/ (accessed on 19 August 2024).
- Omasits, U.; Ahrens, C.H.; Müller, S.; Wollscheid, B. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 2014, 30, 884–886. [Google Scholar] [CrossRef]
- Phobius. A Combined Transmembrane Topology and Signal Peptide Predictor. Available online: https://phobius.sbc.su.se/ (accessed on 13 August 2024).
- Thomas, S. Mapping the accessory proteins (Orfs) of Severe Acute Respiratory Syndrome Coronavirus 2. Re:GEN Open 2023, 3, 1–10. [Google Scholar] [CrossRef]
- Thomas, S. Plotting the major proteins of borealpox virus. Front. Virol. 2024, 4, 1451810. [Google Scholar] [CrossRef]
- Alphafold2. Available online: https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb (accessed on 12 August 2024).
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Phyre2. Protein Fold Recognition Server. Available online: http://www.sbg.bio.ic.ac.uk/phyre2 (accessed on 15 August 2024).
- I-TASSER. Protein Structure and Function Predictions. Available online: https://zhanggroup.org/I-TASSER/ (accessed on 12 November 2024).
- Yang, J.; Wang, Y.; Zhang, Y. ResQ: An approach to unified estimation of B-factor and residue-specific error in protein structure prediction. J. Mol. Biol. 2016, 428, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y. Protein Structure and Function Prediction Using I-TASSER. Curr. Protoc. Bioinform. 2015, 52, 5.8.1–5.8.15. [Google Scholar] [CrossRef]
- Zhang, Y.; Skolnick, J. TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005, 33, 2302–2309. [Google Scholar] [CrossRef]
- Kufareva, I.; Abagyan, R. Methods of protein structure comparison. Methods Mol. Biol. 2012, 857, 231–257. [Google Scholar]
- IEDB Analysis Resource. Available online: http://tools.iedb.org/main/ (accessed on 5 September 2024).
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef]
- Department of Health Technology. NetSurfP-3.0. Protein Secondary Structure and Relative Solvent Accessibility. Available online: https://services.healthtech.dtu.dk/services/NetSurfP-3.0/ (accessed on 22 October 2024).
- Høie, M.H.; Kiehl, E.N.; Petersen, B.; Nielsen, M.; Winther, O.; Nielsen, H.; Hallgren, J.; Marcatili, P. NetSurfP-3.0: Accurate and fast prediction of protein structural features by protein language models and deep learning. Nucleic Acids Res. 2022, 50, W510–W515. [Google Scholar] [CrossRef]
- UniProt. Available online: https://www.uniprot.org/align (accessed on 11 September 2025).
- Zhang, Y.; Liu, X.; Wu, Z.; Feng, S.; Lu, K.; Zhu, W.; Sun, H.; Niu, G. Oropouche virus: A neglected global arboviral threat. Virus Res. 2024, 341, 199318. [Google Scholar] [CrossRef]
- Endalew, A.D.; Faburay, B.; Wilson, W.C.; Richt, J.A. Schmallenberg disease—A newly emerged culicoides-borne viral disease of ruminants. Viruses 2019, 11, 1065. [Google Scholar] [CrossRef]
- Pinheiro, F.P.; Pinheiro, M.; Bensabath, G.; Causey, O.R.; Shope, R.R. Epidemia de vírus Oropouche em Belém. Rev. Serv. Esp. Saúde Públ. 1962, 12, 15–23. [Google Scholar]
- Henry, R.; Murphy, F.A. Etymologia: Oropouche virus. Emerg. Infect. Dis. 2018, 24, 937. [Google Scholar] [CrossRef]
- Sah, R.; Srivastava, S.; Kumar, S.; Golmei, P.; Rahaman, S.A.; Mehta, R.; Ferraz, C.; Apostolopoulos, V.; Rodriguez-Morales, A.J. Oropouche fever outbreak in Brazil: An emerging concern in Latin America. Lancet Microbe 2024, 3, 100904. [Google Scholar] [CrossRef]
- World Health Organization. Oropouche Virus Disease—Cuba. 2024. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2024-DON521#:~:text=Oropouche%20virus%20disease%20is%20an,to%2Dhuman%20Oropouche%20virus%20transmission (accessed on 15 August 2024).
- European Centre for Disease Prevention and Control. Oropouche Virus Disease Cases Imported into the European Union—9 August 2024; ECDC: Stockholm, Sweden, 2024. [Google Scholar]
- World Health Organization. Oropouche Virus Disease. 2024. Available online: https://www.who.int/news-room/fact-sheets/detail/oropouche-virus-disease#:~:text=In%202024%20concerns%20arose%20about,that%20requires%20further%20investigation%20and (accessed on 15 August 2024).
- Adhikari, U.K.; Tayebi, M.; Rahman, M.M. Immunoinformatics approach for epitope-based peptide vaccine design and active site prediction against polyprotein of emerging Oropouche virus. J. Immunol. Res. 2018, 2018, 6718083. [Google Scholar] [CrossRef]
- Stubbs, S.H.; Cornejo Pontelli, M.; Mishra, N.; Zhou, C.; de Paula Souza, J.; Mendes Viana, R.M.; Lipkin, W.I.; Knipe, D.M.; Arruda, E.; Whelan, S.P.J. Vesicular stomatitis virus chimeras expressing the Oropouche virus glycoproteins elicit protective immune responses in mice. mBio 2021, 12, e0046321. [Google Scholar] [CrossRef] [PubMed]
- Scheiblhofer, S.; Laimer, J.; Machado, Y.; Weiss, R.; Thalhamer, J. Influence of protein fold stability on immunogenicity and its implications for vaccine design. Expert Rev. Vaccines 2017, 16, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, T.A.; Eweida, A.E.; Sheweita, S.A. B-cell epitope mapping for the design of vaccines and effective diagnostics. Trials Vaccinol. 2016, 5, 71–83. [Google Scholar] [CrossRef]
- Wong, D.A.; Shaver, Z.M.; Cabezas, M.D.; Daniel-Ivad, M.; Warfel, K.F.; Prasanna, D.V.; Sobol, S.E.; Fernandez, R.; Tobias, F.; Filip, S.K.; et al. Characterizing and engineering post-translational modifications with high-throughput cell-free expression. Nat. Commun. 2025, 16, 7215. [Google Scholar] [CrossRef]
- Kumar, R.; Mehta, D.; Mishra, N.; Nayak, D.; Sunil, S. Role of host-mediated post-translational modifications (PTMs) in RNA virus pathogenesis. Int. J. Mol. Sci. 2020, 22, 323. [Google Scholar] [CrossRef]
- Thomas, S.; Thirumalapura, N.; Crossley, E.C.; Ismail, N.; Walker, D.H. Antigenic protein modifications in Ehrlichia. Parasite Immunol. 2009, 31, 296–303. [Google Scholar] [CrossRef]
- Thomas, S.; Thirumalapura, N.R.; Crocquet-Valdes, P.A.; Luxon, B.A.; Walker, D.H. Structure-based vaccines provide protection in a mouse model of ehrlichiosis. PLoS ONE 2011, 6, e27981. [Google Scholar] [CrossRef]
- Yee, D.S.; Davies, J.P.; VanBlargan, L.A.; Rahmberg, A.R.; Burgomaster, K.E.; Brooks, K.; Flynn, J.K.; Shue, B.; Ortiz, A.M.; Schaughency, P.; et al. Oropouche virus efficiently replicates and is immunostimulatory in vivo in nonhuman primate species. Sci. Adv. 2025, 11, eadx9405. [Google Scholar] [CrossRef]
- Navarro, J.C.; Giambalvo, D.; Hernandez, R.; Auguste, A.J.; Tesh, R.B.; Weaver, S.C.; Montañez, H.; Liria, J.; Lima, A.; Travassos da Rosa, J.F.; et al. Isolation of Madre de Dios Virus (Orthobunyavirus; Bunyaviridae), an Oropouche virus species reassortant, from a monkey in Venezuela. Am. J. Trop. Med. Hyg. 2016, 95, 328–338. [Google Scholar] [CrossRef]
| OROV Protein | Accession Number (Amino Acid) | Function |
|---|---|---|
| Gn glycoprotein | UYI36405 (1–350) | Viral entry and exit |
| Gc glycoprotein | UYI36405 (482–1420) | Interaction with host cells |
| NSm non-structural protein | UYI36405 (351–481) | Virus assembly and budding |
| NSs non-structural protein | AEH03002 | Evade host immunity |
| Nucleocapsid protein | AJE24680 | Encapsulates viral RNA |
| RNA polymerase | AJE24678 | Replication |
| Protein | C-Score | TM-Score | RMSD |
|---|---|---|---|
| Gn glycoprotein | −2.21 | 0.45 ± 0.15 | 11.7 ± 4.5 Å |
| Gc glycoprotein | −2.85 | 0.39 ± 0.13 | 16.1 ± 3.1 Å |
| NSm non-structural protein | −4.30 | 0.26 ± 0.08 | 14.7 ± 3.6 Å |
| NSs non-structural protein | −3.25 | 0.35 ± 0.12 | 11.0 ± 4.6 Å |
| Nucleocapsid protein | 1.57 | 0.93 ± 0.06 | 2.6 ± 1.9 Å |
| RNA polymerase | 0.52 | 0.78 ± 0.09 | 8.6 ± 4.5 Å |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thomas, S. Charting the Proteins of Oropouche Virus. Viruses 2025, 17, 1434. https://doi.org/10.3390/v17111434
Thomas S. Charting the Proteins of Oropouche Virus. Viruses. 2025; 17(11):1434. https://doi.org/10.3390/v17111434
Chicago/Turabian StyleThomas, Sunil. 2025. "Charting the Proteins of Oropouche Virus" Viruses 17, no. 11: 1434. https://doi.org/10.3390/v17111434
APA StyleThomas, S. (2025). Charting the Proteins of Oropouche Virus. Viruses, 17(11), 1434. https://doi.org/10.3390/v17111434






