Manipulation of Nuclear-Related Pathways During Kaposi’s Sarcoma-Associated Herpesvirus Lytic Replication
Abstract
1. Introduction
2. What Impact Does KSHV Lytic Replication Have on the Nuclear Architecture?
2.1. Formation of KSHV Replication Centres

2.2. How Does vRTC Formation Affect Sub-Nuclear Organelles?
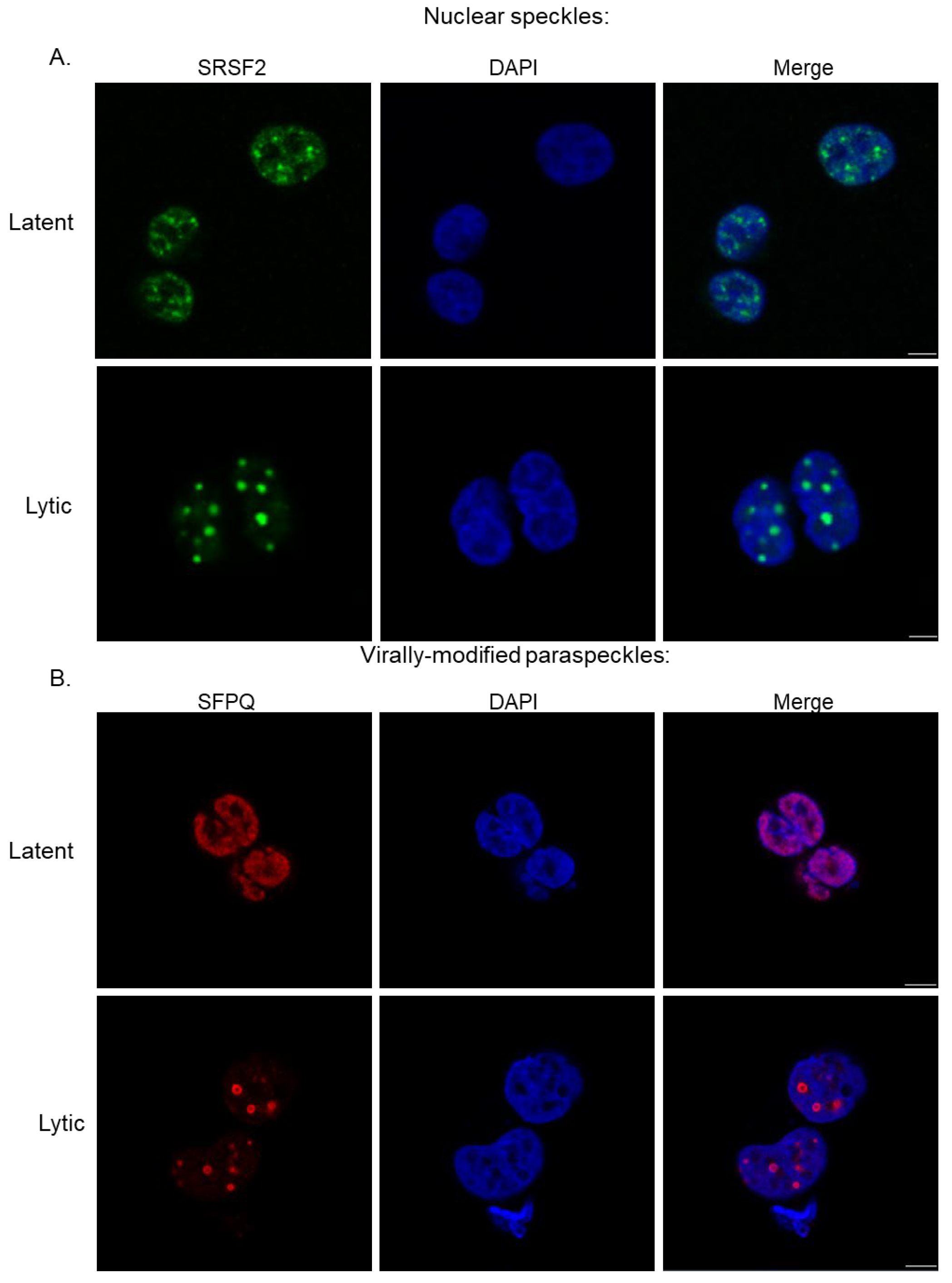
2.2.1. Nuclear Speckles and Paraspeckles
2.2.2. PML Nuclear Bodies
2.2.3. Stress Granule Modulation
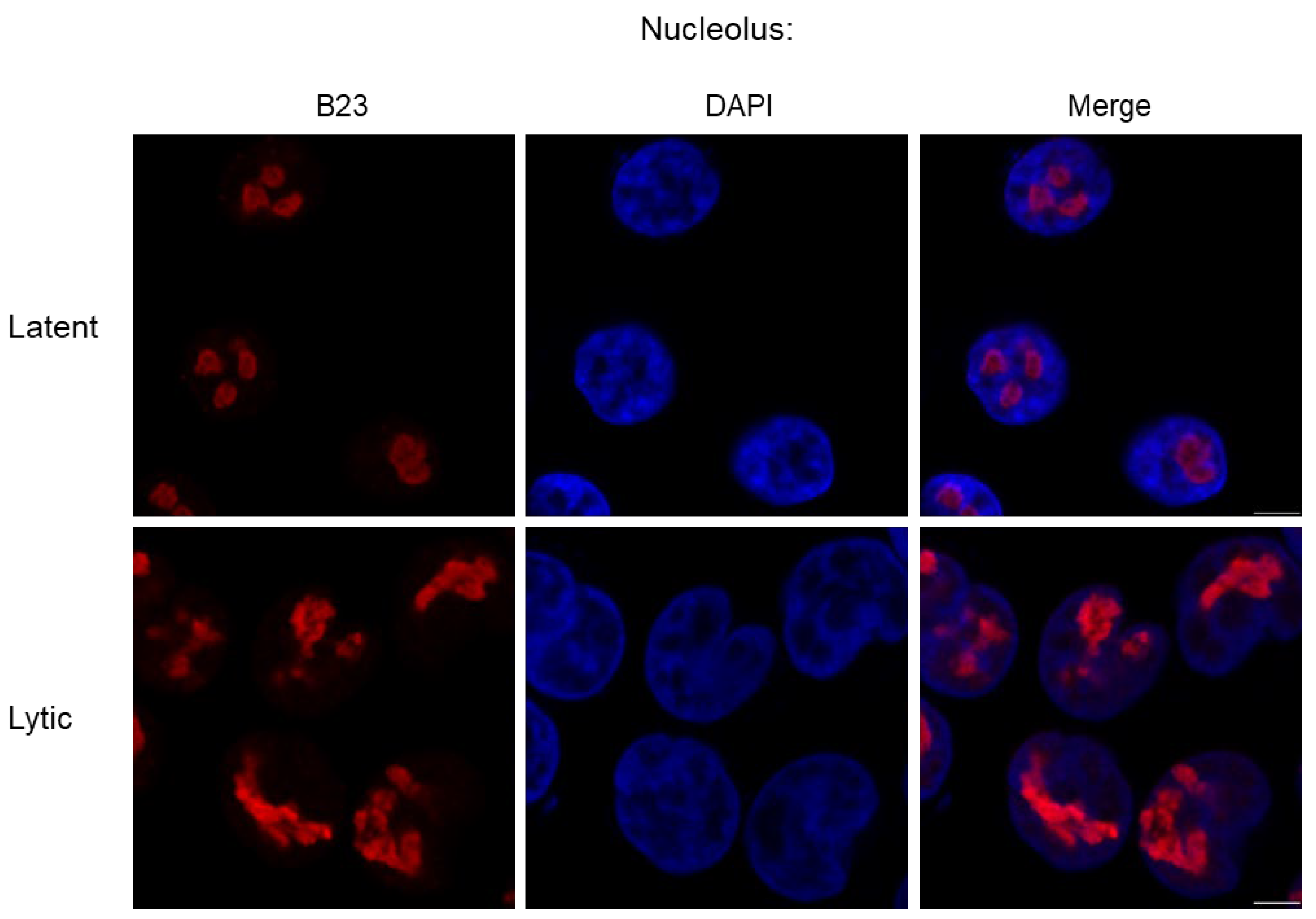
2.2.4. Nucleolus
3. What Impact Does KSHV Lytic Replication Have on Nuclear-Related Pathways?
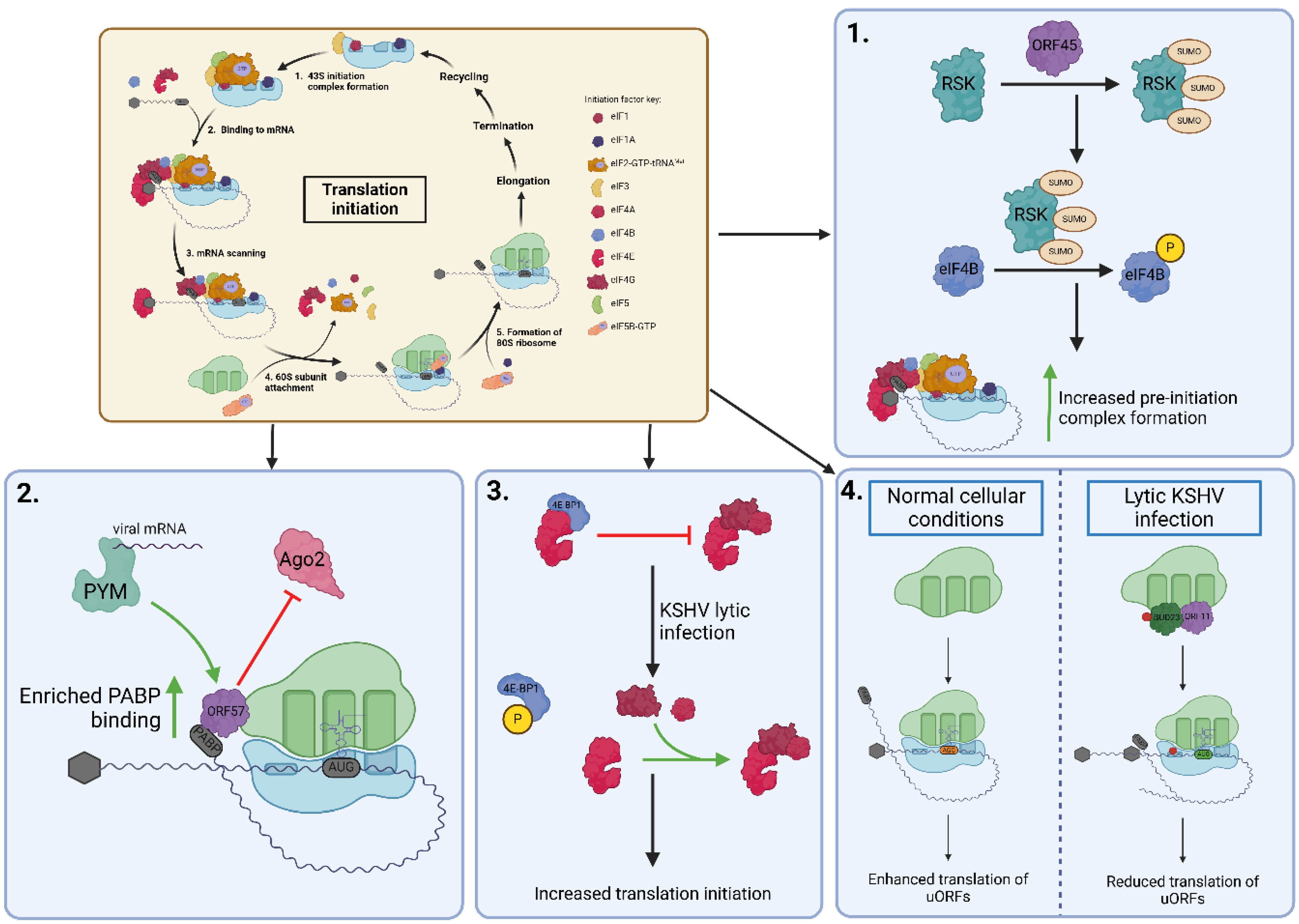
3.1. Ribosomal Biogenesis and Translation Control
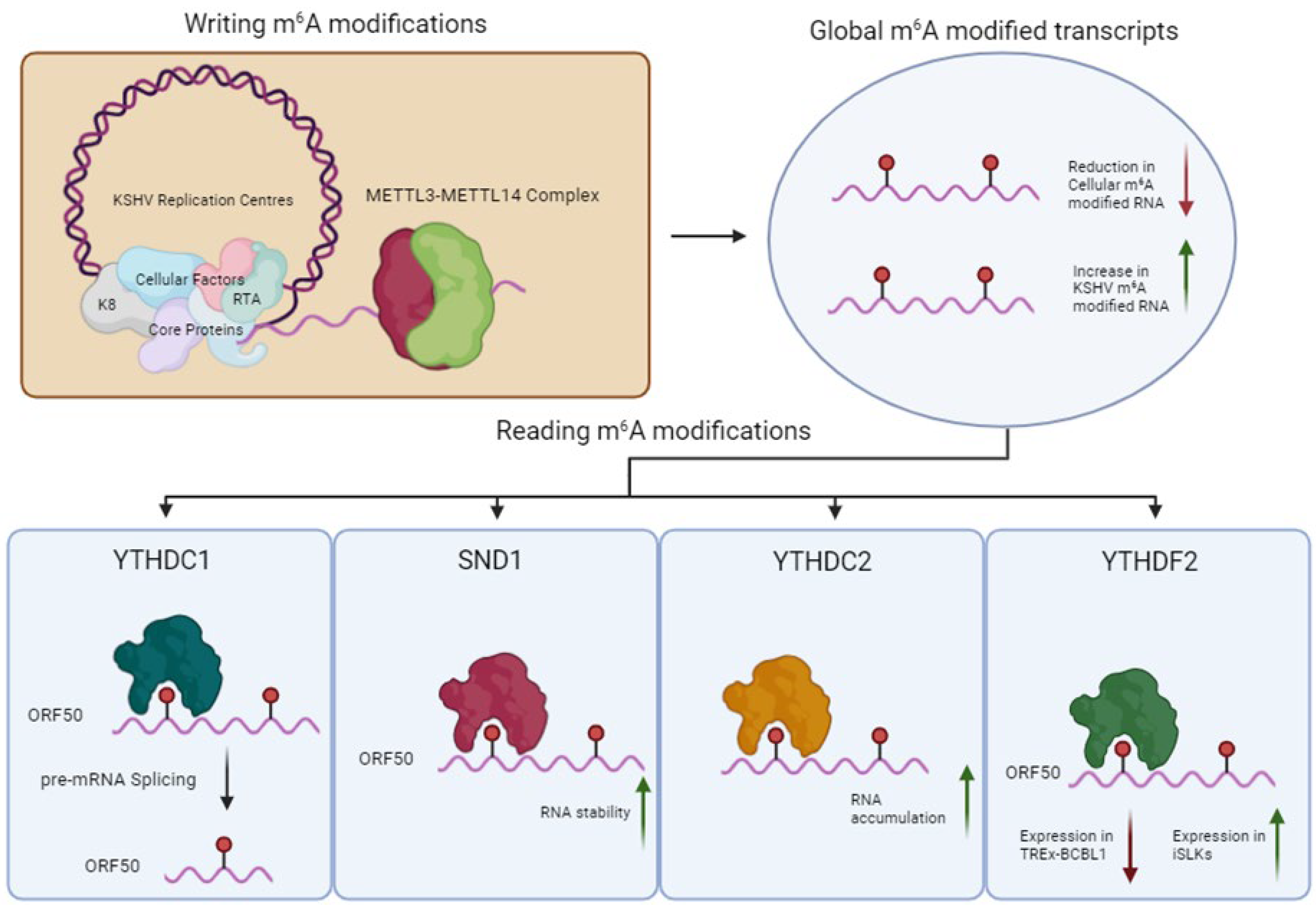
3.2. RNA Modification Pathways
3.3. Epigenetic Control Mechanisms
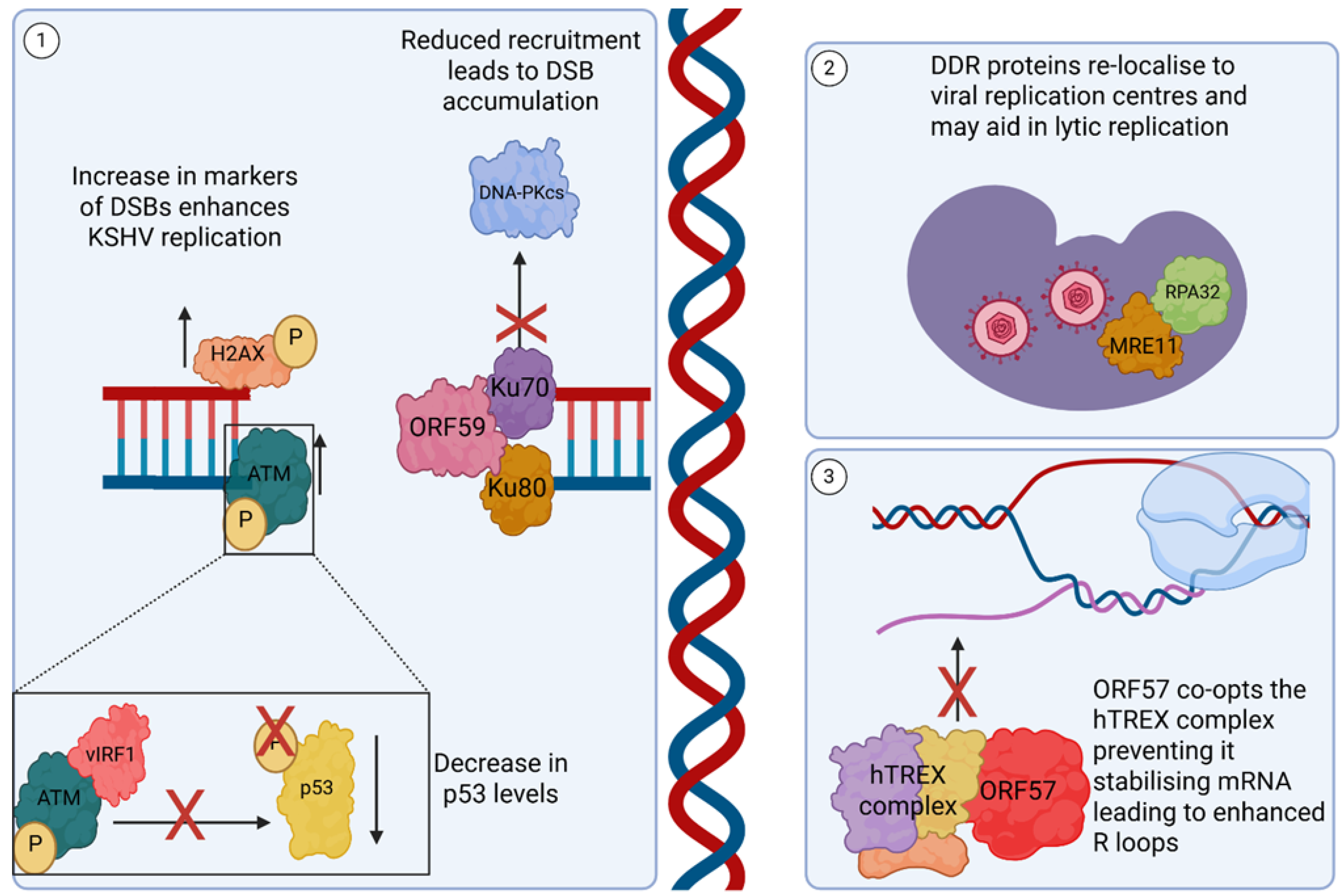
3.4. DNA Damage-Related Pathways
3.5. Tumourigenesis-Related Pathways
4. Summary
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Manners, O.; Murphy, J.C.; Coleman, A.; Hughes, D.J.; Whitehouse, A. Contribution of the KSHV and EBV lytic cycles to tumourigenesis. Curr. Opin. Virol. 2018, 32, 60–70. [Google Scholar] [CrossRef]
- Carden, H.; Harper, K.L.; Mottram, T.J.; Manners, O.; Allott, K.L.; Dallas, M.L.; Hughes, D.J.; Lippiat, J.D.; Mankouri, J.; Whitehouse, A. Kv1. 3-induced hyperpolarization is required for efficient Kaposi’s sarcoma–associated herpesvirus lytic replication. Sci. Signal 2024, 17, eadg4124. [Google Scholar] [CrossRef] [PubMed]
- Dittmer, D.; Lagunoff, M.; Renne, R.; Staskus, K.; Haase, A.; Ganem, D. A cluster of latently expressed genes in Kaposi’s sarcoma-associated herpesvirus. J. Virol. 1998, 72, 8309–8315. [Google Scholar] [CrossRef] [PubMed]
- Staskus, K.A.; Zhong, W.; Gebhard, K.; Herndier, B.; Wang, H.; Renne, R.; Beneke, J.; Pudney, J.; Anderson, D.J.; Ganem, D.; et al. Kaposi’s sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 1997, 71, 715–719. [Google Scholar] [CrossRef]
- McClure, L.V.; Sullivan, C.S. Kaposi’s sarcoma herpes virus taps into a host microRNA regulatory network. Cell Host Microbe 2008, 3, 1–3. [Google Scholar] [CrossRef]
- Broussard, G.; Damania, B. Regulation of KSHV Latency and Lytic Reactivation. Viruses 2020, 12, 1034. [Google Scholar] [CrossRef]
- Arias, C.; Weisburd, B.; Stern-Ginossar, N.; Mercier, A.; Madrid, A.S.; Bellare, P.; Holdorf, M.; Weissman, J.S.; Ganem, D. KSHV 2.0: A comprehensive annotation of the Kaposi’s sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog. 2014, 10, e1003847. [Google Scholar] [CrossRef]
- Baquero-Pérez, B.; Whitehouse, A. Hsp70 Isoforms Are Essential for the Formation of Kaposi’s Sarcoma-Associated Herpesvirus Replication and Transcription Compartments. PLoS Pathog. 2015, 11, e1005274. [Google Scholar] [CrossRef]
- McNamee, E.E.; Taylor, T.J.; Knipe, D.M. A dominant-negative herpesvirus protein inhibits intranuclear targeting of viral proteins: Effects on DNA replication and late gene expression. J. Virol. 2000, 74, 10122–10131. [Google Scholar] [CrossRef]
- Monier, K.; Armas, J.C.; Etteldorf, S.; Ghazal, P.; Sullivan, K.F. Annexation of the interchromosomal space during viral infection. Nat. Cell Biol. 2000, 2, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Taylor, T.J.; McNamee, E.E.; Day, C.; Knipe, D.M. Herpes simplex virus replication compartments can form by coalescence of smaller compartments. Virology 2003, 309, 232–247. [Google Scholar] [CrossRef]
- Chen, L.-W.; Wang, S.-S.; Chen, L.-Y.; Huang, H.-Y.; He, S.-M.; Hung, C.-H.; Lin, C.-L.; Chang, P.-J. Interaction and assembly of the DNA replication core proteins of Kaposi’s sarcoma-associated herpesvirus. Microbiol. Spectr. 2023, 11, e0225423. [Google Scholar] [CrossRef]
- Wu, F.Y.; Ahn, J.H.; Alcendor, D.J.; Jang, W.J.; Xiao, J.; Hayward, S.D.; Hayward, G.S. Origin-independent assembly of Kaposi’s sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 2001, 75, 1487–1506. [Google Scholar] [CrossRef]
- Romero-Brey, I. 3D Electron Microscopy (EM) and Correlative Light Electron Microscopy (CLEM) Methods to Study Virus-Host Interactions. Methods Mol. Biol. 2018, 1836, 213–236. [Google Scholar]
- Dahmane, S.; Kerviel, A.; Morado, D.R.; Shankar, K.; Ahlman, B.; Lazarou, M.; Altan-Bonnet, N.; Carlson, L.A. Membrane-assisted assembly and selective secretory autophagy of enteroviruses. Nat. Commun. 2022, 13, 5986. [Google Scholar] [CrossRef]
- Harper, K.L.; Harrington, E.M.; Hayward, C.; Anene, C.A.; Wongwiwat, W.; White, R.E.; Whitehouse, A. Virus-modified paraspeckle-like condensates are hubs for viral RNA processing and their formation drives genomic instability. Nat. Commun. 2024, 15, 10240. [Google Scholar] [CrossRef]
- Giudice, J.; Jiang, H. Splicing regulation through biomolecular condensates and membraneless organelles. Nat. Rev. Mol. Cell Biol. 2024, 25, 683–700. [Google Scholar] [CrossRef] [PubMed]
- Ingram, H.B.; Fox, A.H. Unveiling the intricacies of paraspeckle formation and function. Curr. Opin. Cell Biol. 2024, 90, 102399. [Google Scholar] [CrossRef] [PubMed]
- McCluggage, F.; Fox, A.H. Paraspeckle nuclear condensates: Global sensors of cell stress? Bioessays 2021, 43, e2000245. [Google Scholar] [CrossRef] [PubMed]
- Majerciak, V.; Alvarado-Hernandez, B.; Lobanov, A.; Cam, M.; Zheng, Z.M. Genome-wide regulation of KSHV RNA splicing by viral RNA-binding protein ORF57. PLoS Pathog. 2022, 18, e1010311. [Google Scholar] [CrossRef]
- Alkalay, E.; Gam Ze Letova Refael, C.; Shoval, I.; Kinor, N.; Sarid, R.; Shav-Tal, Y. The Sub-Nuclear Localization of RNA-Binding Proteins in KSHV-Infected Cells. Cells 2020, 9, 1958. [Google Scholar] [CrossRef]
- Kleer, M.; Johnston, M.J.; Corcoran, J.A. Nuclear body reorganization by the viral RNA kaposin promotes Kaposi’s sarcoma-associated herpesvirus gene expression. bioRxiv 2024. [Google Scholar] [CrossRef]
- Harper, K.L.; Mottram, T.J.; Anene, C.A.; Foster, B.; Patterson, M.R.; McDonnell, E.; Macdonald, A.; Westhead, D.; Whitehouse, A. Dysregulation of the miR-30c/DLL4 axis by circHIPK3 is essential for KSHV lytic replication. EMBO Rep. 2022, 23, e54117. [Google Scholar] [CrossRef] [PubMed]
- Jackson, B.R.; Noerenberg, M.; Whitehouse, A. A novel mechanism inducing genome instability in Kaposi’s sarcoma-associated herpesvirus infected cells. PLoS Pathog. 2014, 10, e1004098. [Google Scholar] [CrossRef] [PubMed]
- Scherer, M.; Stamminger, T. Emerging Role of PML Nuclear Bodies in Innate Immune Signaling. J. Virol. 2016, 90, 5850–5854. [Google Scholar] [CrossRef]
- Everett, R.D.; Chelbi-Alix, M.K. PML and PML nuclear bodies: Implications in antiviral defence. Biochimie 2007, 89, 819–830. [Google Scholar] [CrossRef]
- Neerukonda, S.N. Interplay between RNA Viruses and Promyelocytic Leukemia Nuclear Bodies. Vet. Sci. 2021, 8, 57. [Google Scholar] [CrossRef]
- Geoffroy, M.C.; Chelbi-Alix, M.K. Role of promyelocytic leukemia protein in host antiviral defense. J. Interferon Cytokine Res. 2011, 31, 145–158. [Google Scholar] [CrossRef]
- Tavalai, N.; Stamminger, T. Interplay between Herpesvirus Infection and Host Defense by PML Nuclear Bodies. Viruses 2009, 1, 1240–1264. [Google Scholar] [CrossRef]
- Chee, A.V.; Lopez, P.; Pandolfi, P.P.; Roizman, B. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J. Virol. 2003, 77, 7101–7105. [Google Scholar] [CrossRef]
- Scherer, M.; Read, C.; Neusser, G.; Kranz, C.; Kuderna, A.K.; Müller, R.; Full, F.; Wörz, S.; Reichel, A.; Schilling, E.M.; et al. Dual signaling via interferon and DNA damage response elicits entrapment by giant PML nuclear bodies. Elife 2022, 11, e73006. [Google Scholar] [CrossRef]
- Full, F.; Jungnickl, D.; Reuter, N.; Bogner, E.; Brulois, K.; Scholz, B.; Stürzl, M.; Myoung, J.; Jung, J.U.; Stamminger, T.; et al. Kaposi’s sarcoma associated herpesvirus tegument protein ORF75 is essential for viral lytic replication and plays a critical role in the antagonization of ND10-instituted intrinsic immunity. PLoS Pathog. 2014, 10, e1003863. [Google Scholar] [CrossRef]
- Reichelt, M.; Wang, L.; Sommer, M.; Perrino, J.; Nour, A.M.; Sen, N.; Baiker, A.; Zerboni, L.; Arvin, A.M. Entrapment of viral capsids in nuclear PML cages is an intrinsic antiviral host defense against varicella-zoster virus. PLoS Pathog. 2011, 7, e1001266. [Google Scholar] [CrossRef]
- Paget, M.; Cadena, C.; Ahmad, S.; Wang, H.T.; Jordan, T.X.; Kim, E.; Koo, B.; Lyons, S.M.; Ivanov, P.; tenOever, B.; et al. Stress granules are shock absorbers that prevent excessive innate immune responses to dsRNA. Mol. Cell 2023, 83, 1180–1196.e8. [Google Scholar] [CrossRef] [PubMed]
- Schumann, S.; Jackson, B.R.; Yule, I.; Whitehead, S.K.; Revill, C.; Foster, R.; Whitehouse, A. Targeting the ATP-dependent formation of herpesvirus ribonucleoprotein particle assembly as an antiviral approach. Nat. Microbiol. 2016, 2, 16201. [Google Scholar] [CrossRef]
- Sharma, N.R.; Majerciak, V.; Kruhlak, M.J.; Zheng, Z.M. KSHV inhibits stress granule formation by viral ORF57 blocking PKR activation. PLoS Pathog. 2017, 13, e1006677. [Google Scholar] [CrossRef]
- An, H.; Tan, J.T.; Shelkovnikova, T.A. Stress granules regulate stress-induced paraspeckle assembly. J. Cell Biol. 2019, 218, 4127–4140. [Google Scholar] [CrossRef]
- Nakajima, K.I.; Guevara-Plunkett, S.; Chuang, F.; Wang, K.H.; Lyu, Y.; Kumar, A.; Luxardi, G.; Izumiya, C.; Soulika, A.; Campbell, M.; et al. Rainbow Kaposi’s Sarcoma-Associated Herpesvirus Revealed Heterogenic Replication with Dynamic Gene Expression. J. Virol. 2020, 94, e01565-19. [Google Scholar] [CrossRef] [PubMed]
- Dubois, M.L.; Boisvert, F.M. The Nucleolus: Structure and Function. In The Functional Nucleus; Springer: Cham, Switzerland, 2016; pp. 29–49. [Google Scholar]
- Yang, K.; Yang, J.; Yi, J. Nucleolar Stress: Hallmarks, sensing mechanism and diseases. Cell Stress 2018, 2, 125–140. [Google Scholar] [CrossRef] [PubMed]
- James, A.; Wang, Y.; Raje, H.; Rosby, R.; DiMario, P. Nucleolar stress with and without p53. Nucleus 2014, 5, 402–426. [Google Scholar] [CrossRef]
- Atari, N.; Rajan, K.S.; Chikne, V.; Cohen-Chalamish, S.; Doniger, T.; Orbaum, O.; Jacob, A.; Kalt, I.; Michaeli, S.; Sarid, R. Lytic Reactivation of the Kaposi’s Sarcoma-Associated Herpesvirus (KSHV) Is Accompanied by Major Nucleolar Alterations. Viruses 2022, 14, 1720. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.C.; Harrington, E.M.; Schumann, S.; Vasconcelos, E.J.R.; Mottram, T.J.; Harper, K.L.; Aspden, J.L.; Whitehouse, A. Kaposi’s sarcoma-associated herpesvirus induces specialised ribosomes to efficiently translate viral lytic mRNAs. Nat. Commun. 2023, 14, 300. [Google Scholar] [CrossRef] [PubMed]
- Harrington, E.M.; Murphy, J.C.; Harper, K.L.; Hayward, C.; Mottram, T.J.; Aspden, J.L.; Whitehouse, A. EMG1 methyltransferase activity affects ribosome occupancy at KSHV uORFs. Cell Rep. 2025, 44, 115516. [Google Scholar] [CrossRef]
- Hiscox, J.A.; Whitehouse, A.; Matthews, D.A. Nucleolar proteomics and viral infection. Proteomics 2010, 10, 4077–4086. [Google Scholar] [CrossRef]
- Boyne, J.R.; Jackson, B.R.; Taylor, A.; Macnab, S.A.; Whitehouse, A. Kaposi’s sarcoma-associated herpesvirus ORF57 protein interacts with PYM to enhance translation of viral intronless mRNAs. EMBO J. 2010, 29, 1851–1864. [Google Scholar] [CrossRef]
- Boyne, J.R.; Whitehouse, A. Nucleolar disruption impairs Kaposi’s sarcoma-associated herpesvirus ORF57-mediated nuclear export of intronless viral mRNAs. FEBS Lett. 2009, 583, 3549–3556. [Google Scholar] [CrossRef]
- Gabaev, I.; Williamson, J.C.; Crozier, T.W.M.; Schulz, T.F.; Lehner, P.J. Quantitative Proteomics Analysis of Lytic KSHV Infection in Human Endothelial Cells Reveals Targets of Viral Immune Modulation. Cell Rep. 2020, 33, 108249. [Google Scholar] [CrossRef]
- Glaunsinger, B.; Ganem, D. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol. Cell 2004, 13, 713–723. [Google Scholar] [CrossRef]
- Schumann, S.; Jackson, B.R.; Baquero-Perez, B.; Whitehouse, A. Kaposi’s sarcoma-associated herpesvirus ORF57 protein: Exploiting all stages of viral mRNA processing. Viruses 2013, 5, 1901–1923. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Jiang, B.; Zhang, Y. 4E-BP1, a multifactor regulated multifunctional protein. Cell Cycle 2016, 15, 781–786. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.; Walsh, D.; Harbell, J.; Wilson, A.C.; Mohr, I. Activation of host translational control pathways by a viral developmental switch. PLoS Pathog. 2009, 5, e1000334. [Google Scholar] [CrossRef] [PubMed]
- Kuang, E.; Tang, Q.; Maul, G.G.; Zhu, F. Activation of p90 ribosomal S6 kinase by ORF45 of Kaposi’s sarcoma-associated herpesvirus and its role in viral lytic replication. J. Virol. 2008, 82, 1838–1850. [Google Scholar] [CrossRef]
- Kuang, E.; Fu, B.; Liang, Q.; Myoung, J.; Zhu, F. Phosphorylation of eukaryotic translation initiation factor 4B (EIF4B) by open reading frame 45/p90 ribosomal S6 kinase (ORF45/RSK) signaling axis facilitates protein translation during Kaposi sarcoma-associated herpesvirus (KSHV) lytic replication. J. Biol. Chem. 2011, 286, 41171–41182. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, C.; Wang, X.; Li, W.; Zhou, J.; Dong, P.; Xiao, M.Z.X.; Wang, C.; Zhang, Y.; Fu, J.; et al. RSK1 SUMOylation is required for KSHV lytic replication. PLoS Pathog. 2021, 17, e1010123. [Google Scholar] [CrossRef]
- Alvarado-Hernandez, B.; Ma, Y.; Sharma, N.R.; Majerciak, V.; Lobanov, A.; Cam, M.; Zhu, J.; Zheng, Z.M. Protein-RNA Interactome Analysis Reveals Wide Association of Kaposi’s Sarcoma-Associated Herpesvirus ORF57 with Host Noncoding RNAs and Polysomes. J. Virol. 2022, 96, e0178221. [Google Scholar] [CrossRef]
- Mendez-Solis, O.; Bendjennat, M.; Naipauer, J.; Theodoridis, P.R.; Ho, J.J.D.; Verdun, R.E.; Hare, J.M.; Cesarman, E.; Lee, S.; Mesri, E.A. Kaposi’s sarcoma herpesvirus activates the hypoxia response to usurp HIF2alpha-dependent translation initiation for replication and oncogenesis. Cell Rep. 2021, 37, 110144. [Google Scholar] [CrossRef]
- Norris, K.; Hopes, T.; Aspden, J.L. Ribosome heterogeneity and specialization in development. Wiley Interdiscip. Rev. RNA 2021, 12, e1644. [Google Scholar] [CrossRef] [PubMed]
- Haag, S.; Kretschmer, J.; Bohnsack, M.T. WBSCR22/Merm1 is required for late nuclear pre-ribosomal RNA processing and mediates N7-methylation of G1639 in human 18S rRNA. RNA 2015, 21, 180–187. [Google Scholar] [CrossRef]
- Wurm, J.P.; Meyer, B.; Bahr, U.; Held, M.; Frolow, O.; Kotter, P.; Engels, J.W.; Heckel, A.; Karas, M.; Entian, K.D.; et al. The ribosome assembly factor Nep1 responsible for Bowen-Conradi syndrome is a pseudouridine-N1-specific methyltransferase. Nucleic Acids Res. 2010, 38, 2387–2398. [Google Scholar] [CrossRef]
- Baquero-Perez, B.; Geers, D.; Díez, J. From A to m6A: The Emerging Viral Epitranscriptome. Viruses 2021, 13, 1049. [Google Scholar] [CrossRef] [PubMed]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef]
- Yang, Y.; Hsu, P.J.; Chen, Y.S.; Yang, Y.G. Dynamic transcriptomic m6A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772. [Google Scholar] [CrossRef]
- Shi, H.; Wei, J.; He, C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 2019, 74, 640–650. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Ke, S.; Pandya-Jones, A.; Saito, Y.; Fak, J.J.; Vågbø, C.B.; Geula, S.; Hanna, J.H.; Black, D.L.; Darnell, J.E.; Darnell, R.B. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes. Dev. 2017, 31, 990–1006. [Google Scholar] [CrossRef]
- Li, F.; Zhao, D.; Wu, J.; Shi, Y. Structure of the YTH domain of human YTHDF2 in complex with an m(6)A mononucleotide reveals an aromatic cage for m(6)A recognition. Cell Res. 2014, 24, 1490–1492. [Google Scholar] [CrossRef]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef]
- Meyer, K.D.; Jaffrey, S.R. Rethinking m(6)A Readers, Writers, and Erasers. Annu. Rev. Cell Dev. Biol. 2017, 33, 319–342. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vagbo, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Mauer, J.; Luo, X.; Blanjoie, A.; Jiao, X.; Grozhik, A.V.; Patil, D.P.; Linder, B.; Pickering, B.F.; Vasseur, J.J.; Chen, Q.; et al. Reversible methylation of m(6)A(m) in the 5’ cap controls mRNA stability. Nature 2017, 541, 371–375. [Google Scholar] [CrossRef]
- Tan, B.; Liu, H.; Zhang, S.; da Silva, S.R.; Zhang, L.; Meng, J.; Cui, X.; Yuan, H.; Sorel, O.; Zhang, S.W.; et al. Viral and Cellular N6-Methyladenosine (m6A) and N6, 2′-O-Dimethyladenosine (m6Am) Epitranscriptomes in KSHV Life Cycle. Nat. Microbiol. 2018, 3, 108–120. [Google Scholar] [CrossRef]
- Ye, F.; Chen, E.R.; Nilsen, T.W. Kaposi’s Sarcoma-Associated Herpesvirus Utilizes and Manipulates RNA N6-Adenosine Methylation To Promote Lytic Replication. J. Virol. 2017, 91, e00466-17. [Google Scholar] [CrossRef]
- Manners, O.; Baquero-Perez, B.; Whitehouse, A. m6A: Widespread regulatory control in virus replication. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 370–381. [Google Scholar] [CrossRef]
- Baquero-Perez, B.; Antanaviciute, A.; Yonchev, I.D.; Carr, I.M.; Wilson, S.A.; Whitehouse, A. The Tudor SND1 protein is an m6A RNA reader essential for replication of Kaposi’s sarcoma-associated herpesvirus. Elife 2019, 8, 47261. [Google Scholar] [CrossRef]
- Hesser, C.R.; Karijolich, J.; Dominissini, D.; He, C.; Glaunsinger, B.A. N6-methyladenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during Kaposi’s sarcoma-associated herpesvirus infection. PLoS Pathog. 2018, 14, e1006995. [Google Scholar] [CrossRef]
- Macveigh-Fierro, D.; Cicerchia, A.; Cadorette, A.; Sharma, V.; Muller, M. The m6A reader YTHDC2 is essential for escape from KSHV SOX-induced RNA decay. Proc. Natl. Acad. Sci. USA 2022, 119, e2116662119. [Google Scholar] [CrossRef]
- Nishikura, K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010, 79, 321–349. [Google Scholar] [CrossRef]
- Deffit, S.N.; Hundley, H.A. To edit or not to edit: Regulation of ADAR editing specificity and efficiency. Wiley Interdiscip. Rev. RNA 2016, 7, 113–127. [Google Scholar] [CrossRef]
- Rajendren, S.; Karijolich, J. The impact of RNA modifications on the biology of DNA virus infection. Eur. J. Cell Biol. 2022, 101, 151239. [Google Scholar] [CrossRef]
- Rajendren, S.; Ye, X.; Dunker, W.; Richardson, A.; Karijolich, J. The cellular and KSHV A-to-I RNA editome in primary effusion lymphoma and its role in the viral lifecycle. Nat. Commun. 2023, 14, 1367. [Google Scholar] [CrossRef]
- Zhang, H.; Ni, G.; Damania, B. ADAR1 Facilitates KSHV Lytic Reactivation by Modulating the RLR-Dependent Signaling Pathway. Cell Rep. 2020, 31, 107564. [Google Scholar] [CrossRef]
- Yan, Q.; Zhou, J.; Wang, Z.; Ding, X.; Ma, X.; Li, W.; Jia, X.; Gao, S.J.; Lu, C. NAT10-dependent N. Nat. Commun. 2023, 14, 6327. [Google Scholar] [CrossRef]
- Mottram, T.J.; Harper, K.L.; Vasconcelos, E.J.R.; Whitehouse, A. Pseudouridine prevalence in Kaposi’s sarcoma-associated herpesvirus transcriptome reveals an essential mechanism for viral replication. Proc. Natl. Acad. Sci. USA 2025, 122, e2508523122. [Google Scholar] [CrossRef]
- Inagaki, T.; Kumar, A.; Komaki, S.; Nakajima, K.I.; Izumiya, Y. An atlas of chromatin landscape in KSHV-infected cells during de novo infection and reactivation. Virology 2024, 597, 110146. [Google Scholar] [CrossRef]
- Campbell, M.; Yang, W.S.; Yeh, W.W.; Kao, C.H.; Chang, P.C. Epigenetic Regulation of Kaposi’s Sarcoma-Associated Herpesvirus Latency. Front. Microbiol. 2020, 11, 850. [Google Scholar] [CrossRef]
- Günther, T.; Grundhoff, A. The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog. 2010, 6, e1000935. [Google Scholar] [CrossRef]
- Kang, H.; Wiedmer, A.; Yuan, Y.; Robertson, E.; Lieberman, P.M. Coordination of KSHV latent and lytic gene control by CTCF-cohesin mediated chromosome conformation. PLoS Pathog. 2011, 7, e1002140. [Google Scholar] [CrossRef]
- Mesri, E.A.; Cesarman, E.; Boshoff, C. Kaposi’s sarcoma and its associated herpesvirus. Nat. Rev. Cancer 2010, 10, 707–719. [Google Scholar] [CrossRef]
- Toth, Z.; Maglinte, D.T.; Lee, S.H.; Lee, H.R.; Wong, L.Y.; Brulois, K.F.; Lee, S.; Buckley, J.D.; Laird, P.W.; Marquez, V.E.; et al. Epigenetic analysis of KSHV latent and lytic genomes. PLoS Pathog. 2010, 6, e1001013. [Google Scholar] [CrossRef]
- Darst, R.P.; Haecker, I.; Pardo, C.E.; Renne, R.; Kladde, M.P. Epigenetic diversity of Kaposi’s sarcoma-associated herpesvirus. Nucleic Acids Res. 2013, 41, 2993–3009. [Google Scholar] [CrossRef]
- Lu, F.; Day, L.; Gao, S.J.; Lieberman, P.M. Acetylation of the latency-associated nuclear antigen regulates repression of Kaposi’s sarcoma-associated herpesvirus lytic transcription. J. Virol. 2006, 80, 5273–5282. [Google Scholar] [CrossRef]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef]
- Weterings, E.; Chen, D.J. The endless tale of non-homologous end-joining. Cell Res. 2008, 18, 114–124. [Google Scholar] [CrossRef]
- Hollingworth, R.; Skalka, G.L.; Stewart, G.S.; Hislop, A.D.; Blackbourn, D.J.; Grand, R.J. Activation of DNA Damage Response Pathways during Lytic Replication of KSHV. Viruses 2015, 7, 2908–2927. [Google Scholar] [CrossRef]
- Xiao, Y.; Chen, J.; Liao, Q.; Wu, Y.; Peng, C.; Chen, X. Lytic infection of Kaposi’s sarcoma-associated herpesvirus induces DNA double-strand breaks and impairs non-homologous end joining. J. Gen. Virol. 2013, 94 Pt 8, 1870–1875. [Google Scholar] [CrossRef]
- Hau, P.M.; Deng, W.; Jia, L.; Yang, J.; Tsurumi, T.; Chiang, A.K.; Huen, M.S.; Tsao, S.W. Role of ATM in the formation of the replication compartment during lytic replication of Epstein-Barr virus in nasopharyngeal epithelial cells. J. Virol. 2015, 89, 652–668. [Google Scholar] [CrossRef]
- Shin, Y.C.; Nakamura, H.; Liang, X.; Feng, P.; Chang, H.; Kowalik, T.F.; Jung, J.U. Inhibition of the ATM/p53 signal transduction pathway by Kaposi’s sarcoma-associated herpesvirus interferon regulatory factor 1. J. Virol. 2006, 80, 2257–2266. [Google Scholar] [CrossRef]
- Hagemeier, S.R.; Barlow, E.A.; Meng, Q.; Kenney, S.C. The cellular ataxia telangiectasia-mutated kinase promotes epstein-barr virus lytic reactivation in response to multiple different types of lytic reactivation-inducing stimuli. J. Virol. 2012, 86, 13360–13370. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhu, J.; Xie, Z.; Liao, G.; Liu, J.; Chen, M.R.; Hu, S.; Woodard, C.; Lin, J.; Taverna, S.D.; et al. Conserved herpesvirus kinases target the DNA damage response pathway and TIP60 histone acetyltransferase to promote virus replication. Cell Host Microbe 2011, 10, 390–400. [Google Scholar] [CrossRef]
- Knott, G.J.; Bond, C.S.; Fox, A.H. The DBHS proteins SFPQ, NONO and PSPC1: A multipurpose molecular scaffold. Nucleic Acids Res. 2016, 44, 3989–4004. [Google Scholar] [CrossRef] [PubMed]
- Bladen, C.L.; Udayakumar, D.; Takeda, Y.; Dynan, W.S. Identification of the polypyrimidine tract binding protein-associated splicing factor.p54(nrb) complex as a candidate DNA double-strand break rejoining factor. J. Biol. Chem. 2005, 280, 5205–5210. [Google Scholar] [CrossRef]
- Udayakumar, D.; Dynan, W.S. Characterization of DNA binding and pairing activities associated with the native SFPQ·NONO DNA repair protein complex. Biochem. Biophys. Res. Commun. 2015, 463, 473–478. [Google Scholar] [CrossRef]
- Domínguez-Sánchez, M.S.; Barroso, S.; Gómez-González, B.; Luna, R.; Aguilera, A. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet. 2011, 7, e1002386. [Google Scholar] [CrossRef]
- Boyne, J.R.; Colgan, K.J.; Whitehouse, A. Recruitment of the complete hTREX complex is required for Kaposi’s sarcoma-associated herpesvirus intronless mRNA nuclear export and virus replication. PLoS Pathog. 2008, 4, e1000194. [Google Scholar] [CrossRef]
- Schulz, T.F.; Cesarman, E. Kaposi Sarcoma-associated Herpesvirus: Mechanisms of oncogenesis. Curr. Opin. Virol. 2015, 14, 116–128. [Google Scholar] [CrossRef]
- Mocroft, A.; Kirk, O.; Clumeck, N.; Gargalianos-Kakolyris, P.; Trocha, H.; Chentsova, N.; Antunes, F.; Stellbrink, H.J.; Phillips, A.N.; Lundgren, J.D. The changing pattern of Kaposi sarcoma in patients with HIV, 1994-2003: The EuroSIDA Study. Cancer 2004, 100, 2644–2654. [Google Scholar] [CrossRef]
- Glesby, M.J.; Hoover, D.R.; Weng, S.; Graham, N.M.; Phair, J.P.; Detels, R.; Ho, M.; Saah, A.J. Use of antiherpes drugs and the risk of Kaposi’s sarcoma: Data from the Multicenter AIDS Cohort Study. J. Infect. Dis. 1996, 173, 1477–1480. [Google Scholar] [CrossRef]
- Aneja, K.K.; Yuan, Y. Reactivation and Lytic Replication of Kaposi’s Sarcoma-Associated Herpesvirus: An Update. Front. Microbiol. 2017, 8, 613. [Google Scholar] [CrossRef]
- Grundhoff, A.; Ganem, D. Inefficient establishment of KSHV latency suggests an additional role for continued lytic replication in Kaposi sarcoma pathogenesis. J. Clin. Investig. 2004, 113, 124–136. [Google Scholar] [CrossRef]
- Cesarman, E.; Damania, B.; Krown, S.E.; Martin, J.; Bower, M.; Whitby, D. Kaposi sarcoma. Nat. Rev. Dis. Primers 2019, 5, 9. [Google Scholar] [CrossRef]
- Cavallin, L.E.; Goldschmidt-Clermont, P.; Mesri, E.A. Molecular and cellular mechanisms of KSHV oncogenesis of Kaposi’s sarcoma associated with HIV/AIDS. PLoS Pathog. 2014, 10, e1004154. [Google Scholar] [CrossRef] [PubMed]
- Medina, M.V.; Agostino, A.D.; Ma, Q.; Eroles, P.; Cavallin, L.; Chiozzini, C.; Sapochnik, D.; Cymeryng, C.; Hyjek, E.; Cesarman, E.; et al. KSHV G-protein coupled receptor vGPCR oncogenic signaling upregulation of Cyclooxygenase-2 expression mediates angiogenesis and tumorigenesis in Kaposi’s sarcoma. PLoS Pathog. 2020, 16, e1009006. [Google Scholar] [CrossRef] [PubMed]
- Bais, C.; Santomasso, B.; Coso, O.; Arvanitakis, L.; Raaka, E.G.; Gutkind, J.S.; Asch, A.S.; Cesarman, E.; Gershengorn, M.C.; Mesri, E.A. G-protein-coupled receptor of Kaposi’s sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature 1998, 391, 86–89. [Google Scholar] [CrossRef]
- Bais, C.; Van Geelen, A.; Eroles, P.; Mutlu, A.; Chiozzini, C.; Dias, S.; Silverstein, R.L.; Rafii, S.; Mesri, E.A. Kaposi’s sarcoma associated herpesvirus G protein-coupled receptor immortalizes human endothelial cells by activation of the VEGF receptor-2/KDR. Cancer Cell 2003, 3, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Chiou, C.J.; Poole, L.J.; Kim, P.S.; Ciufo, D.M.; Cannon, J.S.; ap Rhys, C.M.; Alcendor, D.J.; Zong, J.C.; Ambinder, R.F.; Hayward, G.S. Patterns of gene expression and a transactivation function exhibited by the vGCR (ORF74) chemokine receptor protein of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 2002, 76, 3421–3439. [Google Scholar] [CrossRef]
- Bottero, V.; Kerur, N.; Sadagopan, S.; Patel, K.; Sharma-Walia, N.; Chandran, B. Phosphorylation and polyubiquitination of transforming growth factor beta-activated kinase 1 are necessary for activation of NF-kappaB by the Kaposi’s sarcoma-associated herpesvirus G protein-coupled receptor. J. Virol. 2011, 85, 1980–1993. [Google Scholar] [CrossRef]
- Tamanaha-Nakasone, A.; Uehara, K.; Tanabe, Y.; Ishikawa, H.; Yamakawa, N.; Toyoda, Z.; Kurima, K.; Kina, S.; Tsuneki, M.; Okubo, Y.; et al. K1 gene transformation activities in AIDS-related and classic type Kaposi’s sarcoma: Correlation with clinical presentation. Sci. Rep. 2019, 9, 6416. [Google Scholar] [CrossRef]
- Tomlinson, C.C.; Damania, B. The K1 protein of Kaposi’s sarcoma-associated herpesvirus activates the Akt signaling pathway. J. Virol. 2004, 78, 1918–1927. [Google Scholar] [CrossRef]
- Komaki, S.; Inagaki, T.; Kumar, A.; Izumiya, Y. The Role of vIL-6 in KSHV-Mediated Immune Evasion and Tumorigenesis. Viruses 2024, 16, 1900. [Google Scholar] [CrossRef]
- Lin, R.; Genin, P.; Mamane, Y.; Sgarbanti, M.; Battistini, A.; Harrington, W.J., Jr.; Barber, G.N.; Hiscott, J. HHV-8 encoded vIRF-1 represses the interferon antiviral response by blocking IRF-3 recruitment of the CBP/p300 coactivators. Oncogene 2001, 20, 800–811. [Google Scholar] [CrossRef] [PubMed]
- Burysek, L.; Yeow, W.S.; Lubyova, B.; Kellum, M.; Schafer, S.L.; Huang, Y.Q.; Pitha, P.M. Functional analysis of human herpesvirus 8-encoded viral interferon regulatory factor 1 and its association with cellular interferon regulatory factors and p300. J. Virol. 1999, 73, 7334–7342. [Google Scholar] [CrossRef] [PubMed]
- Mutlu, A.D.; Cavallin, L.E.; Vincent, L.; Chiozzini, C.; Eroles, P.; Duran, E.M.; Asgari, Z.; Hooper, A.T.; La Perle, K.M.; Hilsher, C.; et al. In vivo-restricted and reversible malignancy induced by human herpesvirus-8 KSHV: A cell and animal model of virally induced Kaposi’s sarcoma. Cancer Cell 2007, 11, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Naipauer, J.; Rosario, S.; Gupta, S.; Premer, C.; Mendez-Solis, O.; Schlesinger, M.; Ponzinibbio, V.; Jain, V.; Gay, L.; Renne, R.; et al. PDGFRA defines the mesenchymal stem cell Kaposi’s sarcoma progenitors by enabling KSHV oncogenesis in an angiogenic environment. PLoS Pathog. 2019, 15, e1008221. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hayward, C.; Harper, K.L.; Harrington, E.M.; Mottram, T.J.; Whitehouse, A. Manipulation of Nuclear-Related Pathways During Kaposi’s Sarcoma-Associated Herpesvirus Lytic Replication. Viruses 2025, 17, 1427. https://doi.org/10.3390/v17111427
Hayward C, Harper KL, Harrington EM, Mottram TJ, Whitehouse A. Manipulation of Nuclear-Related Pathways During Kaposi’s Sarcoma-Associated Herpesvirus Lytic Replication. Viruses. 2025; 17(11):1427. https://doi.org/10.3390/v17111427
Chicago/Turabian StyleHayward, Connor, Katherine L. Harper, Elena M. Harrington, Timothy J. Mottram, and Adrian Whitehouse. 2025. "Manipulation of Nuclear-Related Pathways During Kaposi’s Sarcoma-Associated Herpesvirus Lytic Replication" Viruses 17, no. 11: 1427. https://doi.org/10.3390/v17111427
APA StyleHayward, C., Harper, K. L., Harrington, E. M., Mottram, T. J., & Whitehouse, A. (2025). Manipulation of Nuclear-Related Pathways During Kaposi’s Sarcoma-Associated Herpesvirus Lytic Replication. Viruses, 17(11), 1427. https://doi.org/10.3390/v17111427





