Roles of RNA Structures in the Genome Translation of (+) Sense RNA Viruses
Abstract
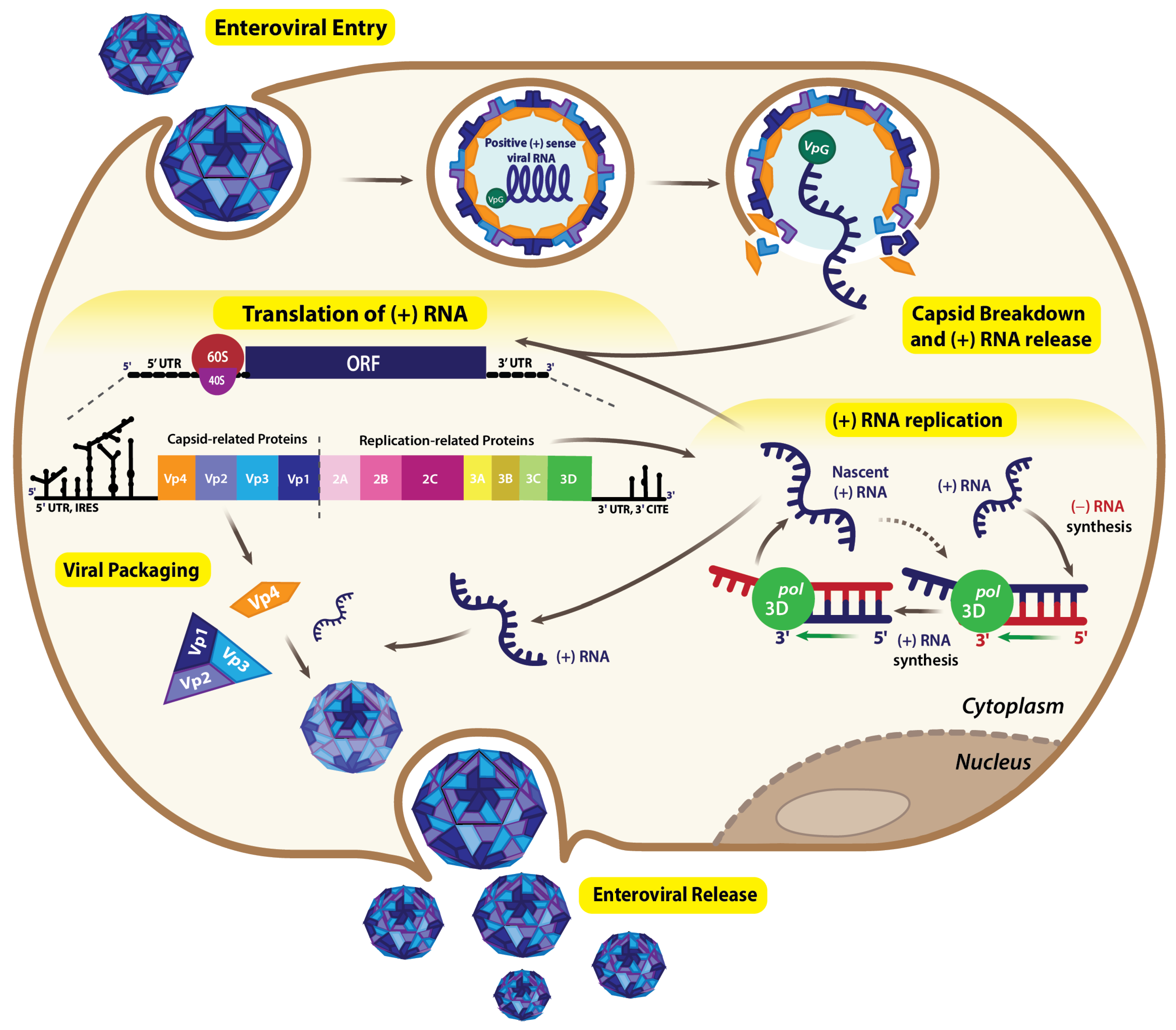
1. Introduction
2. RNA Structures Associated with Viral Genome Translation
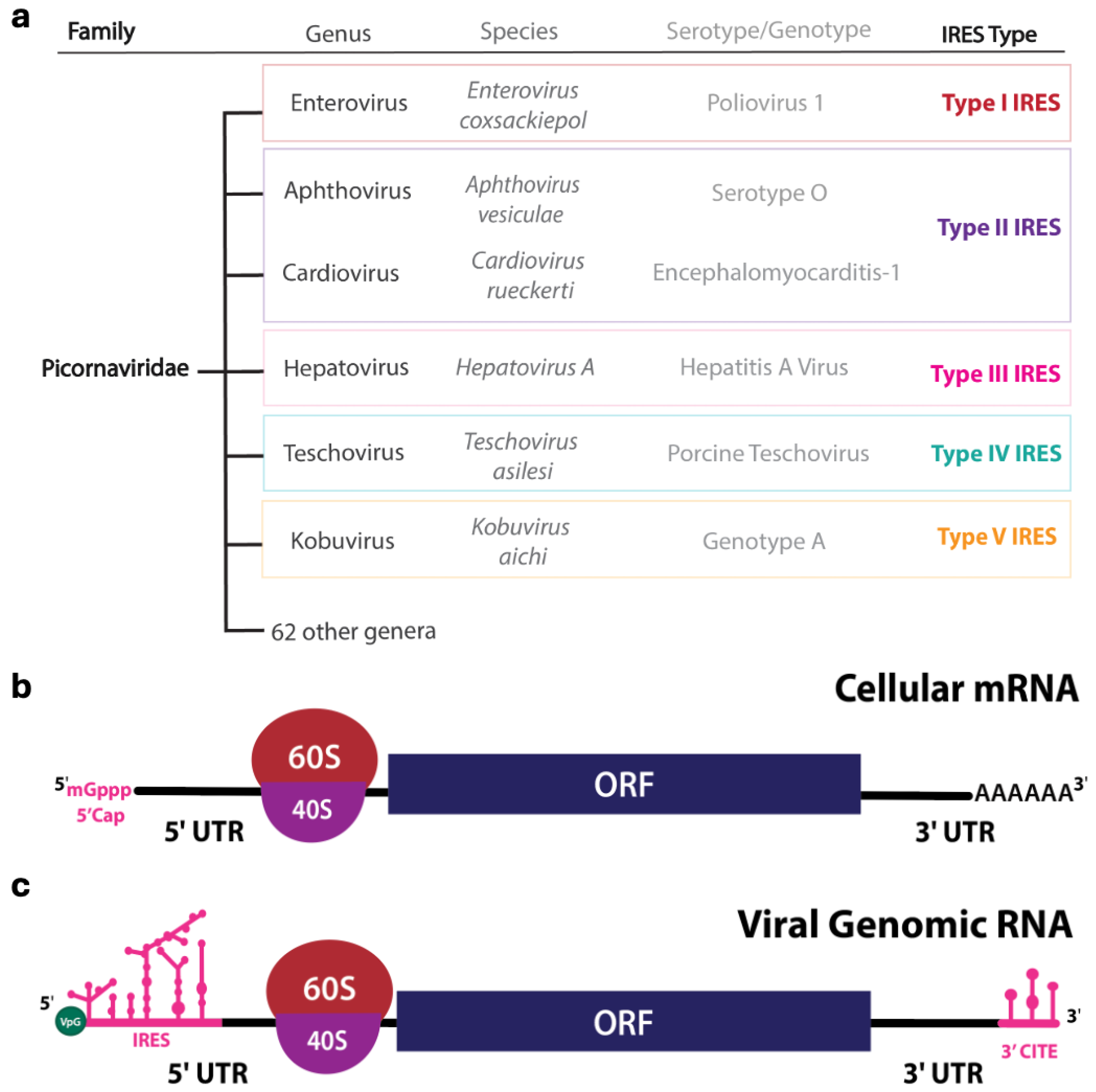
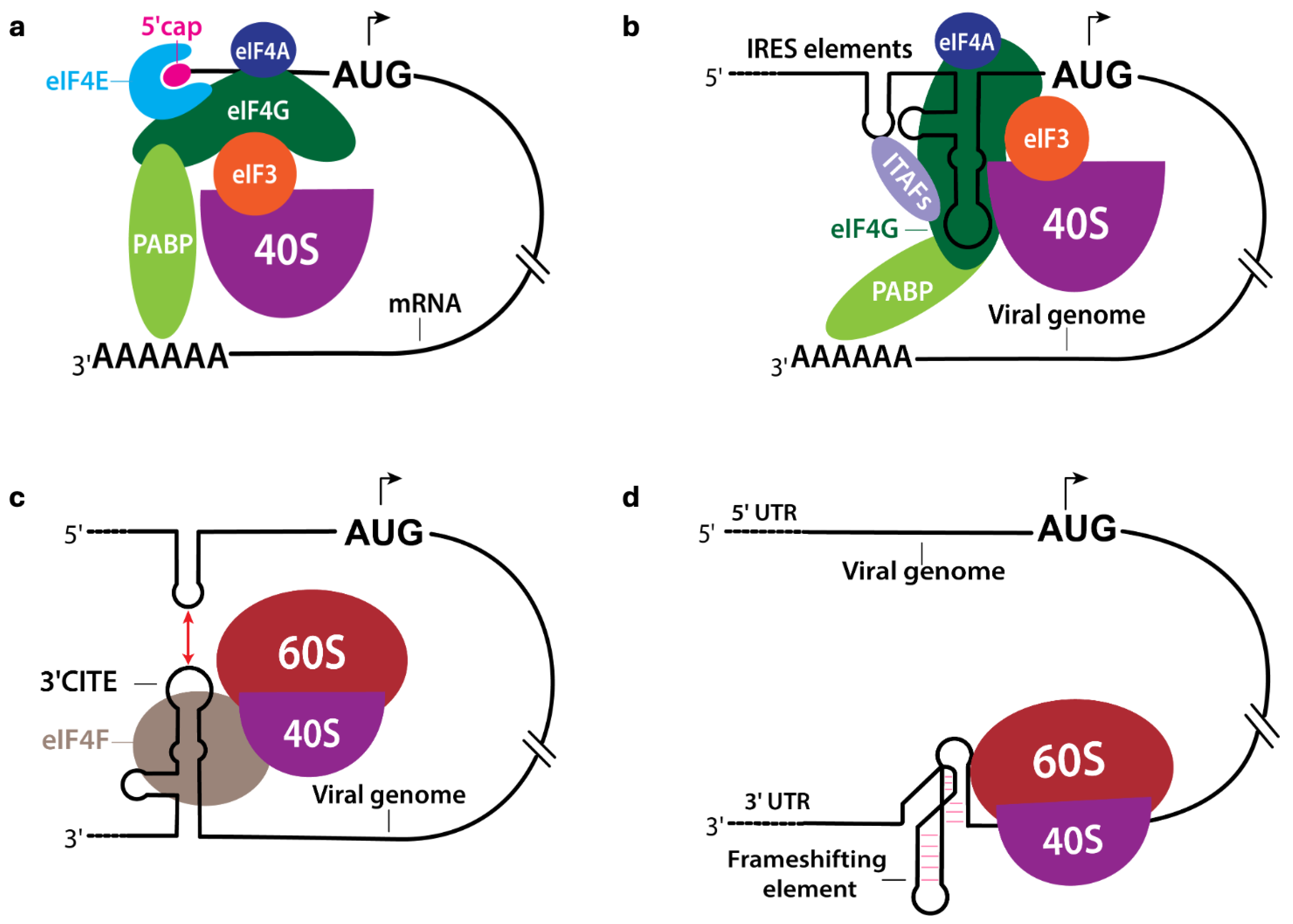
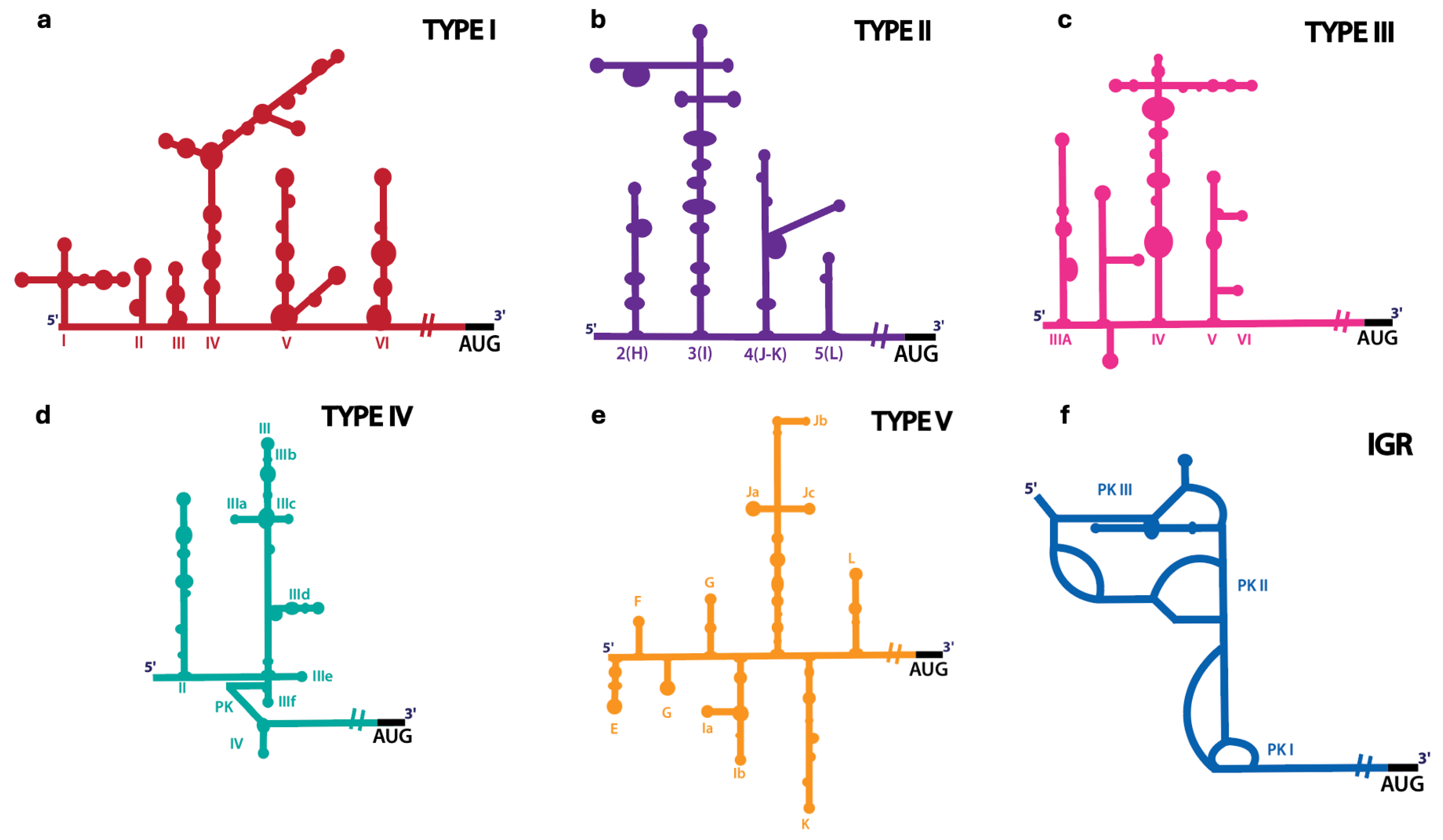
2.1. Internal Ribosome Entry Site (IRES)
| IRES Types | Length | Virus | Secondary Structure | eIFs | ITAFs |
|---|---|---|---|---|---|
| Type I | ~450 nts | Enterovirus coxsackiepol, Enterovirus alpharhino, Enterovirus betarhino, Enterovirus cerhino, Caliciviruses | Five modular domains (II to VI) [81] | eIF1A, eIF2, eIF3, eIF4A, eIF4G, eIF4B, eIF5B [52,53,54] | PCBP2 [53,54,55], PTB [54,56], La [57], Unr [58], Sam68 [82] |
| Type II | ~450 nts | Cardiovirus rueckerti, Aphthovirus vesiculae, Caliciviridae | Five modular domains (H to L) [64,83,84] | eIF1, eIF1A, eIF2, eIF3, eIF4G, eIF4A [67,68,85,86] | PTB [70], PTB + ITAF45/EBP1 [68], Gemin5 [87], Sam68 [88] |
| Type III | ~700 nts | Hepatovirus A | Five modular domains (II to VI) [76,77] | eIF2, eIF3, eIF4A, eIF4G, eIF4B, eIF4E [73,78,79] | PDAP1 [80], PTB [89], PCBP2 [90,91], La [92] |
| Type IV | ~300 nts | Hepatitis C virus, Classical swine fever virus, Penguin megirivirus, Ruddy turnstone calicivirus | Compact structure with pseudoknots (II to III) [93,94] | eIF1A, eIF2, eIF3, eIF5B [95,96,97,98,99,100,101] | La [57,102], hnRNP L [103], hnRNP D [104] |
| Type V | ~450 nts | Kobuvirus aichi, Calicivirus | Eight modular domains (E to L) [105] | eIF2, eIF3, eIF4A, eIF4G [105] | DHX29 [105], PTB [105] |
| IGR | ~200 nts | Cricket paralysis Virus | Three nested pseudoknots (PKI to PKIII) [106] | NA | NA |
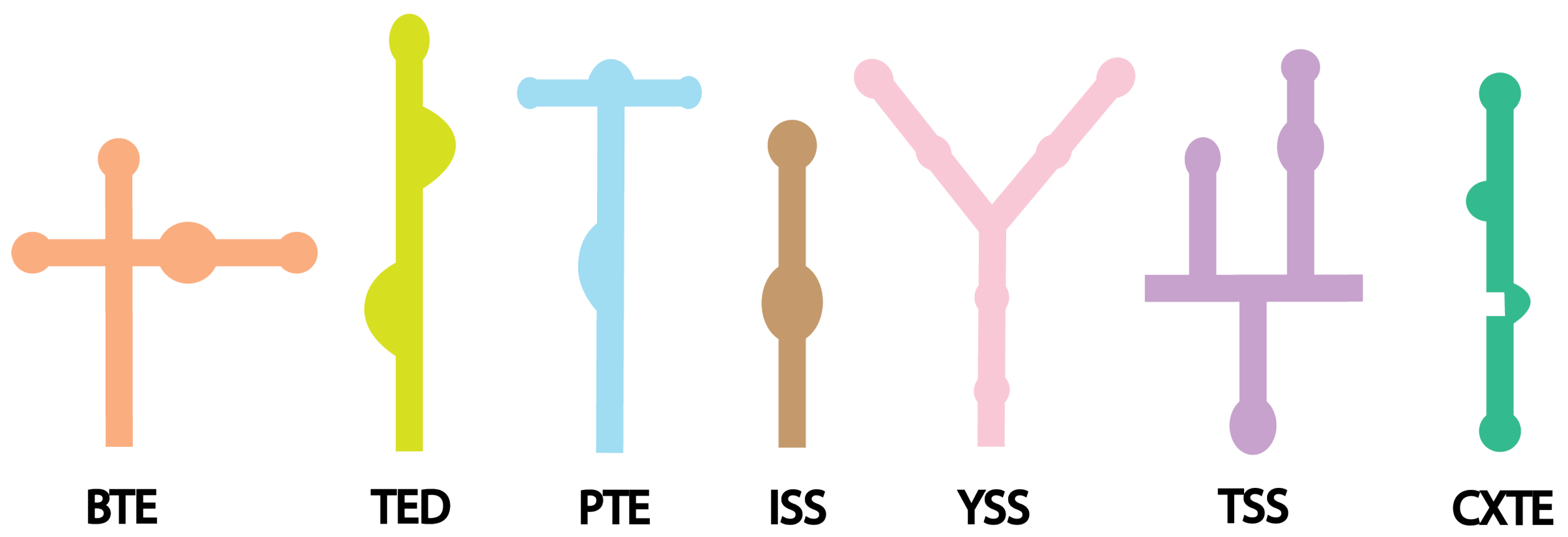
2.2. 3′ Cap-Independent Translation Enhancer (3′ CITE)
| 3′ CITEs | Length | Virus | Secondary Structure | Roles |
|---|---|---|---|---|
| BTE | ~100 nts | Barley Yellow Dwarf Virus [135] | Multiple stem loops [142] | Interacts with eIF4G [144,152] |
| TED | ~120 to150 nts | Satellite Tobacco Necrosis Virus [128] | Complex structure, multiple stem loops [128] | Interacts with eIF4E [38] |
| PTE | ~100 nts | Panicum Mosaic Virus [136], Pea enation mosaic virus [153] | Pseudoknot containing structure [153,154] | Interacts with eIF4E, promotes RNA circularization [153] |
| ISS | ~60 nts | Tombusviruses [134], Maize necrotic streak virus [137] | Complex stem loop structure with multiple bulges [134,137] | Interacts with eIF4E [134,155] |
| YSS | ~130 to 150 nts | Tomato bushy stunt virus [131], Tombusvirus [138] | Y-shaped three-way junction architecture [131] | Interacts with eIF4E [153,156] |
| TSS | ~100 nts | Turnip crinkle Virus [139] | T-shaped structure, tRNA-like [139] | Interacts directly to ribosomal subunits, promotes RNA circularization [140] |
| CXTE | ~55 nts | Cucurbit aphid-borne yellows virus [141], Melon Necrotic Spot Virus [150] | Complex structure with multiple stem loops [150] | Interacts with eIF4G [141] |
2.3. Other Regulatory RNA Elements in Viral Translation
3. Protein-RNA Interactions in Translation-Associated Complexes
4. Structural Studies of Translation-Related RNAs
4.1. Biochemical Probing Methods
4.2. X-Ray Crystallography
4.3. NMR Spectroscopy
4.4. Cryo-Electron Microscopy (Cryo-EM)
5. Regulation of the Replication-Translation Switching
6. Perspectives and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Baltimore, D. Expression of animal virus genomes. Bacteriol. Rev. 1971, 35, 235–241. [Google Scholar] [CrossRef]
- Knipe, D.M.; Howley, P. Fields Virology; Wolters Kluwer Health: Philadelphia, PA, USA, 2013. [Google Scholar]
- Fehr, A.R.; Perlman, S. Coronaviruses: An Overview of Their Replication and Pathogenesis. In Coronaviruses: Methods and Protocols; Maier, H.J., Bickerton, E., Britton, P., Eds.; Springer: New York, NY, USA, 2015; pp. 1–23. [Google Scholar]
- Flather, D.; Semler, B.L. Picornaviruses and nuclear functions: Targeting a cellular compartment distinct from the replication site of a positive-strand RNA virus. Front. Microbiol. 2015, 6, 594. [Google Scholar] [CrossRef]
- Pyle, J.D.; Mandadi, K.K.; Scholthof, K.B.G. Panicum Mosaic Virus and Its Satellites Acquire RNA Modifications Associated with Host-Mediated Antiviral Degradation. mBio 2019, 10, e01900-19. [Google Scholar] [CrossRef]
- Qin, L.; Shen, W.; Tang, Z.; Hu, W.; Shangguan, L.; Wang, Y.; Tuo, D.; Li, Z.; Miao, W.; Valli, A.A.; et al. A Newly Identified Virus in the Family Potyviridae Encodes Two Leader Cysteine Proteases in Tandem That Evolved Contrasting RNA Silencing Suppression Functions. J. Virol. 2020, 95, e01414-20. [Google Scholar] [CrossRef] [PubMed]
- Abram, Q.H.; Landry, B.N.; Wang, A.B.; Kothe, R.F.; Hauch, H.C.H.; Sagan, S.M. The myriad roles of RNA structure in the flavivirus life cycle. RNA Biol. 2024, 21, 14–30. [Google Scholar] [CrossRef]
- Ahlquist, P.; Noueiry, A.O.; Lee, W.M.; Kushner, D.B.; Dye, B.T. Host factors in positive-strand RNA virus genome replication. J. Virol. 2003, 77, 8181–8186. [Google Scholar] [CrossRef]
- Nagy, P.D.; Pogany, J. The dependence of viral RNA replication on co-opted host factors. Nat. Rev. Microbiol. 2011, 10, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Modrow, S.; Falke, D.; Truyen, U.; Schätzl, H. Viruses with Single-Stranded, Positive-Sense RNA Genomes. In Molecular Virology; Springer: Berlin/Heidelberg, Germany, 2013; pp. 185–349. [Google Scholar]
- Cifuente, J.O.; Moratorio, G. Evolutionary and Structural Overview of Human Picornavirus Capsid Antibody Evasion. Front. Cell. Infect. Microbiol. 2019, 9, 283. [Google Scholar] [CrossRef]
- Andino, R.; Kirkegaard, K.; Macadam, A.; Racaniello, V.R.; Rosenfeld, A.B. The Picornaviridae Family: Knowledge Gaps, Animal Models, Countermeasures, and Prototype Pathogens. J. Infect. Dis. 2023, 228 (Suppl. 6), S427–S445. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.G.; Kerrigan, D.; Evans, D.J. Structure and function analysis of the poliovirus cis-acting replication element (CRE). RNA 2003, 9, 124–137. [Google Scholar] [CrossRef]
- Goodfellow, I.; Chaudhry, Y.; Gioldasi, I.; Gerondopoulos, A.; Natoni, A.; Labrie, L.; Laliberte, J.F.; Roberts, L. Calicivirus translation initiation requires an interaction between VPg and eIF 4 E. EMBO Rep. 2005, 6, 968–972. [Google Scholar] [CrossRef]
- Warsaba, R.; Stoynov, N.; Moon, K.M.; Flibotte, S.; Foster, L.; Jan, E. Multiple Viral Protein Genome-Linked Proteins Compensate for Viral Translation in a Positive-Sense Single-Stranded RNA Virus Infection. J. Virol. 2022, 96, e0069922. [Google Scholar] [CrossRef]
- Pelletier, J.; Sonenberg, N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 1988, 334, 320–325. [Google Scholar] [CrossRef]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef]
- Witwer, C.; Rauscher, S.; Hofacker, I.L.; Stadler, P.F. Conserved RNA secondary structures in Picornaviridae genomes. Nucleic Acids Res. 2001, 29, 5079–5089. [Google Scholar] [CrossRef]
- Herold, J.; Andino, R. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 2001, 7, 581–591. [Google Scholar] [CrossRef]
- Goodfellow, I.; Chaudhry, Y.; Richardson, A.; Meredith, J.; Almond, J.W.; Barclay, W.; Evans, D.J. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 2000, 74, 4590–4600. [Google Scholar] [CrossRef]
- Cordey, S.; Gerlach, D.; Junier, T.; Zdobnov, E.M.; Kaiser, L.; Tapparel, C. The cis-acting replication elements define human enterovirus and rhinovirus species. RNA 2008, 14, 1568–1578. [Google Scholar] [CrossRef]
- den Boon, J.A.; Nishikiori, M.; Zhan, H.; Ahlquist, P. Positive-strand RNA virus genome replication organelles: Structure, assembly, control. Trends Genet. 2024, 40, 681–693. [Google Scholar] [CrossRef]
- Bosch, B.J.; van der Zee, R.; de Haan, C.A.; Rottier, P.J. The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. J. Virol. 2003, 77, 8801–8811. [Google Scholar] [CrossRef]
- Machitani, M.; Yasukawa, M.; Nakashima, J.; Furuichi, Y.; Masutomi, K. RNA-dependent RNA polymerase, RdRP, a promising therapeutic target for cancer and potentially COVID-19. Cancer Sci. 2020, 111, 3976–3984. [Google Scholar] [CrossRef]
- Lindenbach, B.D.; Rice, C.M. Unravelling hepatitis C virus replication from genome to function. Nature 2005, 436, 933–938. [Google Scholar] [CrossRef]
- Boersma, S.; Rabouw, H.H.; Bruurs, L.J.M.; Pavlovic, T.; van Vliet, A.L.W.; Beumer, J.; Clevers, H.; van Kuppeveld, F.J.M.; Tanenbaum, M.E. Translation and Replication Dynamics of Single RNA Viruses. Cell 2020, 183, 1930–1945.e23. [Google Scholar] [CrossRef]
- Jackson, R.J.; Hellen, C.U.; Pestova, T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010, 11, 113–127. [Google Scholar] [CrossRef]
- Pestova, T.V.; Kolupaeva, V.G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002, 16, 2906–2922. [Google Scholar] [CrossRef]
- Jaafar, Z.A.; Kieft, J.S. Viral RNA structure-based strategies to manipulate translation. Nat. Rev. Microbiol. 2019, 17, 110–123. [Google Scholar] [CrossRef]
- Pestova, T.; Lorsch, J.; Hellen, C. The mechanism of translation initiation in eukaryotes. In Translational Control in Biology and Medicine; Mathews, M.B., Sonenberg, N., Hershey, J.W.B., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2007; pp. 87–128. [Google Scholar]
- Walsh, D.; Mohr, I. Viral subversion of the host protein synthesis machinery. Nat. Rev. Microbiol. 2011, 9, 860–875. [Google Scholar] [CrossRef]
- Tsukiyama-Kohara, K.; Iizuka, N.; Kohara, M.; Nomoto, A. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 1992, 66, 1476–1483. [Google Scholar] [CrossRef]
- Jang, S.K.; Krausslich, H.G.; Nicklin, M.J.; Duke, G.M.; Palmenberg, A.C.; Wimmer, E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988, 62, 2636–2643. [Google Scholar] [CrossRef]
- Jackson, R.J. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem. Soc. Trans. 2005, 33 Pt 6, 1231–1241. [Google Scholar] [CrossRef]
- Martinez-Salas, E.; Pacheco, A.; Serrano, P.; Fernandez, N. New insights into internal ribosome entry site elements relevant for viral gene expression. J. Gen. Virol. 2008, 89 Pt 3, 611–626. [Google Scholar] [CrossRef]
- Niepmann, M. Internal translation initiation of picornaviruses and hepatitis C virus. Biochim. Biophys. Acta 2009, 1789, 529–541. [Google Scholar]
- Sorokin, I.I.; Vassilenko, K.S.; Terenin, I.M.; Kalinina, N.O.; Agol, V.I.; Dmitriev, S.E. Non-Canonical Translation Initiation Mechanisms Employed by Eukaryotic Viral mRNAs. Biochemistry 2021, 86, 1060–1094. [Google Scholar] [CrossRef]
- Gazo, B.M.; Murphy, P.; Gatchel, J.R.; Browning, K.S. A novel interaction of Cap-binding protein complexes eukaryotic initiation factor (eIF) 4F and eIF(iso)4F with a region in the 3′-untranslated region of satellite tobacco necrosis virus. J. Biol. Chem. 2004, 279, 13584–13592. [Google Scholar] [CrossRef]
- Gao, F.; Kasprzak, W.; Stupina, V.A.; Shapiro, B.A.; Simon, A.E. A ribosome-binding, 3′ translational enhancer has a T-shaped structure and engages in a long-distance RNA-RNA interaction. J. Virol. 2012, 86, 9828–9842. [Google Scholar] [CrossRef]
- Simon, A.E.; Miller, W.A. 3′ cap-independent translation enhancers of plant viruses. Annu. Rev. Microbiol. 2013, 67, 21–42. [Google Scholar] [CrossRef]
- Zhang, K.; Zheludev, I.N.; Hagey, R.J.; Haslecker, R.; Hou, Y.J.; Kretsch, R.; Pintilie, G.D.; Rangan, R.; Kladwang, W.; Li, S.; et al. Cryo-EM and antisense targeting of the 28-kDa frameshift stimulation element from the SARS-CoV-2 RNA genome. Nat. Struct. Mol. Biol. 2021, 28, 747–754. [Google Scholar]
- Roman, C.; Lewicka, A.; Koirala, D.; Li, N.S.; Piccirilli, J.A. The SARS-CoV-2 Programmed −1 Ribosomal Frameshifting Element Crystal Structure Solved to 2.09 Å Using Chaperone-Assisted RNA Crystallography. ACS Chem. Biol. 2021, 16, 1469–1481. [Google Scholar] [CrossRef]
- Bhatt, P.R.; Scaiola, A.; Loughran, G.; Leibundgut, M.; Kratzel, A.; Meurs, R.; Dreos, R.; O’Connor, K.M.; McMillan, A.; Bode, J.W.; et al. Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome. Science 2021, 372, 1306–1313. [Google Scholar] [CrossRef]
- Kneller, E.L.; Rakotondrafara, A.M.; Miller, W.A. Cap-independent translation of plant viral RNAs. Virus Res. 2006, 119, 63–75. [Google Scholar] [CrossRef]
- Martinez-Salas, E.; Francisco-Velilla, R.; Fernandez-Chamorro, J.; Embarek, A.M. Insights into Structural and Mechanistic Features of Viral IRES Elements. Front. Microbiol. 2017, 8, 2629. [Google Scholar]
- Hanson, P.J.; Zhang, H.M.; Hemida, M.G.; Ye, X.; Qiu, Y.; Yang, D. IRES-Dependent Translational Control during Virus-Induced Endoplasmic Reticulum Stress and Apoptosis. Front. Microbiol. 2012, 3, 92. [Google Scholar]
- Godet, A.C.; David, F.; Hantelys, F.; Tatin, F.; Lacazette, E.; Garmy-Susini, B.; Prats, A.C. IRES Trans-Acting Factors, Key Actors of the Stress Response. Int. J. Mol. Sci. 2019, 20, 924. [Google Scholar] [CrossRef]
- Marques, R.; Lacerda, R.; Romao, L. Internal Ribosome Entry Site (IRES)-Mediated Translation and Its Potential for Novel mRNA-Based Therapy Development. Biomedicines 2022, 10, 1865. [Google Scholar] [CrossRef]
- Chen, L.L.; Kung, Y.A.; Weng, K.F.; Lin, J.Y.; Horng, J.T.; Shih, S.R. Enterovirus 71 infection cleaves a negative regulator for viral internal ribosomal entry site-driven translation. J. Virol. 2013, 87, 3828–3838. [Google Scholar] [CrossRef]
- Dave, P.; George, B.; Sharma, D.K.; Das, S. Polypyrimidine tract-binding protein (PTB) and PTB-associated splicing factor in CVB3 infection: An ITAF for an ITAF. Nucleic Acids Res. 2017, 45, 9068–9084. [Google Scholar]
- Thompson, S.R.; Sarnow, P. Enterovirus 71 contains a type I IRES element that functions when eukaryotic initiation factor eIF4G is cleaved. Virology 2003, 315, 259–266. [Google Scholar] [CrossRef]
- White, J.P.; Reineke, L.C.; Lloyd, R.E. Poliovirus switches to an eIF2-independent mode of translation during infection. J. Virol. 2011, 85, 8884–8893. [Google Scholar] [CrossRef]
- Sweeney, T.R.; Abaeva, I.S.; Pestova, T.V.; Hellen, C.U. The mechanism of translation initiation on Type 1 picornavirus IRESs. EMBO J. 2014, 33, 76–92. [Google Scholar] [CrossRef]
- Arhab, Y.; Pestova, T.V.; Hellen, C.U.T. Translation of Overlapping Open Reading Frames Promoted by Type 2 IRESs in Avian Calicivirus Genomes. Viruses 2024, 16, 1413. [Google Scholar] [CrossRef]
- Blyn, L.B.; Towner, J.S.; Semler, B.L.; Ehrenfeld, E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 1997, 71, 6243–6246. [Google Scholar] [CrossRef]
- Kafasla, P.; Morgner, N.; Robinson, C.V.; Jackson, R.J. Polypyrimidine tract-binding protein stimulates the poliovirus IRES by modulating eIF4G binding. EMBO J. 2010, 29, 3710–3722. [Google Scholar] [CrossRef]
- Costa-Mattioli, M.; Svitkin, Y.; Sonenberg, N. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol. Cell. Biol. 2004, 24, 6861–6870. [Google Scholar] [CrossRef]
- Hunt, S.L.; Hsuan, J.J.; Totty, N.; Jackson, R.J. Unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 1999, 13, 437–448. [Google Scholar] [CrossRef]
- de Breyne, S.; Yu, Y.; Unbehaun, A.; Pestova, T.V.; Hellen, C.U. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc. Natl. Acad. Sci. USA 2009, 106, 9197–9202. [Google Scholar] [CrossRef] [PubMed]
- Luz, N.; Beck, E. Interaction of a cellular 57-kilodalton protein with the internal translation initiation site of foot-and-mouth disease virus. J. Virol. 1991, 65, 6486–6494. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Sun, S.Q.; Guo, H.C. Biological function of Foot-and-mouth disease virus non-structural proteins and non-coding elements. Virol. J. 2016, 13, 107. [Google Scholar] [CrossRef] [PubMed]
- Arhab, Y.; Miścicka, A.; Pestova, T.V.; Hellen, C.U.T. Horizontal gene transfer as a mechanism for the promiscuous acquisition of distinct classes of IRES by avian caliciviruses. Nucleic Acids Res. 2022, 50, 1052–1068. [Google Scholar] [CrossRef]
- Asnani, M.; Pestova, T.V.; Hellen, C.U. Initiation on the divergent Type I cadicivirus IRES: Factor requirements and interactions with the translation apparatus. Nucleic Acids Res. 2016, 44, 3390–3407. [Google Scholar] [CrossRef]
- Maloney, A.; Joseph, S. Validating the EMCV IRES Secondary Structure with Structure-Function Analysis. Biochemistry 2024, 63, 107–115. [Google Scholar] [CrossRef]
- Imai, S.; Kumar, P.; Hellen, C.U.; D’Souza, V.M.; Wagner, G. An accurately preorganized IRES RNA structure enables eIF4G capture for initiation of viral translation. Nat. Struct. Mol. Biol. 2016, 23, 859–864. [Google Scholar] [CrossRef] [PubMed]
- Chamond, N.; Deforges, J.; Ulryck, N.; Sargueil, B. 40S recruitment in the absence of eIF4G/4A by EMCV IRES refines the model for translation initiation on the archetype of Type II IRESs. Nucleic Acids Res. 2014, 42, 10373–10384. [Google Scholar] [CrossRef]
- Pestova, T.V.; Hellen, C.U.; Shatsky, I.N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 1996, 16, 6859–6869. [Google Scholar] [CrossRef] [PubMed]
- Pilipenko, E.V.; Pestova, T.V.; Kolupaeva, V.G.; Khitrina, E.V.; Poperechnaya, A.N.; Agol, V.I.; Hellen, C.U. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000, 14, 2028–2045. [Google Scholar] [CrossRef] [PubMed]
- Andreev, D.E.; Fernandez-Miragall, O.; Ramajo, J.; Dmitriev, S.E.; Terenin, I.M.; Martinez-Salas, E.; Shatsky, I.N. Differential factor requirement to assemble translation initiation complexes at the alternative start codons of foot-and-mouth disease virus RNA. RNA 2007, 13, 1366–1374. [Google Scholar] [CrossRef]
- Kaminski, A.; Jackson, R.J. The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA 1998, 4, 626–638. [Google Scholar] [CrossRef]
- Yu, Y.; Abaeva, I.S.; Marintchev, A.; Pestova, T.V.; Hellen, C.U. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Res. 2011, 39, 4851–4865. [Google Scholar] [CrossRef]
- López-Ulloa, B.; Fuentes, Y.; Pizarro-Ortega, M.S.; López-Lastra, M. RNA-Binding Proteins as Regulators of Internal Initiation of Viral mRNA Translation. Viruses 2022, 14, 188. [Google Scholar] [CrossRef]
- Borman, A.M.; Kean, K.M. Intact eukaryotic initiation factor 4G is required for hepatitis A virus internal initiation of translation. Virology 1997, 237, 129–136. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, Z. IRES-mediated cap-independent translation, a path leading to hidden proteome. J. Mol. Cell Biol. 2019, 11, 911–919. [Google Scholar] [CrossRef]
- Cohen, J.I.; Ticehurst, J.R.; Purcell, R.H.; Buckler-White, A.; Baroudy, B.M. Complete nucleotide sequence of wild-type hepatitis A virus: Comparison with different strains of hepatitis A virus and other picornaviruses. J. Virol. 1987, 61, 50–59. [Google Scholar] [CrossRef]
- Brown, E.A.; Day, S.P.; Jansen, R.W.; Lemon, S.M. The 5′ nontranslated region of hepatitis A virus RNA: Secondary structure and elements required for translation in vitro. J. Virol. 1991, 65, 5828–5838. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.A.; Zajac, A.J.; Lemon, S.M. In vitro characterization of an internal ribosomal entry site (IRES) present within the 5′ nontranslated region of hepatitis A virus RNA: Comparison with the IRES of encephalomyocarditis virus. J. Virol. 1994, 68, 1066–1074. [Google Scholar] [CrossRef]
- Ali, I.K.; McKendrick, L.; Morley, S.J.; Jackson, R.J. Activity of the hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J. Virol. 2001, 75, 7854–7863. [Google Scholar] [PubMed]
- Borman, A.M.; Michel, Y.M.; Kean, K.M. Detailed analysis of the requirements of hepatitis A virus internal ribosome entry segment for the eukaryotic initiation factor complex eIF4F. J. Virol. 2001, 75, 7864–7871. [Google Scholar] [CrossRef]
- Shirasaki, T.; Lenarcic, E.; Misumi, I.; Xie, L.; Fusco, W.G.; Yonish, B.; Das, A.; Kim, H.; Cameron, C.E.; Leger-Abraham, M.; et al. Hepatovirus translation requires PDGFA-associated protein 1, an eIF4E-binding protein regulating endoplasmic reticulum stress responses. Sci. Adv. 2024, 10, eadq6342. [Google Scholar] [CrossRef]
- Burrill, C.P.; Westesson, O.; Schulte, M.B.; Strings, V.R.; Segal, M.; Andino, R. Global RNA structure analysis of poliovirus identifies a conserved RNA structure involved in viral replication and infectivity. J. Virol. 2013, 87, 11670–11683. [Google Scholar] [CrossRef]
- Zhang, H.; Song, L.; Cong, H.; Tien, P. Nuclear Protein Sam68 Interacts with the Enterovirus 71 Internal Ribosome Entry Site and Positively Regulates Viral Protein Translation. J. Virol. 2015, 89, 10031–10043. [Google Scholar] [CrossRef]
- Duke, G.M.; Hoffman, M.A.; Palmenberg, A.C. Sequence and structural elements that contribute to efficient encephalomyocarditis virus RNA translation. J. Virol. 1992, 66, 1602–1609. [Google Scholar] [CrossRef]
- Bochkov, Y.A.; Palmenberg, A.C. Translational efficiency of EMCV IRES in bicistronic vectors is dependent upon IRES sequence and gene location. Biotechniques 2006, 41, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Kolupaeva, V.G.; Pestova, T.V.; Hellen, C.U.; Shatsky, I.N. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J. Biol. Chem. 1998, 273, 18599–18604. [Google Scholar] [CrossRef]
- Pestova, T.V.; Borukhov, S.I.; Hellen, C.U. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 1998, 394, 854–859. [Google Scholar] [CrossRef]
- Pacheco, A.; Lopez de Quinto, S.; Ramajo, J.; Fernandez, N.; Martinez-Salas, E. A novel role for Gemin5 in mRNA translation. Nucleic Acids Res. 2009, 37, 582–590. [Google Scholar] [CrossRef]
- Rai, D.K.; Lawrence, P.; Kloc, A.; Schafer, E.; Rieder, E. Analysis of the interaction between host factor Sam68 and viral elements during foot-and-mouth disease virus infections. Virol. J. 2015, 12, 224. [Google Scholar] [CrossRef]
- Venkatramana, M.; Ray, P.S.; Chadda, A.; Das, S. A 25 kDa cleavage product of polypyrimidine tract binding protein (PTB) present in mouse tissues prevents PTB binding to the 5′ untranslated region and inhibits translation of hepatitis A virus RNA. Virus Res. 2003, 98, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Graff, J.; Cha, J.; Blyn, L.B.; Ehrenfeld, E. Interaction of poly(rC) binding protein 2 with the 5′ noncoding region of hepatitis A virus RNA and its effects on translation. J. Virol. 1998, 72, 9668–9675. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Seitz, S.; Kusov, Y.; Zell, R.; Gauss-Muller, V. RNA interaction and cleavage of poly(C)-binding protein 2 by hepatitis A virus protease. Biochem. Biophys. Res. Commun. 2007, 364, 725–730. [Google Scholar] [CrossRef]
- Jiang, X.; Kanda, T.; Wu, S.; Nakamoto, S.; Saito, K.; Shirasawa, H.; Kiyohara, T.; Ishii, K.; Wakita, T.; Okamoto, H.; et al. Suppression of La antigen exerts potential antiviral effects against hepatitis A virus. PLoS ONE 2014, 9, e101993. [Google Scholar] [CrossRef]
- Rijnbrand, R.; van der Straaten, T.; van Rijn, P.A.; Spaan, W.J.; Bredenbeek, P.J. Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initiation codon. J. Virol. 1997, 71, 451–457. [Google Scholar] [CrossRef]
- Lukavsky, P.J.; Otto, G.A.; Lancaster, A.M.; Sarnow, P.; Puglisi, J.D. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat. Struct. Biol. 2000, 7, 1105–1110. [Google Scholar] [PubMed]
- Pestova, T.V.; Shatsky, I.N.; Fletcher, S.P.; Jackson, R.J.; Hellen, C.U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998, 12, 67–83. [Google Scholar]
- Locker, N.; Easton, L.E.; Lukavsky, P.J. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 2007, 26, 795–805. [Google Scholar] [CrossRef] [PubMed]
- Pestova, T.V.; de Breyne, S.; Pisarev, A.V.; Abaeva, I.S.; Hellen, C.U. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: A common role of domain II. EMBO J. 2008, 27, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Terenin, I.M.; Dmitriev, S.E.; Andreev, D.E.; Shatsky, I.N. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat. Struct. Mol. Biol. 2008, 15, 836–841. [Google Scholar] [CrossRef] [PubMed]
- Hashem, Y.; des Georges, A.; Dhote, V.; Langlois, R.; Liao, H.Y.; Grassucci, R.A.; Pestova, T.V.; Hellen, C.U.; Frank, J. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature 2013, 503, 539–543. [Google Scholar] [CrossRef]
- Jaafar, Z.A.; Oguro, A.; Nakamura, Y.; Kieft, J.S. Translation initiation by the hepatitis C virus IRES requires eIF1A and ribosomal complex remodeling. eLife 2016, 5, e21198. [Google Scholar] [CrossRef]
- Brown, Z.P.; Abaeva, I.S.; De, S.; Hellen, C.U.T.; Pestova, T.V.; Frank, J. Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES. EMBO J. 2022, 41, e110581. [Google Scholar] [CrossRef]
- Ali, N.; Pruijn, G.J.; Kenan, D.J.; Keene, J.D.; Siddiqui, A. Human La antigen is required for the hepatitis C virus internal ribosome entry site-mediated translation. J. Biol. Chem. 2000, 275, 27531–27540. [Google Scholar] [CrossRef]
- Hahm, B.; Kim, Y.K.; Kim, J.H.; Kim, T.Y.; Jang, S.K. Heterogeneous nuclear ribonucleoprotein L interacts with the 3′ border of the internal ribosomal entry site of hepatitis C virus. J. Virol. 1998, 72, 8782–8788. [Google Scholar] [CrossRef]
- Paek, K.Y.; Kim, C.S.; Park, S.M.; Kim, J.H.; Jang, S.K. RNA-binding protein hnRNP D modulates internal ribosome entry site-dependent translation of hepatitis C virus RNA. J. Virol. 2008, 82, 12082–12093. [Google Scholar] [CrossRef]
- Yu, Y.; Sweeney, T.R.; Kafasla, P.; Jackson, R.J.; Pestova, T.V.; Hellen, C.U. The mechanism of translation initiation on Aichivirus RNA mediated by a novel type of picornavirus IRES. EMBO J. 2011, 30, 4423–4436. [Google Scholar] [CrossRef] [PubMed]
- Schuler, M.; Connell, S.R.; Lescoute, A.; Giesebrecht, J.; Dabrowski, M.; Schroeer, B.; Mielke, T.; Penczek, P.A.; Westhof, E.; Spahn, C.M. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat. Struct. Mol. Biol. 2006, 13, 1092–1096. [Google Scholar] [CrossRef] [PubMed]
- Willcocks, M.M.; Locker, N.; Gomwalk, Z.; Royall, E.; Bakhshesh, M.; Belsham, G.J.; Idamakanti, N.; Burroughs, K.D.; Reddy, P.S.; Hallenbeck, P.L.; et al. Structural features of the Seneca Valley virus internal ribosome entry site (IRES) element: A picornavirus with a pestivirus-like IRES. J. Virol. 2011, 85, 4452–4461. [Google Scholar] [CrossRef]
- Asnani, M.; Kumar, P.; Hellen, C.U. Widespread distribution and structural diversity of Type IV IRESs in members of Picornaviridae. Virology 2015, 478, 61–74. [Google Scholar] [CrossRef]
- Arhab, Y.; Bulakhov, A.G.; Pestova, T.V.; Hellen, C.U.T. Dissemination of Internal Ribosomal Entry Sites (IRES) Between Viruses by Horizontal Gene Transfer. Viruses 2020, 12, 612. [Google Scholar] [CrossRef]
- Yinda, C.K.; Esefeld, J.; Peter, H.U.; Matthijnssens, J.; Zell, R. Penguin megrivirus, a novel picornavirus from an Adelie penguin (Pygoscelis adeliae). Arch. Virol. 2019, 164, 2887–2890. [Google Scholar] [CrossRef] [PubMed]
- Vernygora, O.; Sullivan, D.; Nielsen, O.; Huntington, K.B.; Rouse, N.; Popov, V.L.; Lung, O. Senecavirus cetus a novel picornavirus isolated from cetaceans represents a major host switching to the marine environment. npj Viruses 2024, 2, 33. [Google Scholar] [CrossRef]
- Liu, F.; Wang, N.; Wang, Q.; Shan, H. Motif mutations in pseudoknot stem I upstream of start codon in Senecavirus A genome: Impacts on activity of viral IRES and on rescue of recombinant virus. Vet. Microbiol. 2021, 262, 109223. [Google Scholar] [CrossRef]
- Easton, L.E.; Locker, N.; Lukavsky, P.J. Conserved functional domains and a novel tertiary interaction near the pseudoknot drive translational activity of hepatitis C virus and hepatitis C virus-like internal ribosome entry sites. Nucleic Acids Res. 2009, 37, 5537–5549. [Google Scholar] [CrossRef]
- Malygin, A.A.; Kossinova, O.A.; Shatsky, I.N.; Karpova, G.G. HCV IRES interacts with the 18S rRNA to activate the 40S ribosome for subsequent steps of translation initiation. Nucleic Acids Res. 2013, 41, 8706–8714. [Google Scholar] [CrossRef]
- Pisarev, A.V.; Chard, L.S.; Kaku, Y.; Johns, H.L.; Shatsky, I.N.; Belsham, G.J. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J. Virol. 2004, 78, 4487–4497. [Google Scholar] [CrossRef]
- Willcocks, M.M.; Zaini, S.; Chamond, N.; Ulryck, N.; Allouche, D.; Rajagopalan, N.; Davids, N.A.; Fahnoe, U.; Hadsbjerg, J.; Rasmussen, T.B.; et al. Distinct roles for the IIId2 sub-domain in pestivirus and picornavirus internal ribosome entry sites. Nucleic Acids Res. 2017, 45, 13016–13028. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, T.; Machida, K.; Iwasaki, W.; Shigeta, T.; Nishimoto, M.; Takahashi, M.; Sakamoto, A.; Yonemochi, M.; Harada, Y.; Shigematsu, H.; et al. HCV IRES Captures an Actively Translating 80S Ribosome. Mol. Cell 2019, 74, 1205–1214.e8. [Google Scholar] [CrossRef] [PubMed]
- Abaeva, I.S.; Pestova, T.V.; Hellen, C.U.T. Genetic mechanisms underlying the structural elaboration and dissemination of viral internal ribosomal entry sites. bioRxiv 2024, 590008. [Google Scholar] [CrossRef] [PubMed]
- Pestova, T.V.; Lomakin, I.B.; Hellen, C.U. Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep. 2004, 5, 906–913. [Google Scholar] [CrossRef]
- Miścicka, A.; Lu, K.; Abaeva, I.S.; Pestova, T.V.; Hellen, C.U.T. Initiation of translation on nedicistrovirus and related intergenic region IRESs by their factor-independent binding to the P site of 80S ribosomes. RNA 2023, 29, 1051–1068. [Google Scholar] [CrossRef]
- Wilson, J.E.; Pestova, T.V.; Hellen, C.U.; Sarnow, P. Initiation of protein synthesis from the A site of the ribosome. Cell 2000, 102, 511–520. [Google Scholar] [CrossRef]
- Kerr, C.H.; Ma, Z.W.; Jang, C.J.; Thompson, S.R.; Jan, E. Molecular analysis of the factorless internal ribosome entry site in Cricket Paralysis virus infection. Sci. Rep. 2016, 6, 37319. [Google Scholar] [CrossRef]
- Abaeva, I.S.; Young, C.; Warsaba, R.; Khan, N.; Tran, L.V.; Jan, E.; Pestova, T.V.; Hellen, C.U.T. The structure and mechanism of action of a distinct class of dicistrovirus intergenic region IRESs. Nucleic Acids Res. 2023, 51, 9294–9313. [Google Scholar] [CrossRef]
- Fernandez, I.S.; Bai, X.C.; Murshudov, G.; Scheres, S.H.; Ramakrishnan, V. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell 2014, 157, 823–831. [Google Scholar] [CrossRef]
- Pisareva, V.P.; Pisarev, A.V.; Fernandez, I.S. Dual tRNA mimicry in the Cricket Paralysis Virus IRES uncovers an unexpected similarity with the Hepatitis C Virus IRES. eLife 2018, 7, e34062. [Google Scholar] [CrossRef]
- Abaeva, I.S.; Vicens, Q.; Bochler, A.; Soufari, H.; Simonetti, A.; Pestova, T.V.; Hashem, Y.; Hellen, C.U.T. The Halastavi árva Virus Intergenic Region IRES Promotes Translation by the Simplest Possible Initiation Mechanism. Cell Rep. 2020, 33, 108476. [Google Scholar] [CrossRef]
- Chapagain, S.; Salcedo-Porras, N.; Abdolahzadeh, A.; Zhang, Y.; Pereira, H.S.; Flibotte, S.; Low, K.; Young, C.; Wu, Y.; Wang, S.; et al. Discovery of functional factorless internal ribosome entry site-like structures through virome mining. PLoS Pathog. 2025, 21, e1013255. [Google Scholar] [CrossRef]
- Danthinne, X.; Seurinck, J.; Meulewaeter, F.; Van Montagu, M.; Cornelissen, M. The 3′ untranslated region of satellite tobacco necrosis virus RNA stimulates translation in vitro. Mol. Cell. Biol. 1993, 13, 3340–3349. [Google Scholar]
- Truniger, V.; Pechar, G.S.; Aranda, M.A. Advances in Understanding the Mechanism of Cap-Independent Cucurbit Aphid-Borne Yellows Virus Protein Synthesis. Int. J. Mol. Sci. 2023, 24, 17598. [Google Scholar] [CrossRef]
- Paudel, D.B.; Sanfacon, H. Mapping of sequences in the 5′ region and 3′ UTR of tomato ringspot virus RNA2 that facilitate cap-independent translation of reporter transcripts in vitro. PLoS ONE 2021, 16, e0249928. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; White, K.A. 5′-3′ RNA-RNA interaction facilitates cap- and poly(A) tail-independent translation of tomato bushy stunt virus mrna: A potential common mechanism for tombusviridae. J. Biol. Chem. 2004, 279, 28862–28872. [Google Scholar] [CrossRef] [PubMed]
- Rakotondrafara, A.M.; Polacek, C.; Harris, E.; Miller, W.A. Oscillating kissing stem-loop interactions mediate 5′ scanning-dependent translation by a viral 3′-cap-independent translation element. RNA 2006, 12, 1893–1906. [Google Scholar] [CrossRef]
- Nicholson, B.L.; White, K.A. 3′ Cap-independent translation enhancers of positive-strand RNA plant viruses. Curr. Opin. Virol. 2011, 1, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, B.L.; Wu, B.; Chevtchenko, I.; White, K.A. Tombusvirus recruitment of host translational machinery via the 3′ UTR. RNA 2010, 16, 1402–1419. [Google Scholar] [CrossRef]
- Wang, S.; Browning, K.S.; Miller, W.A. A viral sequence in the 3′-untranslated region mimics a 5′ cap in facilitating translation of uncapped mRNA. EMBO J. 1997, 16, 4107–4116. [Google Scholar] [CrossRef]
- Batten, J.S.; Desvoyes, B.; Yamamura, Y.; Scholthof, K.B. A translational enhancer element on the 3′-proximal end of the Panicum mosaic virus genome. FEBS Lett. 2006, 580, 2591–2597. [Google Scholar] [CrossRef]
- Scheets, K.; Redinbaugh, M.G. Infectious cDNA transcripts of Maize necrotic streak virus: Infectivity and translational characteristics. Virology 2006, 350, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; White, K.A. Analysis of a 3′-translation enhancer in a tombusvirus: A dynamic model for RNA-RNA interactions of mRNA termini. RNA 2006, 12, 1304–1314. [Google Scholar] [CrossRef]
- McCormack, J.C.; Yuan, X.; Yingling, Y.G.; Kasprzak, W.; Zamora, R.E.; Shapiro, B.A.; Simon, A.E. Structural domains within the 3′ untranslated region of Turnip crinkle virus. J. Virol. 2008, 82, 8706–8720. [Google Scholar] [CrossRef] [PubMed]
- Stupina, V.A.; Meskauskas, A.; McCormack, J.C.; Yingling, Y.G.; Shapiro, B.A.; Dinman, J.D.; Simon, A.E. The 3′ proximal translational enhancer of Turnip crinkle virus binds to 60S ribosomal subunits. RNA 2008, 14, 2379–2393. [Google Scholar] [CrossRef]
- Truniger, V.; Miras, M.; Aranda, M.A. Structural and Functional Diversity of Plant Virus 3′-Cap-Independent Translation Enhancers (3′-CITEs). Front. Plant Sci. 2017, 8, 2047. [Google Scholar] [CrossRef]
- Guo, L.; Allen, E.; Miller, W.A. Structure and function of a cap-independent translation element that functions in either the 3′ or the 5′ untranslated region. RNA 2000, 6, 1808–1820. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Kraft, J.J.; Hui, A.Y.; Miller, W.A. Structural plasticity of Barley yellow dwarf virus-like cap-independent translation elements in four genera of plant viral RNAs. Virology 2010, 402, 177–186. [Google Scholar]
- Zhao, P.; Liu, Q.; Miller, W.A.; Goss, D.J. Eukaryotic translation initiation factor 4G (eIF4G) coordinates interactions with eIF4A, eIF4B, and eIF4E in binding and translation of the barley yellow dwarf virus 3′ cap-independent translation element (BTE). J. Biol. Chem. 2017, 292, 5921–5931. [Google Scholar] [CrossRef]
- Ilyas, M.; Du, Z.; Simon, A.E. Opium Poppy Mosaic Virus Has an Xrn-Resistant, Translated Subgenomic RNA and a BTE 3′ CITE. J. Virol. 2021, 95, e02109-20. [Google Scholar] [CrossRef]
- Chattopadhyay, M.; Kuhlmann, M.M.; Kumar, K.; Simon, A.E. Position of the kissing-loop interaction associated with PTE-type 3′CITEs can affect enhancement of cap-independent translation. Virology 2014, 458–459, 43–52. [Google Scholar] [CrossRef]
- van Lipzig, R.; Van Montagu, M.; Cornelissen, M.; Meulewaeter, F. Functionality of the STNV translational enhancer domain correlates with affinity for two wheat germ factors. Nucleic Acids Res. 2001, 29, 1080–1086. [Google Scholar] [CrossRef]
- Chattopadhyay, M.; Shi, K.; Yuan, X.; Simon, A.E. Long-distance kissing loop interactions between a 3′ proximal Y-shaped structure and apical loops of 5′ hairpins enhance translation of Saguaro cactus virus. Virology 2011, 417, 113–125. [Google Scholar] [PubMed]
- Miras, M.; Truniger, V.; Querol-Audi, J.; Aranda, M.A. Analysis of the interacting partners eIF4F and 3′-CITE required for Melon necrotic spot virus cap-independent translation. Mol. Plant Pathol. 2017, 18, 635–648. [Google Scholar] [PubMed]
- Miras, M.; Rodríguez-Hernández, A.M.; Romero-López, C.; Berzal-Herranz, A.; Colchero, J.; Aranda, M.A.; Truniger, V. A Dual Interaction Between the 5′- and 3′-Ends of the Melon Necrotic Spot Virus (MNSV) RNA Genome Is Required for Efficient Cap-Independent Translation. Front. Plant Sci. 2018, 9, 625. [Google Scholar] [CrossRef]
- Zuo, X.; Wang, J.; Yu, P.; Eyler, D.; Xu, H.; Starich, M.R.; Tiede, D.M.; Simon, A.E.; Kasprzak, W.; Schwieters, C.D.; et al. Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3′ UTR of turnip crinkle virus. Proc. Natl. Acad. Sci. USA 2010, 107, 1385–1390. [Google Scholar] [CrossRef]
- Treder, K.; Kneller, E.L.; Allen, E.M.; Wang, Z.; Browning, K.S.; Miller, W.A. The 3′ cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA 2008, 14, 134–147. [Google Scholar] [PubMed]
- Wang, Z.; Treder, K.; Miller, W.A. Structure of a viral cap-independent translation element that functions via high affinity binding to the eIF4E subunit of eIF4F. J. Biol. Chem. 2009, 284, 14189–14202. [Google Scholar] [CrossRef]
- Du, Z.; Alekhina, O.M.; Vassilenko, K.S.; Simon, A.E. Concerted action of two 3′ cap-independent translation enhancers increases the competitive strength of translated viral genomes. Nucleic Acids Res. 2017, 45, 9558–9572. [Google Scholar] [CrossRef]
- Liu, Q.; Goss, D.J. The 3′ mRNA I-shaped structure of maize necrotic streak virus binds to eukaryotic translation factors for eIF4F-mediated translation initiation. J. Biol. Chem. 2018, 293, 9486–9495. [Google Scholar] [CrossRef]
- Nicholson, B.L.; Zaslaver, O.; Mayberry, L.K.; Browning, K.S.; White, K.A. Tombusvirus Y-shaped translational enhancer forms a complex with eIF4F and can be functionally replaced by heterologous translational enhancers. J. Virol. 2013, 87, 1872–1883. [Google Scholar] [CrossRef]
- Miras, M.; Sempere, R.N.; Kraft, J.J.; Miller, W.A.; Aranda, M.A.; Truniger, V. Interfamilial recombination between viruses led to acquisition of a novel translation-enhancing RNA element that allows resistance breaking. New Phytol. 2014, 202, 233–246. [Google Scholar]
- Harger, J.W.; Meskauskas, A.; Dinman, J.D. An “integrated model” of programmed ribosomal frameshifting. Trends Biochem. Sci. 2002, 27, 448–454. [Google Scholar] [CrossRef]
- Plant, E.P. Ribosomal Frameshift Signals in Viral Genomes. In Viral Genomes—Molecular Structure, Diversity, Gene Expression Mechanisms and Host-Virus Interactions; Garcia, M.L., Romanowski, V., Eds.; IntechOpen: Rijeka, Croatia, 2012. [Google Scholar]
- Dinman, J.D.; Wickner, R.B. Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J. Virol. 1992, 66, 3669–3676. [Google Scholar]
- Karasev, A.V.; Boyko, V.P.; Gowda, S.; Nikolaeva, O.V.; Hilf, M.E.; Koonin, E.V.; Niblett, C.L.; Cline, K.; Gumpf, D.J.; Lee, R.F.; et al. Complete sequence of the citrus tristeza virus RNA genome. Virology 1995, 208, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Plant, E.P.; Rakauskaite, R.; Taylor, D.R.; Dinman, J.D. Achieving a golden mean: Mechanisms by which coronaviruses ensure synthesis of the correct stoichiometric ratios of viral proteins. J. Virol. 2010, 84, 4330–4340. [Google Scholar] [CrossRef] [PubMed]
- Giedroc, D.P.; Cornish, P.V. Frameshifting RNA pseudoknots: Structure and mechanism. Virus Res. 2009, 139, 193–208. [Google Scholar] [CrossRef]
- Jones, C.P.; Ferre-D’Amare, A.R. Crystal structure of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) frameshifting pseudoknot. RNA 2022, 28, 239–249. [Google Scholar] [PubMed]
- Peterson, J.M.; Becker, S.T.; O’Leary, C.A.; Juneja, P.; Yang, Y.; Moss, W.N. Structure of the SARS-CoV-2 Frameshift Stimulatory Element with an Upstream Multibranch Loop. Biochemistry 2024, 63, 1287–1296. [Google Scholar] [CrossRef]
- Poyry, T.A.; Kaminski, A.; Connell, E.J.; Fraser, C.S.; Jackson, R.J. The mechanism of an exceptional case of reinitiation after translation of a long ORF reveals why such events do not generally occur in mammalian mRNA translation. Genes Dev. 2007, 21, 3149–3162. [Google Scholar] [CrossRef]
- Khan, D.; Fox, P.L. Host-like RNA Elements Regulate Virus Translation. Viruses 2024, 16, 468. [Google Scholar] [CrossRef] [PubMed]
- Walter, B.L.; Nguyen, J.H.; Ehrenfeld, E.; Semler, B.L. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA 1999, 5, 1570–1585. [Google Scholar] [CrossRef]
- Bedard, K.M.; Daijogo, S.; Semler, B.L. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 2007, 26, 459–467. [Google Scholar] [CrossRef]
- Beckham, S.A.; Matak, M.Y.; Belousoff, M.J.; Venugopal, H.; Shah, N.; Vankadari, N.; Elmlund, H.; Nguyen, J.H.C.; Semler, B.L.; Wilce, M.C.J.; et al. Structure of the PCBP2/stem-loop IV complex underlying translation initiation mediated by the poliovirus type I IRES. Nucleic Acids Res. 2020, 48, 8006–8021. [Google Scholar] [CrossRef] [PubMed]
- Joachims, M.; Van Breugel, P.C.; Lloyd, R.E. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 1999, 73, 718–727. [Google Scholar] [CrossRef]
- Hellen, C.U.; Witherell, G.W.; Schmid, M.; Shin, S.H.; Pestova, T.V.; Gil, A.; Wimmer, E. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA 1993, 90, 7642–7646. [Google Scholar] [CrossRef] [PubMed]
- Berry, K.E.; Waghray, S.; Doudna, J.A. The HCV IRES pseudoknot positions the initiation codon on the 40S ribosomal subunit. RNA 2010, 16, 1559–1569. [Google Scholar] [CrossRef]
- Berry, K.E.; Waghray, S.; Mortimer, S.A.; Bai, Y.; Doudna, J.A. Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning. Structure 2011, 19, 1456–1466. [Google Scholar] [CrossRef]
- Koirala, D.; Lewicka, A.; Koldobskaya, Y.; Huang, H.; Piccirilli, J.A. Synthetic Antibody Binding to a Preorganized RNA Domain of Hepatitis C Virus Internal Ribosome Entry Site Inhibits Translation. ACS Chem. Biol. 2020, 15, 205–216. [Google Scholar] [CrossRef]
- Hertz, M.I.; Thompson, S.R. Mechanism of translation initiation by Dicistroviridae IGR IRESs. Virology 2011, 411, 355–361. [Google Scholar] [CrossRef]
- Cammas, A.; Pileur, F.; Bonnal, S.; Lewis, S.M.; Leveque, N.; Holcik, M.; Vagner, S. Cytoplasmic relocalization of heterogeneous nuclear ribonucleoprotein A1 controls translation initiation of specific mRNAs. Mol. Biol. Cell 2007, 18, 5048–5059. [Google Scholar] [CrossRef]
- Meerovitch, K.; Svitkin, Y.V.; Lee, H.S.; Lejbkowicz, F.; Kenan, D.J.; Chan, E.K.; Agol, V.I.; Keene, J.D.; Sonenberg, N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 1993, 67, 3798–3807. [Google Scholar] [CrossRef] [PubMed]
- Ali, N.; Siddiqui, A. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc. Natl. Acad. Sci. USA 1997, 94, 2249–2254. [Google Scholar] [PubMed]
- Yu, Y.; Ji, H.; Doudna, J.A.; Leary, J.A. Mass spectrometric analysis of the human 40S ribosomal subunit: Native and HCV IRES-bound complexes. Protein Sci. 2005, 14, 1438–1446. [Google Scholar] [CrossRef] [PubMed]
- Majzoub, K.; Hafirassou, M.L.; Meignin, C.; Goto, A.; Marzi, S.; Fedorova, A.; Verdier, Y.; Vinh, J.; Hoffmann, J.A.; Martin, F.; et al. RACK1 controls IRES-mediated translation of viruses. Cell 2014, 159, 1086–1095. [Google Scholar] [CrossRef]
- Pineiro, D.; Fernandez, N.; Ramajo, J.; Martinez-Salas, E. Gemin5 promotes IRES interaction and translation control through its C-terminal region. Nucleic Acids Res. 2013, 41, 1017–1028. [Google Scholar]
- Lawrence, P.; Schafer, E.A.; Rieder, E. The nuclear protein Sam68 is cleaved by the FMDV 3C protease redistributing Sam68 to the cytoplasm during FMDV infection of host cells. Virology 2012, 425, 40–52. [Google Scholar] [CrossRef]
- Komar, A.A.; Hatzoglou, M. Cellular IRES-mediated translation: The war of ITAFs in pathophysiological states. Cell Cycle 2011, 10, 229–240. [Google Scholar]
- Iwakawa, H.O.; Tajima, Y.; Taniguchi, T.; Kaido, M.; Mise, K.; Tomari, Y.; Taniguchi, H.; Okuno, T. Poly(A)-binding protein facilitates translation of an uncapped/nonpolyadenylated viral RNA by binding to the 3′ untranslated region. J. Virol. 2012, 86, 7836–7849. [Google Scholar]
- Michon, T.; Estevez, Y.; Walter, J.; German-Retana, S.; Le Gall, O. The potyviral virus genome-linked protein VPg forms a ternary complex with the eukaryotic initiation factors eIF4E and eIF4G and reduces eIF4E affinity for a mRNA cap analogue. FEBS J. 2006, 273, 1312–1322. [Google Scholar] [CrossRef] [PubMed]
- Pathak, H.B.; Oh, H.S.; Goodfellow, I.G.; Arnold, J.J.; Cameron, C.E. Picornavirus genome replication: Roles of precursor proteins and rate-limiting steps in oriI-dependent VPg uridylylation. J. Biol. Chem. 2008, 283, 30677–30688. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.V.; Wimmer, E. Initiation of protein-primed picornavirus RNA synthesis. Virus Res. 2015, 206, 12–26. [Google Scholar] [CrossRef]
- Chaudhry, Y.; Nayak, A.; Bordeleau, M.E.; Tanaka, J.; Pelletier, J.; Belsham, G.J.; Roberts, L.O.; Goodfellow, I.G. Caliciviruses differ in their functional requirements for eIF4F components. J. Biol. Chem. 2006, 281, 25315–25325. [Google Scholar] [CrossRef]
- Rieder, E.; Paul, A.V.; Kim, D.W.; van Boom, J.H.; Wimmer, E. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 2000, 74, 10371–10380. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, Y.; Shan, C.; Chen, C.; Xu, P.; Song, M.; Zhou, H.; Yang, C.; Xu, W.; Shi, P.Y.; et al. Enterovirus 71 VPg uridylation uses a two-molecular mechanism of 3D polymerase. J. Virol. 2012, 86, 13662–13671. [Google Scholar] [CrossRef]
- Hanson, W.A.; Romero Agosto, G.A.; Rouskin, S. Viral RNA Interactome: The Ultimate Researcher’s Guide to RNA-Protein Interactions. Viruses 2024, 16, 1702. [Google Scholar]
- Perera, R.; Daijogo, S.; Walter, B.L.; Nguyen, J.H.; Semler, B.L. Cellular protein modification by poliovirus: The two faces of poly(rC)-binding protein. J. Virol. 2007, 81, 8919–8932. [Google Scholar]
- Steil, B.P.; Barton, D.J. Cis-active RNA elements (CREs) and picornavirus RNA replication. Virus Res. 2009, 139, 240–252. [Google Scholar] [CrossRef]
- Koirala, D.; Shao, Y.; Koldobskaya, Y.; Fuller, J.R.; Watkins, A.M.; Shelke, S.A.; Pilipenko, E.V.; Das, R.; Rice, P.A.; Piccirilli, J.A. A conserved RNA structural motif for organizing topology within picornaviral internal ribosome entry sites. Nat. Commun. 2019, 10, 3629. [Google Scholar] [CrossRef]
- Muhs, M.; Hilal, T.; Mielke, T.; Skabkin, M.A.; Sanbonmatsu, K.Y.; Pestova, T.V.; Spahn, C.M. Cryo-EM of ribosomal 80S complexes with termination factors reveals the translocated cricket paralysis virus IRES. Mol. Cell 2015, 57, 422–432. [Google Scholar] [CrossRef]
- Murray, J.; Savva, C.G.; Shin, B.-S.; Dever, T.E.; Ramakrishnan, V.; Fernández, I.S. Structural characterization of ribosome recruitment and translocation by type IV IRES. eLife 2016, 5, e13567. [Google Scholar] [CrossRef]
- Ojha, M.; Vogt, J.; Das, N.K.; Redmond, E.; Singh, K.; Banna, H.A.; Sadat, T.; Koirala, D. Structure of saguaro cactus virus 3′ translational enhancer mimics 5′ cap for eIF4E binding. Proc. Natl. Acad. Sci. USA 2024, 121, e2313677121. [Google Scholar] [CrossRef]
- Lewicka, A.; Roman, C.; Jones, S.; Disare, M.; Rice, P.A.; Piccirilli, J.A. Crystal structure of a cap-independent translation enhancer RNA. Nucleic Acids Res. 2023, 51, 8891–8907. [Google Scholar] [CrossRef]
- Pekarek, L.; Zimmer, M.M.; Gribling-Burrer, A.S.; Buck, S.; Smyth, R.; Caliskan, N. Cis-mediated interactions of the SARS-CoV-2 frameshift RNA alter its conformations and affect function. Nucleic Acids Res. 2023, 51, 728–743. [Google Scholar] [CrossRef]
- Akiyama, B.M.; Graham, M.E.; Z, O.D.; Beckham, J.D.; Kieft, J.S. Three-dimensional structure of a flavivirus dumbbell RNA reveals molecular details of an RNA regulator of replication. Nucleic Acids Res. 2021, 49, 7122–7138. [Google Scholar] [CrossRef] [PubMed]
- Dibrov, S.M.; Ding, K.; Brunn, N.D.; Parker, M.A.; Bergdahl, B.M.; Wyles, D.L.; Hermann, T. Structure of a hepatitis C virus RNA domain in complex with a translation inhibitor reveals a binding mode reminiscent of riboswitches. Proc. Natl. Acad. Sci. USA 2012, 109, 5223–5228. [Google Scholar] [CrossRef] [PubMed]
- Banna, H.A.; Berg, K.; Sadat, T.; Das, N.K.; Paudel, R.; D’Souza, V.; Koirala, D. Synthetic anti-RNA antibody derivatives for RNA visualization in mammalian cells. Nucleic Acids Res. 2025, 53, gkae1275. [Google Scholar] [CrossRef] [PubMed]
- Costantino, D.A.; Pfingsten, J.S.; Rambo, R.P.; Kieft, J.S. tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nat. Struct. Mol. Biol. 2008, 15, 57–64. [Google Scholar] [CrossRef]
- Imai, S.; Suzuki, H.; Fujiyoshi, Y.; Shimada, I. Dynamically regulated two-site interaction of viral RNA to capture host translation initiation factor. Nat. Commun. 2023, 14, 4977. [Google Scholar] [CrossRef]
- Davila-Calderon, J.; Patwardhan, N.N.; Chiu, L.Y.; Sugarman, A.; Cai, Z.; Penutmutchu, S.R.; Li, M.L.; Brewer, G.; Hargrove, A.E.; Tolbert, B.S. IRES-targeting small molecule inhibits enterovirus 71 replication via allosteric stabilization of a ternary complex. Nat. Commun. 2020, 11, 4775. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, S.; Phelan, M.M.; Rasul, U.; Ramesh, V. NMR elucidation of the role of Mg2+ in the structure and stability of the conserved RNA motifs of the EMCV IRES element. Org. Biomol. Chem. 2014, 12, 1495–1509. [Google Scholar] [PubMed]
- Dorn, G.; Gmeiner, C.; de Vries, T.; Dedic, E.; Novakovic, M.; Damberger, F.F.; Maris, C.; Finol, E.; Sarnowski, C.P.; Kohlbrecher, J.; et al. Integrative solution structure of PTBP1-IRES complex reveals strong compaction and ordering with residual conformational flexibility. Nat. Commun. 2023, 14, 6429. [Google Scholar] [CrossRef]
- Perard, J.; Leyrat, C.; Baudin, F.; Drouet, E.; Jamin, M. Structure of the full-length HCV IRES in solution. Nat. Commun. 2013, 4, 1612. [Google Scholar] [CrossRef] [PubMed]
- Lukavsky, P.J.; Kim, I.; Otto, G.A.; Puglisi, J.D. Structure of HCV IRES domain II determined by NMR. Nat. Struct. Biol. 2003, 10, 1033–1038. [Google Scholar] [CrossRef]
- Neupane, R.; Pisareva, V.P.; Rodriguez, C.F.; Pisarev, A.V.; Fernandez, I.S. A complex IRES at the 5′-UTR of a viral mRNA assembles a functional 48S complex via an uAUG intermediate. eLife 2020, 9, e54575. [Google Scholar]
- Johnson, P.Z.; Kasprzak, W.K.; Shapiro, B.A.; Simon, A.E. Structural characterization of a new subclass of panicum mosaic virus-like 3′ cap-independent translation enhancer. Nucleic Acids Res. 2022, 50, 1601–1619. [Google Scholar]
- Bera, S.; Ilyas, M.; Mikkelsen, A.A.; Simon, A.E. Conserved Structure Associated with Different 3′CITEs Is Important for Translation of Umbraviruses. Viruses 2023, 15, 638. [Google Scholar] [CrossRef]
- Venkatesan, A.; Dasgupta, A. Novel fluorescence-based screen to identify small synthetic internal ribosome entry site elements. Mol. Cell. Biol. 2001, 21, 2826–2837. [Google Scholar] [CrossRef]
- Lozano, G.; Francisco-Velilla, R.; Martinez-Salas, E. Ribosome-dependent conformational flexibility changes and RNA dynamics of IRES domains revealed by differential SHAPE. Sci. Rep. 2018, 8, 5545. [Google Scholar] [CrossRef]
- García-Sacristán, A.; Moreno, M.; Ariza-Mateos, A.; López-Camacho, E.; Jáudenes, R.M.; Vázquez, L.; Gómez, J.; Martín-Gago, J.; Briones, C. A magnesium-induced RNA conformational switch at the internal ribosome entry site of hepatitis C virus genome visualized by atomic force microscopy. Nucleic Acids Res. 2015, 43, 565–580. [Google Scholar] [PubMed]
- Fernandez-Miragall, O.; Martinez-Salas, E. Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA 2003, 9, 1333–1344. [Google Scholar] [CrossRef] [PubMed]
- Gross, L.; Vicens, Q.; Einhorn, E.; Noireterre, A.; Schaeffer, L.; Kuhn, L.; Imler, J.L.; Eriani, G.; Meignin, C.; Martin, F. The IRES5’UTR of the dicistrovirus cricket paralysis virus is a type III IRES containing an essential pseudoknot structure. Nucleic Acids Res. 2017, 45, 8993–9004. [Google Scholar] [PubMed]
- Mahmud, B.; Horn, C.M.; Tapprich, W.E. Structure of the 5′ Untranslated Region of Enteroviral Genomic RNA. J. Virol. 2019, 93, e01288-19. [Google Scholar] [CrossRef]
- Merino, E.J.; Wilkinson, K.A.; Coughlan, J.L.; Weeks, K.M. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 2005, 127, 4223–4231. [Google Scholar]
- Low, J.T.; Garcia-Miranda, P.; Mouzakis, K.D.; Gorelick, R.J.; Butcher, S.E.; Weeks, K.M. Structure and dynamics of the HIV-1 frameshift element RNA. Biochemistry 2014, 53, 4282–4291. [Google Scholar] [CrossRef]
- Boerneke, M.A.; Ehrhardt, J.E.; Weeks, K.M. Physical and Functional Analysis of Viral RNA Genomes by SHAPE. Annu. Rev. Virol. 2019, 6, 93–117. [Google Scholar] [CrossRef]
- Madden, E.A.; Plante, K.S.; Morrison, C.R.; Kutchko, K.M.; Sanders, W.; Long, K.M.; Taft-Benz, S.; Cruz Cisneros, M.C.; White, A.M.; Sarkar, S.; et al. Using SHAPE-MaP To Model RNA Secondary Structure and Identify 3′UTR Variation in Chikungunya Virus. J. Virol. 2020, 94, e00701-20. [Google Scholar] [CrossRef]
- Diaz-Toledano, R.; Lozano, G.; Martinez-Salas, E. In-cell SHAPE uncovers dynamic interactions between the untranslated regions of the foot-and-mouth disease virus RNA. Nucleic Acids Res. 2017, 45, 1416–1432. [Google Scholar]
- Gosavi, D.; Wower, I.; Beckmann, I.K.; Hofacker, I.L.; Wower, J.; Wolfinger, M.T.; Sztuba-Solinska, J. Insights into the secondary and tertiary structure of the Bovine Viral Diarrhea Virus Internal Ribosome Entry Site. RNA Biol. 2022, 19, 496–506. [Google Scholar] [CrossRef]
- Gamarnik, A.V.; Andino, R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998, 12, 2293–2304. [Google Scholar] [CrossRef]
- Etchison, D.; Milburn, S.C.; Edery, I.; Sonenberg, N.; Hershey, J.W. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 1982, 257, 14806–14810. [Google Scholar] [CrossRef]
- Gradi, A.; Svitkin, Y.V.; Imataka, H.; Sonenberg, N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. USA 1998, 95, 11089–11094. [Google Scholar]
- Sun, D.; Chen, S.; Cheng, A.; Wang, M. Roles of the Picornaviral 3C Proteinase in the Viral Life Cycle and Host Cells. Viruses 2016, 8, 82. [Google Scholar] [CrossRef]
- Yamamoto, H.; Unbehaun, A.; Spahn, C.M.T. Ribosomal Chamber Music: Toward an Understanding of IRES Mechanisms. Trends Biochem. Sci. 2017, 42, 655–668. [Google Scholar] [CrossRef]
- Gamarnik, A.V.; Andino, R. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 2000, 74, 2219–2226. [Google Scholar] [CrossRef]
- Das, N.K.; Vogt, J.; Patel, A.; Banna, H.A.; Koirala, D. Structural basis for a highly conserved RNA-mediated enteroviral genome replication. Nucleic Acids Res. 2024, 52, 11218–11233. [Google Scholar] [CrossRef] [PubMed]
- Zust, R.; Miller, T.B.; Goebel, S.J.; Thiel, V.; Masters, P.S. Genetic interactions between an essential 3′ cis-acting RNA pseudoknot, replicase gene products, and the extreme 3′ end of the mouse coronavirus genome. J. Virol. 2008, 82, 1214–1228. [Google Scholar] [CrossRef] [PubMed]
- Brierley, I.; Dos Ramos, F.J. Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV. Virus Res. 2006, 119, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Alexandersen, S.; Chamings, A.; Bhatta, T.R. SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication. Nat. Commun. 2020, 11, 6059. [Google Scholar] [CrossRef]
- Romero-Lopez, C.; Roda-Herreros, M.; Berzal-Herranz, B.; Ramos-Lorente, S.E.; Berzal-Herranz, A. Inter- and Intramolecular RNA-RNA Interactions Modulate the Regulation of Translation Mediated by the 3′ UTR in West Nile Virus. Int. J. Mol. Sci. 2023, 24, 5337. [Google Scholar] [CrossRef] [PubMed]
- Barrera, A.; Ramos, H.; Vera-Otarola, J.; Fernandez-Garcia, L.; Angulo, J.; Olguin, V.; Pino, K.; Mouland, A.J.; Lopez-Lastra, M. Post-translational modifications of hnRNP A1 differentially modulate retroviral IRES-mediated translation initiation. Nucleic Acids Res. 2020, 48, 10479–10499. [Google Scholar] [CrossRef]
- Soto-Rifo, R.; Rubilar, P.S.; Limousin, T.; de Breyne, S.; Decimo, D.; Ohlmann, T. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J. 2012, 31, 3745–3756. [Google Scholar] [CrossRef]
- Su, Y.S.; Tsai, A.H.; Ho, Y.F.; Huang, S.Y.; Liu, Y.C.; Hwang, L.H. Stimulation of the Internal Ribosome Entry Site (IRES)-Dependent Translation of Enterovirus 71 by DDX3X RNA Helicase and Viral 2A and 3C Proteases. Front. Microbiol. 2018, 9, 1324. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Sun, S.; Li, P.; Liu, Q.; Zhang, Z.; Dong, H.; Sun, M.; Wu, W.; Wang, X.; Guo, H. Ribosomal Protein L13 Promotes IRES-Driven Translation of Foot-and-Mouth Disease Virus in a Helicase DDX3-Dependent Manner. J. Virol. 2020, 94, e01679-19. [Google Scholar] [CrossRef] [PubMed]
| ITAF | IRESs | ITAF’s Roles | IRES Activities | References |
|---|---|---|---|---|
| PTB (hnRNP I) | PV, HRV | RNA chaperone and stabilizer; alters IRES secondary structure | Activation, inhibition (context-dependent) | References [56,70,172] |
| PCBP2 (hnRNP E) | PV, HAV | Binds IRES sites; restricts conformational flexibility; assists eIFs | Activation | References [168,169] |
| hnRNP A1 | HRV-2 | RNA chaperone; nucleocytoplasmic shuttling; affects structure | Activation, inhibition (context-dependent) | Reference [177] |
| La autoantigen | PV, HCV | Assists translation initiation; stabilizes IRES conformation | Activation, inhibition (HAV IRES) | References [57,178,179] |
| Unr | HRV | RNA chaperone; alters IRES secondary structure | Activation | Reference [58] |
| RACK1 | HCV, DCV, CrPV | 40S ribosomal subunit-associated protein, mediates IRES activity | Activation | References [180,181] |
| Gemin5 | FMDV, HCV | Ribosome-associated protein, competes for ITAF binding | Inhibition | Reference [182] |
| Sam68 | FMDV, EV71 | Nucleocytoplasmic shuttling, stimulates IRES activity | Activation | References [82,183] |
| Methods | Strengths | Limitations | Structures |
|---|---|---|---|
| X-ray Crystallography | Atomic-resolution 3D structures; precise protein-RNA contact sites | Requires crystal formation (difficult for large, flexible RNAs); static nature | PEMV2 PTE [199], SCV PTE [198], Donggang virus dumbbells [201] HAV IRES domain V [195], HCV IRES subdomains [174,202,203], IGR IRES [204] |
| Nuclear Magnetic Resonance (NMR) | Probes dynamic conformational changes; solution-state interactions; binding affinities | Size limitations for large complexes; spectral complexity | EMCV IRES + eIF4G/eIF4A complex [205], EV71 IRES SLII + DMA-135 [206], EMCV IRES J-K domain [65,207], PTBP1-EMCV IRES fragment [208], HCV IRES [209,210] |
| Cryo-Electron Microscopy (Cryo-EM) | High-resolution structures of large complexes; captures multiple conformational states | Requires large complexes; data processing complexity | HCV IRES-40S [101], CrPV IRES-40S [211], EMCV IRES-eIF4G/eIF4A [205] |
| Biochemical Methods | Maps RNA secondary structures in solution; identifies flexible regions; guides 3D modeling | Does not provide atomic resolution; mostly limited to secondary structure information | PTE structures [199,212], BTE 3′ CITE [213], Poliovirus IRES [214,215], HCV IRES [216], FMDV IRES [217], CrPV IRES [218], Coxsackievirus B3 IRES [219] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, G.; Beyene, B.G.; Camacho, J.M.; Koirala, D. Roles of RNA Structures in the Genome Translation of (+) Sense RNA Viruses. Viruses 2025, 17, 1404. https://doi.org/10.3390/v17111404
Lu G, Beyene BG, Camacho JM, Koirala D. Roles of RNA Structures in the Genome Translation of (+) Sense RNA Viruses. Viruses. 2025; 17(11):1404. https://doi.org/10.3390/v17111404
Chicago/Turabian StyleLu, Guangming, Bethel G. Beyene, Joshua Miguele Camacho, and Deepak Koirala. 2025. "Roles of RNA Structures in the Genome Translation of (+) Sense RNA Viruses" Viruses 17, no. 11: 1404. https://doi.org/10.3390/v17111404
APA StyleLu, G., Beyene, B. G., Camacho, J. M., & Koirala, D. (2025). Roles of RNA Structures in the Genome Translation of (+) Sense RNA Viruses. Viruses, 17(11), 1404. https://doi.org/10.3390/v17111404






