Dissecting the Unique Self-Assembly Landscape of the HIV-2 Capsid Protein
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Expression and Purification
2.2. In Vitro Assembly of CA Structures
2.3. Sedimentation of Assembled CA Structures
2.4. Monitoring CA Assembly by Optical Density at 350 nm
2.5. Electron Microscopy
2.6. Quantification of CA Assembly Morphologies
2.7. Atomic Model Analysis
3. Results
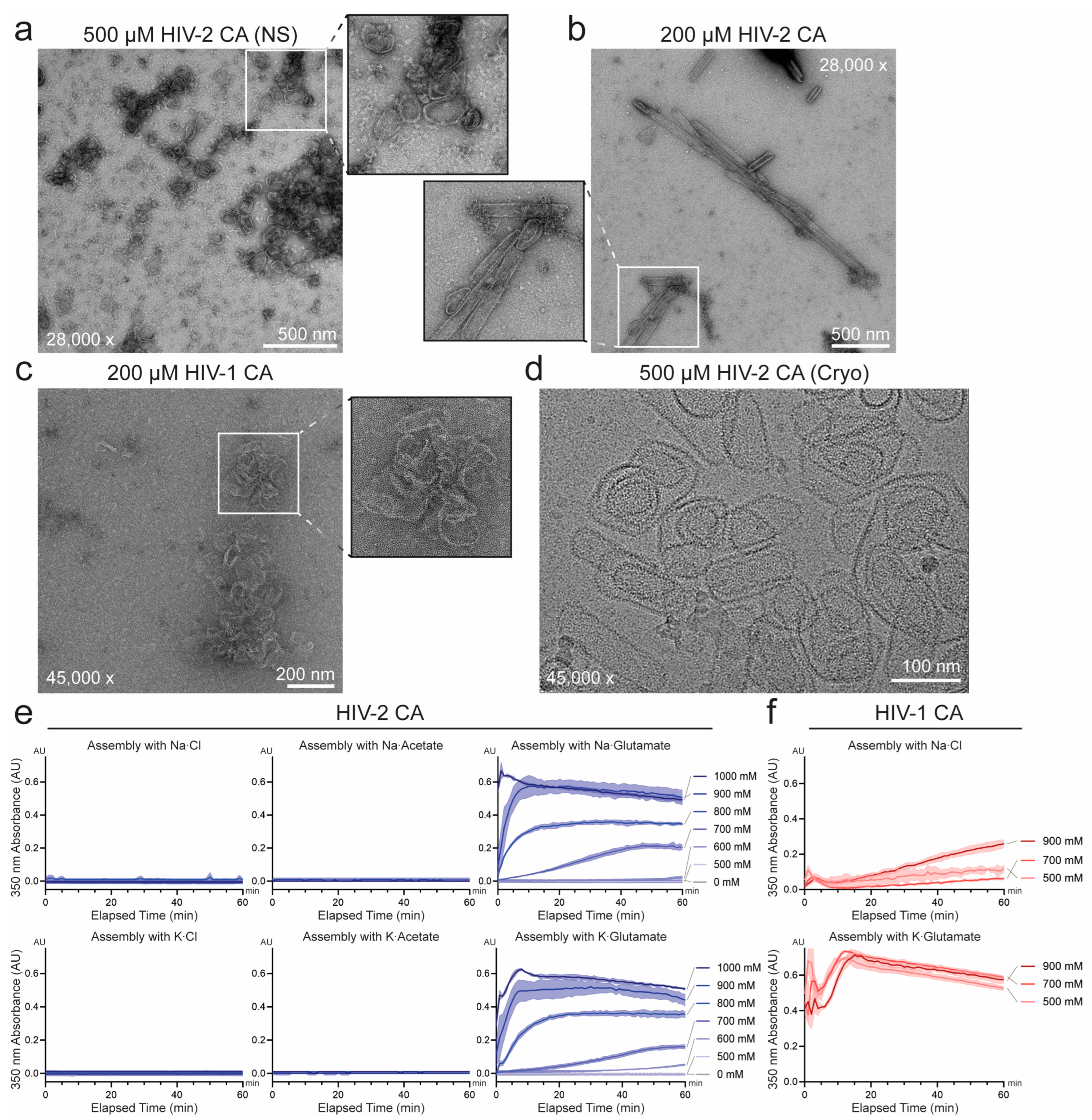
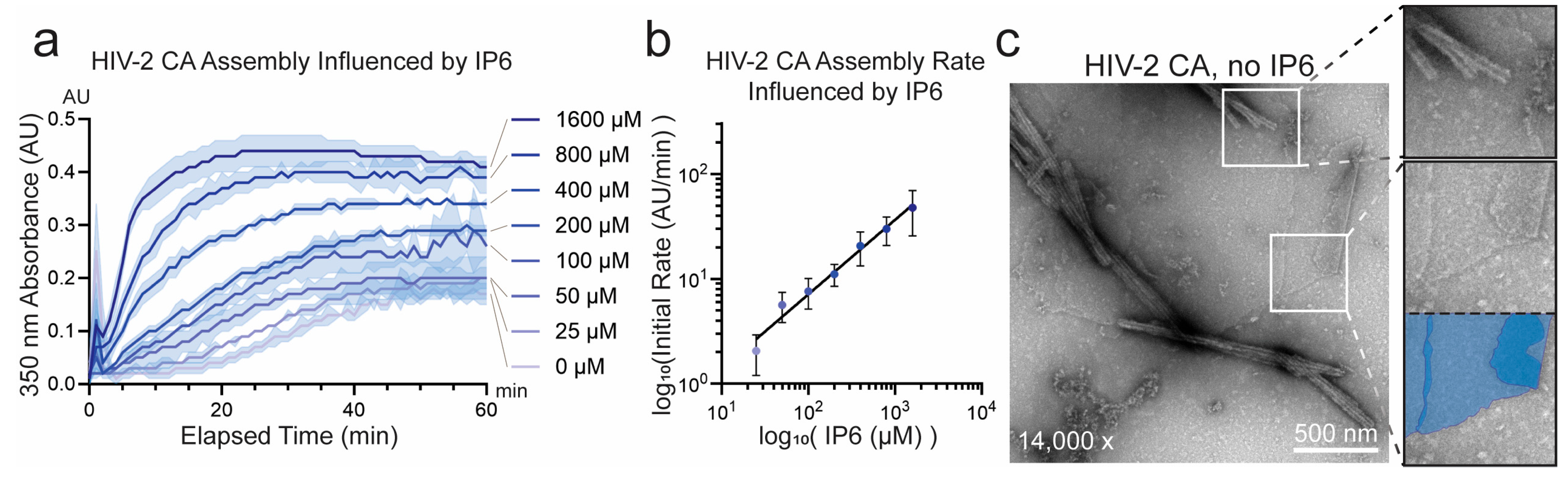
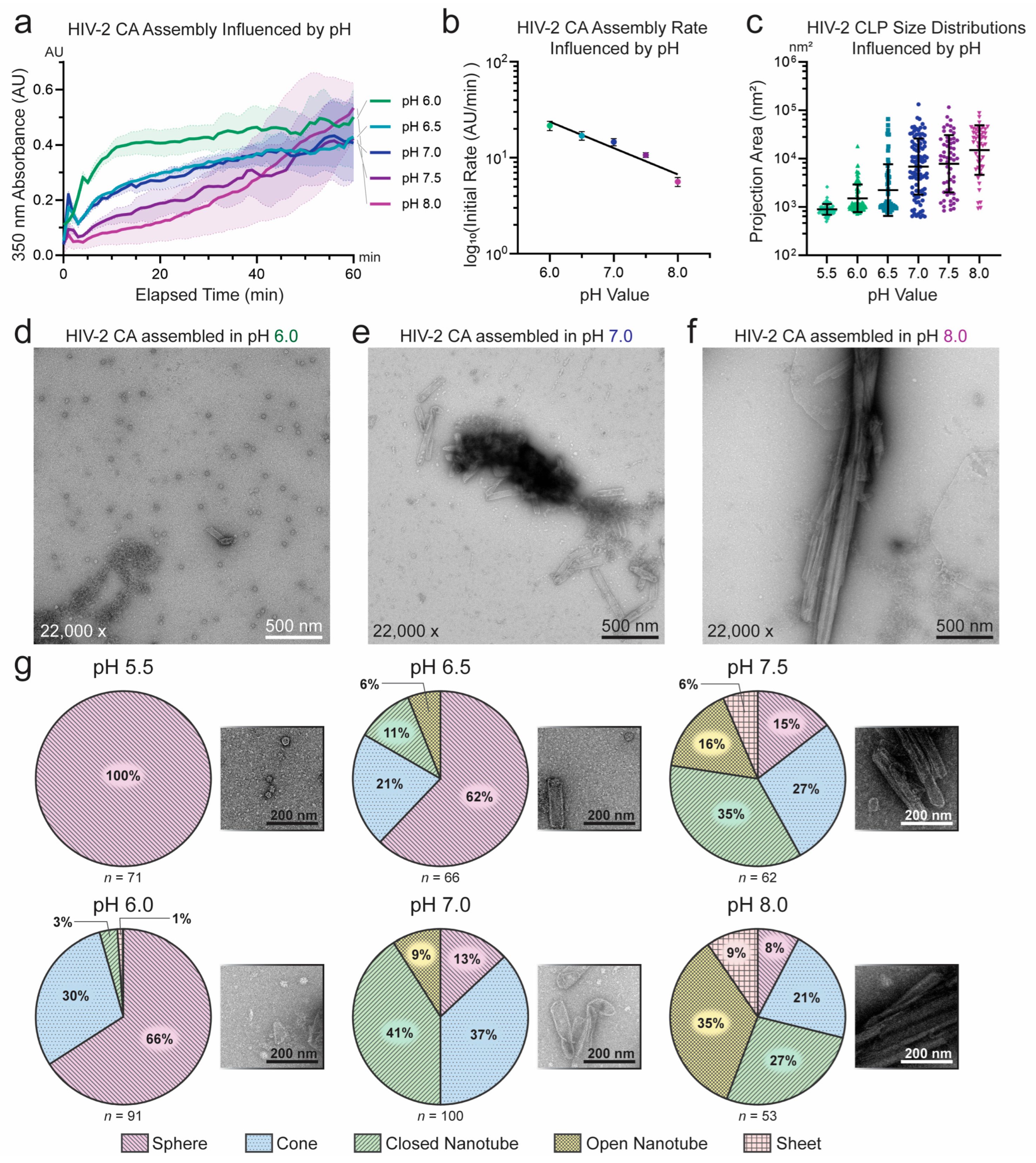
3.1. Unique Self-Assembly Properties of HIV-2 CA Revealed by Robust In Vitro Assembly
3.2. Sequence Divergence at Inter-Protomer Interfaces Modulates CA Assembly Kinetics and Particle Morphology
4. Discussion
Limitations of the Study
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BME | β-mercaptoethanol |
| CA | Capsid protein |
| CLP | Capsid-like particle |
| CTD | C-terminal domain |
| CypA | Cyclophilin A |
| EM | Electron microscopy |
| HIV | Human immunodeficiency virus |
| IP6 | Inositol hexakisphosphate |
| IPTG | Isopropyl β-d-1 thiogalactopyranoside |
| KGlu | Potassium glutamate |
| LUV | Large unilamellar vesicle |
| Ni-NTA | Nickel–nitrilotriacetic acid |
| NTD | N-terminal domain |
| RT | Room temperature |
| SEC | Size-exclusion chromatography |
| TCEP | Tris(2-carboxyethyl)phosphine |
| UA | Uranyl acetate |
References
- Dwivedi, R.; Prakash, P.; Kumbhar, B.V.; Balasubramaniam, M.; Dash, C. HIV-1 capsid and viral DNA integration. mBio 2024, 15, e0021222. [Google Scholar] [CrossRef]
- Link, J.O.; Rhee, M.S.; Tse, W.C.; Zheng, J.; Somoza, J.R.; Rowe, W.; Begley, R.; Chiu, A.; Mulato, A.; Hansen, D.; et al. Clinical targeting of HIV capsid protein with a long-acting small molecule. Nature 2020, 584, 614–618. [Google Scholar] [CrossRef]
- McGraw, A.; Hillmer, G.; Medehincu, S.M.; Hikichi, Y.; Gagliardi, S.; Narayan, K.; Tibebe, H.; Marquez, D.; Bose, L.M.; Keating, A.; et al. Exploring HIV-1 Maturation: A New Frontier in Antiviral Development. Viruses 2024, 16, 1423. [Google Scholar] [CrossRef]
- Hirsch, V.M.; Olmsted, R.A.; Murphey-Corb, M.; Purcell, R.H.; Johnson, P.R. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 1989, 339, 389–392. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Bailes, E.; Robertson, D.L.; Chen, Y.; Rodenburg, C.M.; Michael, S.F.; Cummins, L.B.; Arthur, L.O.; Peeters, M.; Shaw, G.M.; et al. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 1999, 397, 436–441. [Google Scholar] [CrossRef] [PubMed]
- Sauter, D.; Kirchhoff, F. Key Viral Adaptations Preceding the AIDS Pandemic. Cell Host Microbe 2019, 25, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Marlink, R.; Kanki, P.; Thior, I.; Travers, K.; Eisen, G.; Siby, T.; Traore, I.; Hsieh, C.-C.; Dia, M.C.; Gueye, E.-H.; et al. Reduced Rate of Disease Development After HIV-2 Infection as Compared to HIV-1. Science 1994, 265, 1587–1590. [Google Scholar] [CrossRef]
- Popper, S.J.; Sarr, A.D.; Travers, K.U.; Guèye–Ndiaye, A.; Mboup, S.; Essex, M.E.; Kanki, P.J. Lower Human Immunodeficiency Virus (HIV) Type 2 Viral Load Reflects the Difference in Pathogenicity of HIV–1 and HIV–2. J. Infect. Dis. 1999, 180, 1116–1121. [Google Scholar] [CrossRef]
- Ganser, B.K.; Li, S.; Klishko, V.Y.; Finch, J.T.; Sundquist, W.I. Assembly and Analysis of Conical Models for the HIV-1 Core. Science 1999, 283, 80–83. [Google Scholar] [CrossRef]
- Li, S.; Hill, C.P.; Sundquist, W.I.; Finch, J.T. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 2000, 407, 409–413. [Google Scholar] [CrossRef]
- Briggs, J.A.G.; Wilk, T.; Welker, R.; Kräusslich, H.-G.; Fuller, S.D. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 2003, 22, 1707–1715. [Google Scholar] [CrossRef]
- Ni, T.; Zhu, Y.; Yang, Z.; Xu, C.; Chaban, Y.; Nesterova, T.; Ning, J.; Böcking, T.; Parker, M.W.; Monnie, C.; et al. Structure of native HIV-1 cores and their interactions with IP6 and CypA. Sci. Adv. 2021, 7, eabj5715. [Google Scholar] [CrossRef]
- Pornillos, O.; Ganser-Pornillos, B.K.; Kelly, B.N.; Hua, Y.; Whitby, F.G.; Stout, C.D.; Sundquist, W.I.; Hill, C.P.; Yeager, M. X-Ray Structures of the Hexameric Building Block of the HIV Capsid. Cell 2009, 137, 1282–1292. [Google Scholar] [CrossRef] [PubMed]
- Gres, A.T.; Kirby, K.A.; KewalRamani, V.N.; Tanner, J.J.; Pornillos, O.; Sarafianos, S.G. X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability. Science 2015, 349, 99–103. [Google Scholar] [CrossRef]
- Mattei, S.; Glass, B.; Hagen, W.J.H.; Kräusslich, H.-G.; Briggs, J.A.G. The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science 2016, 354, 1434–1437. [Google Scholar] [CrossRef]
- Stacey, J.C.V.; Tan, A.; Lu, J.M.; James, L.C.; Dick, R.A.; Briggs, J.A.G. Two structural switches in HIV-1 capsid regulate capsid curvature and host factor binding. Proc. Natl. Acad. Sci. USA 2023, 120, e2220557120. [Google Scholar] [CrossRef] [PubMed]
- Schirra, R.T.; dos Santos, N.F.B.; Zadrozny, K.K.; Kucharska, I.; Ganser-Pornillos, B.K.; Pornillos, O. A molecular switch modulates assembly and host factor binding of the HIV-1 capsid. Nat. Struct. Mol. Biol. 2023, 30, 383–390. [Google Scholar] [CrossRef]
- Gamble, T.R.; Yoo, S.; Vajdos, F.F.; von Schwedler, U.K.; Worthylake, D.K.; Wang, H.; McCutcheon, J.P.; Sundquist, W.I.; Hill, C.P. Structure of the Carboxyl-Terminal Dimerization Domain of the HIV-1 Capsid Protein. Science 1997, 278, 849–853. [Google Scholar] [CrossRef]
- Byeon, I.-J.L.; Meng, X.; Jung, J.; Zhao, G.; Yang, R.; Ahn, J.; Shi, J.; Concel, J.; Aiken, C.; Zhang, P.; et al. Structural Convergence between Cryo-EM and NMR Reveals Intersubunit Interactions Critical for HIV-1 Capsid Function. Cell 2009, 139, 780–790. [Google Scholar] [CrossRef]
- Deshmukh, L.; Schwieters, C.D.; Grishaev, A.; Ghirlando, R.; Baber, J.L.; Clore, G.M. Structure and Dynamics of Full-Length HIV-1 Capsid Protein in Solution. J. Am. Chem. Soc. 2013, 135, 16133–16147. [Google Scholar] [CrossRef]
- Lu, M.; Russell, R.W.; Bryer, A.J.; Quinn, C.M.; Hou, G.; Zhang, H.; Schwieters, C.D.; Perilla, J.R.; Gronenborn, A.M.; Polenova, T. Atomic-resolution structure of HIV-1 capsid tubes by magic-angle spinning NMR. Nat. Struct. Mol. Biol. 2020, 27, 863–869. [Google Scholar] [CrossRef]
- Ehrlich, L.S.; Agresta, B.E.; Carter, C.A. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J. Virol. 1992, 66, 4874–4883. [Google Scholar] [CrossRef]
- del Álamo, M.; Neira, J.L.; Mateu, M.G. Thermodynamic Dissection of a Low Affinity Protein-Protein Interface Involved in Human Immunodeficiency Virus Assembly. J. Biol. Chem. 2003, 278, 27923–27929. [Google Scholar] [CrossRef]
- Byeon, I.-J.L.; Hou, G.; Han, Y.; Suiter, C.L.; Ahn, J.; Jung, J.; Byeon, C.-H.; Gronenborn, A.M.; Polenova, T. Motions on the Millisecond Time Scale and Multiple Conformations of HIV-1 Capsid Protein: Implications for Structural Polymorphism of CA Assemblies. J. Am. Chem. Soc. 2012, 134, 6455–6466. [Google Scholar] [CrossRef]
- Gross, I.; Hohenberg, H.; Kräusslich, H. In Vitro Assembly Properties of Purified Bacterially Expressed Capsid Proteins of Human Immunodeficiency Virus. Eur. J. Biochem. 1997, 249, 592–600. [Google Scholar] [CrossRef] [PubMed]
- Gross, I.; Hohenberg, H.; Wilk, T.; Wiegers, K.; Grättinger, M.; Müller, B.; Fuller, S.; Kräusslich, H.-G. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 2000, 19, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, L.S.; Liu, T.; Scarlata, S.; Chu, B.; Carter, C.A. HIV-1 Capsid Protein Forms Spherical (Immature-Like) and Tubular (Mature-Like) Particles in Vitro: Structure Switching by pH-induced Conformational Changes. Biophys. J. 2001, 81, 586–594. [Google Scholar] [CrossRef] [PubMed]
- Summers, B.J.; Digianantonio, K.M.; Smaga, S.S.; Huang, P.-T.; Zhou, K.; Gerber, E.E.; Wang, W.; Xiong, Y. Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms. Cell Host Microbe 2019, 26, 203–216.e6. [Google Scholar] [CrossRef]
- Dick, R.A.; Zadrozny, K.K.; Xu, C.; Schur, F.K.M.; Lyddon, T.D.; Ricana, C.L.; Wagner, J.M.; Perilla, J.R.; Ganser-Pornillos, B.K.; Johnson, M.C.; et al. Inositol phosphates are assembly co-factors for HIV-1. Nature 2018, 560, 509–512. [Google Scholar] [CrossRef]
- Mallery, D.L.; Márquez, C.L.; A McEwan, W.; Dickson, C.F.; A Jacques, D.; Anandapadamanaban, M.; Bichel, K.; Towers, G.J.; Saiardi, A.; Böcking, T.; et al. IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis. eLife 2018, 7, e35335. [Google Scholar] [CrossRef]
- Renner, N.; Mallery, D.L.; Faysal, K.M.R.; Peng, W.; Jacques, D.A.; Böcking, T.; James, L.C. A lysine ring in HIV capsid pores coordinates IP6 to drive mature capsid assembly. PLoS Pathog. 2021, 17, e1009164. [Google Scholar] [CrossRef]
- Highland, C.M.; Tan, A.; Ricaña, C.L.; Briggs, J.A.G.; Dick, R.A. Structural insights into HIV-1 polyanion-dependent capsid lattice formation revealed by single particle cryo-EM. Proc. Natl. Acad. Sci. USA 2023, 120, e2220545120. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Pak, A.J.; Voth, G.A. Critical mechanistic features of HIV-1 viral capsid assembly. Sci. Adv. 2023, 9, eadd7434. [Google Scholar] [CrossRef]
- Temple, J.; Tripler, T.N.; Shen, Q.; Xiong, Y. A snapshot of HIV-1 capsid–host interactions. Curr. Res. Struct. Biol. 2020, 2, 222–228. [Google Scholar] [CrossRef]
- Toccafondi, E.; Lener, D.; Negroni, M. HIV-1 Capsid Core: A Bullet to the Heart of the Target Cell. Front. Microbiol. 2021, 12, 652486. [Google Scholar] [CrossRef]
- Goujon, C.; Moncorgé, O.; Bauby, H.; Doyle, T.; Ward, C.C.; Schaller, T.; Hué, S.; Barclay, W.S.; Schulz, R.; Malim, M.H. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature 2013, 502, 559–562. [Google Scholar] [CrossRef]
- Kane, M.; Rebensburg, S.V.; A Takata, M.; Zang, T.M.; Yamashita, M.; Kvaratskhelia, M.; Bieniasz, P.D. Nuclear pore heterogeneity influences HIV-1 infection and the antiviral activity of MX2. eLife 2018, 7, e35738. [Google Scholar] [CrossRef] [PubMed]
- Ylinen, L.M.J.; Keckesova, Z.; Wilson, S.J.; Ranasinghe, S.; Towers, G.J. Differential Restriction of Human Immunodeficiency Virus Type 2 and Simian Immunodeficiency Virus SIVmac by TRIM5α Alleles. J. Virol. 2005, 79, 11580–11587. [Google Scholar] [CrossRef] [PubMed]
- Price, A.J.; Marzetta, F.; Lammers, M.; Ylinen, L.M.J.; Schaller, T.; Wilson, S.J.; Towers, G.J.; James, L.C. Active site remodeling switches HIV specificity of antiretroviral TRIMCyp. Nat. Struct. Mol. Biol. 2009, 16, 1036–1042. [Google Scholar] [CrossRef]
- Takeuchi, J.S.; Perche, B.; Migraine, J.; Mercier-Delarue, S.; Ponscarme, D.; Simon, F.; Clavel, F.; Labrosse, B. High level of susceptibility to human TRIM5α conferred by HIV-2 capsid sequences. Retrovirology 2013, 10, 50. [Google Scholar] [CrossRef]
- Boswell, M.T.; Rowland-Jones, S.L. Delayed disease progression in HIV-2: The importance of TRIM5α and the retroviral capsid. Clin. Exp. Immunol. 2019, 196, 305–317. [Google Scholar] [CrossRef]
- Lahaye, X.; Gentili, M.; Silvin, A.; Conrad, C.; Picard, L.; Jouve, M.; Zueva, E.; Maurin, M.; Nadalin, F.; Knott, G.J.; et al. NONO Detects the Nuclear HIV Capsid to Promote cGAS-Mediated Innate Immune Activation. Cell 2018, 175, 488–501.e22. [Google Scholar] [CrossRef]
- Ni, T.; Gerard, S.; Zhao, G.; Dent, K.; Ning, J.; Zhou, J.; Shi, J.; Anderson-Daniels, J.; Li, W.; Jang, S.; et al. Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A. Nat. Struct. Mol. Biol. 2020, 27, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Padron, A.; Prakash, P.; Pandhare, J.; Luban, J.; Aiken, C.; Balasubramaniam, M.; Dash, C. Emerging role of cyclophilin A in HIV-1 infection: From producer cell to the target cell nucleus. J. Virol. 2023, 97, e0073223. [Google Scholar] [CrossRef] [PubMed]
- Franke, E.K.; Yuan, H.E.H.; Luban, J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 1994, 372, 359–362. [Google Scholar] [CrossRef]
- Colgan, J.; E Yuan, H.; Franke, E.K.; Luban, J. Binding of the human immunodeficiency virus type 1 Gag polyprotein to cyclophilin A is mediated by the central region of capsid and requires Gag dimerization. J. Virol. 1996, 70, 4299–4310. [Google Scholar] [CrossRef]
- Schaller, T.; E Ocwieja, K.; Rasaiyaah, J.; Price, A.J.; Brady, T.L.; Roth, S.L.; Hué, S.; Fletcher, A.J.; Lee, K.; KewalRamani, V.N.; et al. HIV-1 Capsid-Cyclophilin Interactions Determine Nuclear Import Pathway, Integration Targeting and Replication Efficiency. PLoS Pathog. 2011, 7, e1002439. [Google Scholar] [CrossRef] [PubMed]
- Balasubramaniam, M.; Davids, B.-O.; Bryer, A.; Xu, C.; Thapa, S.; Shi, J.; Aiken, C.; Pandhare, J.; Perilla, J.R.; Dash, C. HIV-1 mutants that escape the cytotoxic T-lymphocytes are defective in viral DNA integration. PNAS Nexus 2022, 1, pgac064. [Google Scholar] [CrossRef]
- Thali, M.; Bukovsky, A.; Kondo, E.; Rosenwlrth, B.; Walsh, C.T.; Sodroski, J.; Göttlinger, H.G. Functional association of cyclophilin A with HIV-1 virions. Nature 1994, 372, 363–365. [Google Scholar] [CrossRef]
- Braaten, D.; Franke, E.K.; Luban, J. Cyclophilin A is required for the replication of group M human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus SIV(CPZ)GAB but not group O HIV-1 or other primate immunodeficiency viruses. J. Virol. 1996, 70, 4220–4227. [Google Scholar] [CrossRef]
- Billich, A.; Hammerschmid, F.; Peichl, P.; Wenger, R.; Zenke, G.; Quesniaux, V.; Rosenwirth, B. Mode of action of SDZ NIM 811, a nonimmunosuppressive cyclosporin A analog with activity against human immunodeficiency virus (HIV) type 1: Interference with HIV protein-cyclophilin A interactions. J. Virol. 1995, 69, 2451–2461. [Google Scholar] [CrossRef]
- Lahaye, X.; Satoh, T.; Gentili, M.; Cerboni, S.; Silvin, A.; Conrad, C.; Ahmed-Belkacem, A.; Rodriguez, E.C.; Guichou, J.-F.; Bosquet, N.; et al. Nuclear Envelope Protein SUN2 Promotes Cyclophilin-A-Dependent Steps of HIV Replication. Cell Rep. 2016, 15, 879–892. [Google Scholar] [CrossRef]
- Mamede, J.I.; Damond, F.; de Bernardo, A.; Matheron, S.; Descamps, D.; Battini, J.-L.; Sitbon, M.; Courgnaud, V. Cyclophilins and nucleoporins are required for infection mediated by capsids from circulating HIV-2 primary isolates. Sci. Rep. 2017, 7, srep45214. [Google Scholar] [CrossRef] [PubMed]
- Lahaye, X.; Satoh, T.; Gentili, M.; Cerboni, S.; Conrad, C.; Hurbain, I.; El Marjou, A.; Lacabaratz, C.; Lelièvre, J.-D.; Manel, N. The Capsids of HIV-1 and HIV-2 Determine Immune Detection of the Viral cDNA by the Innate Sensor cGAS in Dendritic Cells. Immunity 2013, 39, 1132–1142. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, Y.; Miyake, A.; Doi, N.; Koma, T.; Uchiyama, T.; Adachi, A.; Nomaguchi, M. Comparison of Biochemical Properties of HIV-1 and HIV-2 Capsid Proteins. Front. Microbiol. 2017, 8, 1082. [Google Scholar] [CrossRef]
- Cook, M.; Freniere, C.; Wu, C.; Lozano, F.; Xiong, Y. Structural insights into HIV-2 CA lattice formation and FG-pocket binding revealed by single-particle cryo-EM. Cell Rep. 2025, 44, 115245. [Google Scholar] [CrossRef]
- Talledge, N.; Yang, H.; Shi, K.; Coray, R.; Yu, G.; Arndt, W.G.; Meng, S.; Baxter, G.C.; Mendonça, L.M.; Castaño-Díez, D.; et al. HIV-2 Immature Particle Morphology Provides Insights into Gag Lattice Stability and Virus Maturation. J. Mol. Biol. 2023, 435, 168143. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, M.; Sakai, H.; Adachi, A. Human Immunodeficiency Virus Vpx Is Required for the Early Phase of Replication in Peripheral Blood Mononuclear Cells. Microbiol. Immunol. 1994, 38, 871–878. [Google Scholar] [CrossRef]
- Adachi, A.; E Gendelman, H.; Koenig, S.; Folks, T.; Willey, R.; Rabson, A.; A Martin, M. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 1986, 59, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Mastronarde, D.N. SerialEM: A Program for Automated Tilt Series Acquisition on Tecnai Microscopes Using Prediction of Specimen Position. Microsc. Microanal. 2003, 9, 1182–1183. [Google Scholar] [CrossRef]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2017, 27, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2020, 30, 70–82. [Google Scholar] [CrossRef]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]
- Leirmo, S.; Harrison, C.; Cayley, D.S.; Burgess, R.R.; Record, M.T. Replacement of potassium chloride by potassium glutamate dramatically enhances protein-DNA interactions in vitro. Biochemistry 1987, 26, 2095–2101. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Guinn, E.J.; Buechel, E.; Wong, R.; Sengupta, R.; Shkel, I.A.; Record, M.T. Basis of Protein Stabilization by K Glutamate: Unfavorable Interactions with Carbon, Oxygen Groups. Biophys. J. 2016, 111, 1854–1865. [Google Scholar] [CrossRef] [PubMed]
- Purohit, A.; Douma, L.G.; Bloom, L.B.; Levitus, M. Potassium Glutamate and Glycine Betaine Induce Self-Assembly of the PCNA and β-Sliding Clamps. Biophys. J. 2021, 120, 73–85. [Google Scholar] [CrossRef]
- Lanman, J.; Sexton, J.; Sakalian, M.; Prevelige, P.E. Kinetic Analysis of the Role of Intersubunit Interactions in Human Immunodeficiency Virus Type 1 Capsid Protein Assembly In Vitro. J. Virol. 2002, 76, 6900–6908. [Google Scholar] [CrossRef]
- Chen, B.; Tycko, R. Simulated Self-Assembly of the HIV-1 Capsid: Protein Shape and Native Contacts Are Sufficient for Two-Dimensional Lattice Formation. Biophys. J. 2011, 100, 3035–3044. [Google Scholar] [CrossRef]
- Valbuena, A.; Maity, S.; Mateu, M.G.; Roos, W.H. Visualization of Single Molecules Building a Viral Capsid Protein Lattice through Stochastic Pathways. ACS Nano 2020, 14, 8724–8734. [Google Scholar] [CrossRef]
- Pornillos, O.; Ganser-Pornillos, B.K.; Banumathi, S.; Hua, Y.; Yeager, M. Disulfide Bond Stabilization of the Hexameric Capsomer of Human Immunodeficiency Virus. J. Mol. Biol. 2010, 401, 985–995. [Google Scholar] [CrossRef]
- Troyano-Hernáez, P.; Reinosa, R.; Holguín, Á. HIV Capsid Protein Genetic Diversity Across HIV-1 Variants and Impact on New Capsid-Inhibitor Lenacapavir. Front. Microbiol. 2022, 13, 854974. [Google Scholar] [CrossRef]
- del Álamo, M.; Mateu, M.G. Electrostatic repulsion, compensatory mutations, and long-range non-additive effects at the dimerization interface of the HIV capsid protein. J. Mol. Biol. 2005, 345, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Aiken, C.; Rousso, I. The HIV-1 capsid and reverse transcription. Retrovirology 2021, 18, 29. [Google Scholar] [CrossRef] [PubMed]
- Zuliani-Alvarez, L.; Govasli, M.L.; Rasaiyaah, J.; Monit, C.; Perry, S.O.; Sumner, R.P.; McAlpine-Scott, S.; Dickson, C.; Faysal, K.M.R.; Hilditch, L.; et al. Evasion of cGAS and TRIM5 defines pandemic HIV. Nat. Microbiol. 2022, 7, 1762–1776. [Google Scholar] [CrossRef]
- Tsiang, M.; Niedziela-Majka, A.; Hung, M.; Jin, D.; Hu, E.; Yant, S.; Samuel, D.; Liu, X.; Sakowicz, R. A Trimer of Dimers Is the Basic Building Block for Human Immunodeficiency Virus-1 Capsid Assembly. Biochemistry 2012, 51, 4416–4428. [Google Scholar] [CrossRef]
- Grime, J.M.A.; Dama, J.F.; Ganser-Pornillos, B.K.; Woodward, C.L.; Jensen, G.J.; Yeager, M.; Voth, G.A. Coarse-grained simulation reveals key features of HIV-1 capsid self-assembly. Nat. Commun. 2016, 7, 11568. [Google Scholar] [CrossRef]
- Escrig, J.; Marcos-Alcalde, Í.; Domínguez-Zotes, S.; Abia, D.; Gómez-Puertas, P.; Valbuena, A.; Mateu, M.G. Structural Basis for Alternative Self-Assembly Pathways Leading to Different Human Immunodeficiency Virus Capsid-Like Nanoparticles. ACS Nano 2024, 18, 27465–27478. [Google Scholar] [CrossRef]
- Rihn, S.J.; Wilson, S.J.; Loman, N.J.; Alim, M.; Bakker, S.E.; Bhella, D.; Gifford, R.J.; Rixon, F.J.; Bieniasz, P.D. Extreme genetic fragility of the HIV-1 capsid. PLoS Pathog 2013, 9, e1003461. [Google Scholar] [CrossRef] [PubMed]
- de Silva, T.I.; Leligdowicz, A.; Carlson, J.; Garcia-Knight, M.; Onyango, C.; Miller, N.; Yindom, L.-M.; Hué, S.; Jaye, A.; Dong, T.; et al. HLA-associated polymorphisms in the HIV-2 capsid highlight key differences between HIV-1 and HIV-2 immune adaptation. AIDS 2018, 32, 709–714. [Google Scholar] [CrossRef]
- I Mamede, J.; Sitbon, M.; Battini, J.-L.; Courgnaud, V. Heterogeneous susceptibility of circulating SIV isolate capsids to HIV-interacting factors. Retrovirology 2013, 10, 77. [Google Scholar] [CrossRef]
- Meyerson, N.R.; Warren, C.J.; Vieira, D.A.S.A.; Diaz-Griferro, F.; Sawyer, S.L. Species-specific vulnerability of RanBP2 shaped the evolution of SIV as it transmitted in African apes. PLoS Pathog 2018, 14, e1006906. [Google Scholar] [CrossRef] [PubMed]
- Yufenyuy, E.L.; Aiken, C. The NTD-CTD intersubunit interface plays a critical role in assembly and stabilization of the HIV-1 capsid. Retrovirology 2013, 10, 29. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Zotes, S.; Fuertes, M.A.; Rodríguez-Huete, A.; Valbuena, A.; Mateu, M.G. A Genetically Engineered, Chain Mail-Like Nanostructured Protein Material with Increased Fatigue Resistance and Enhanced Self-Healing. Small 2022, 18, 2105456. [Google Scholar] [CrossRef] [PubMed]
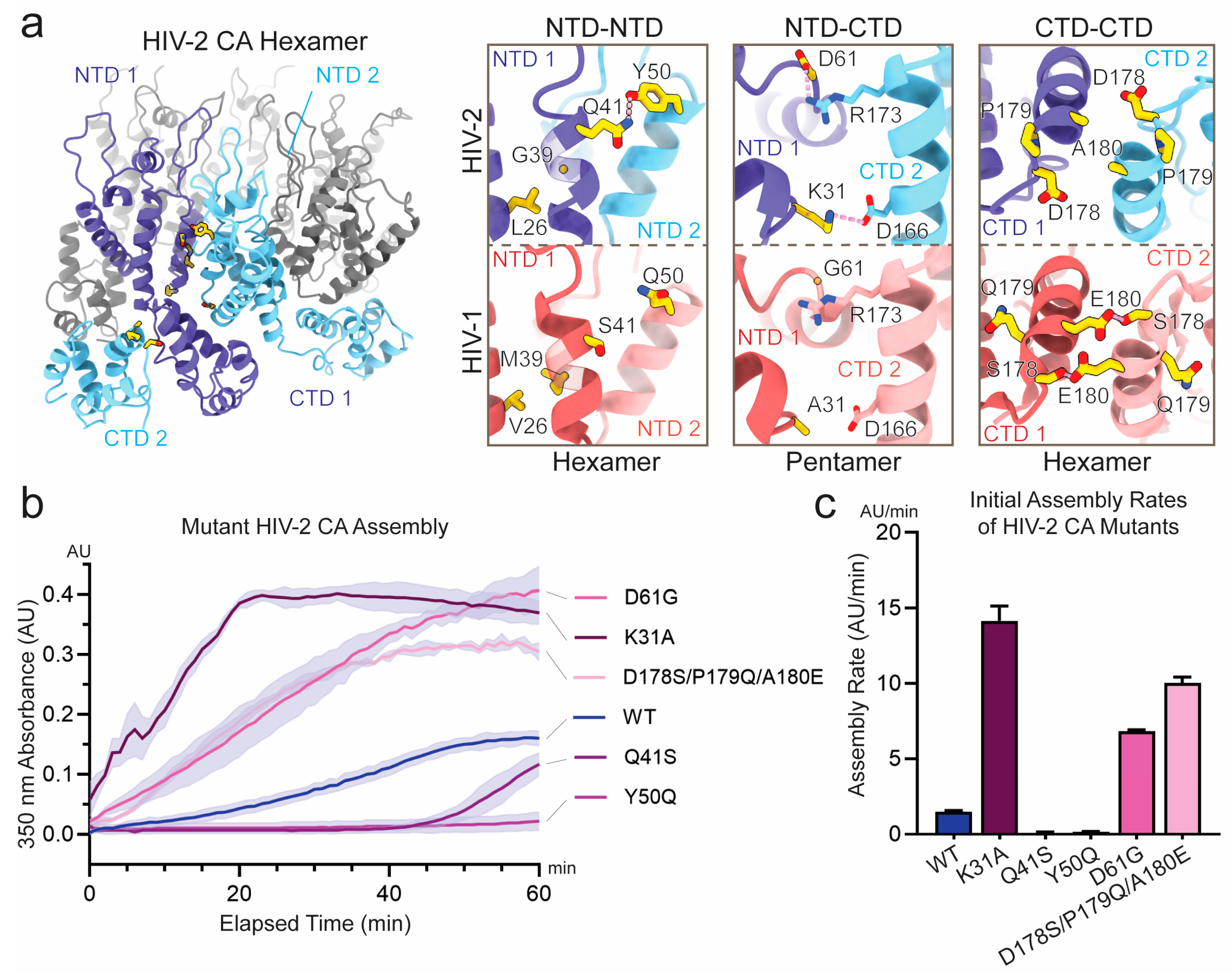

| Position Number | Residues with >10% Frequency | CA Inter-Protomer Interface Engaged | |
|---|---|---|---|
| HIV-2 | HIV-1 | ||
| 26 | L | V | NTD-NTD |
| 31 | K | A/G | NTD-CTD, pentamer only |
| 39 | G | M | NTD-NTD |
| 41 | Q | S/T | NTD-NTD |
| 50 | Y | Q | NTD-NTD, hexamer only |
| 61 | D | G | NTD-CTD |
| 178 | D | T/S | CTD-CTD |
| 179 | P | Q | N/A |
| 180 | A | E/D | CTD-CTD |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cook, M.; Bhardwaj, P.; Lozano, F.; Freniere, C.; Malonis, R.J.; Xiong, Y. Dissecting the Unique Self-Assembly Landscape of the HIV-2 Capsid Protein. Viruses 2025, 17, 1384. https://doi.org/10.3390/v17101384
Cook M, Bhardwaj P, Lozano F, Freniere C, Malonis RJ, Xiong Y. Dissecting the Unique Self-Assembly Landscape of the HIV-2 Capsid Protein. Viruses. 2025; 17(10):1384. https://doi.org/10.3390/v17101384
Chicago/Turabian StyleCook, Matthew, Pushpanjali Bhardwaj, Faith Lozano, Christian Freniere, Ryan J. Malonis, and Yong Xiong. 2025. "Dissecting the Unique Self-Assembly Landscape of the HIV-2 Capsid Protein" Viruses 17, no. 10: 1384. https://doi.org/10.3390/v17101384
APA StyleCook, M., Bhardwaj, P., Lozano, F., Freniere, C., Malonis, R. J., & Xiong, Y. (2025). Dissecting the Unique Self-Assembly Landscape of the HIV-2 Capsid Protein. Viruses, 17(10), 1384. https://doi.org/10.3390/v17101384






