Genomic Characterization of a Novel Yezo Virus Revealed in Ixodes pavlovskyi Tick Virome in Western Siberia
Abstract
1. Introduction
2. Materials and Methods
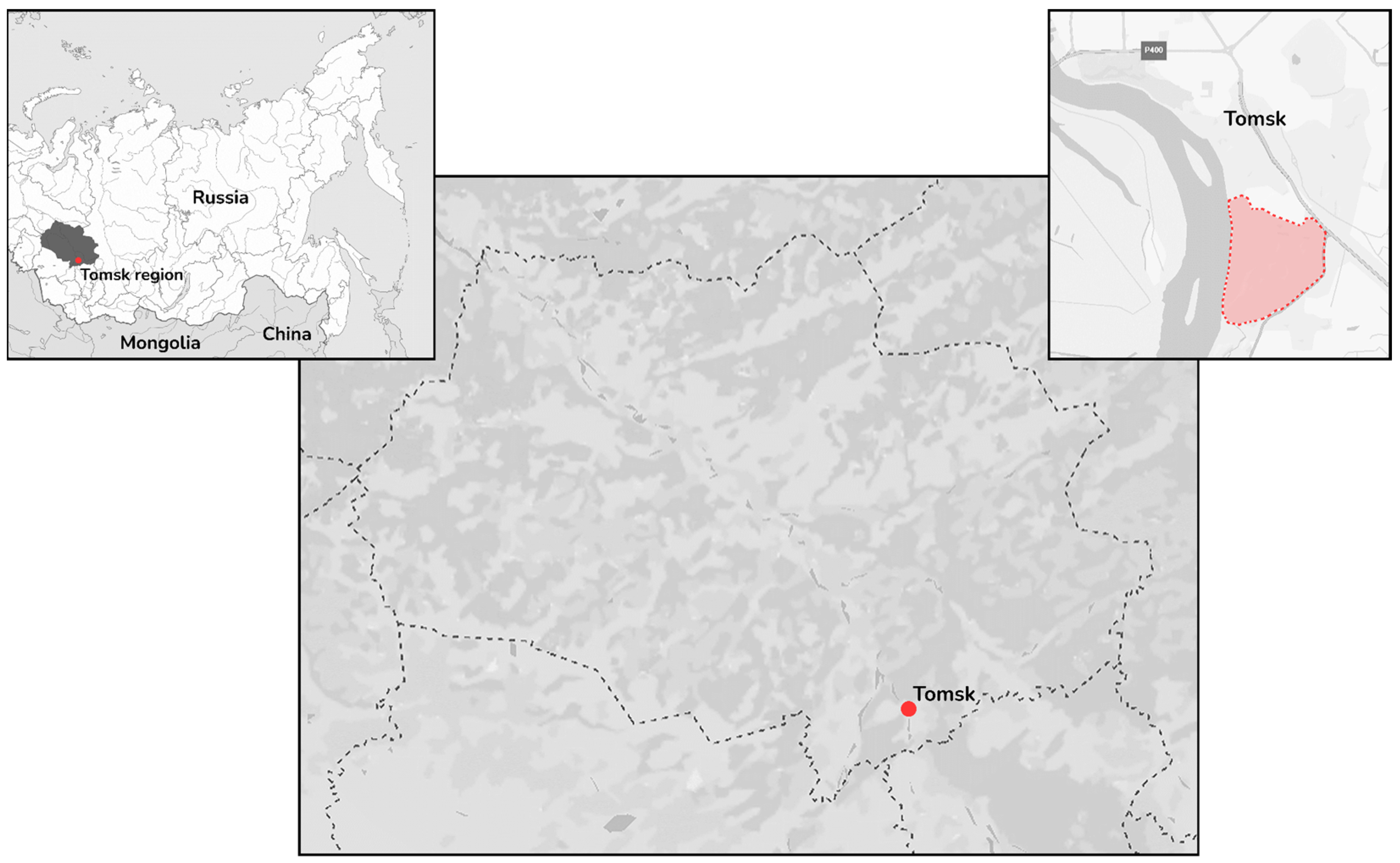
2.1. Sampling
2.2. Enrichment of Virus-like Particles, Nucleic Acid Extraction, Library Preparation and Metagenomic Sequencing
2.3. Virome Data Analysis
2.4. Sequence Analysis
2.5. Phylogenetic Analysis
3. Results
3.1. Characterization of the Metagenomic Library
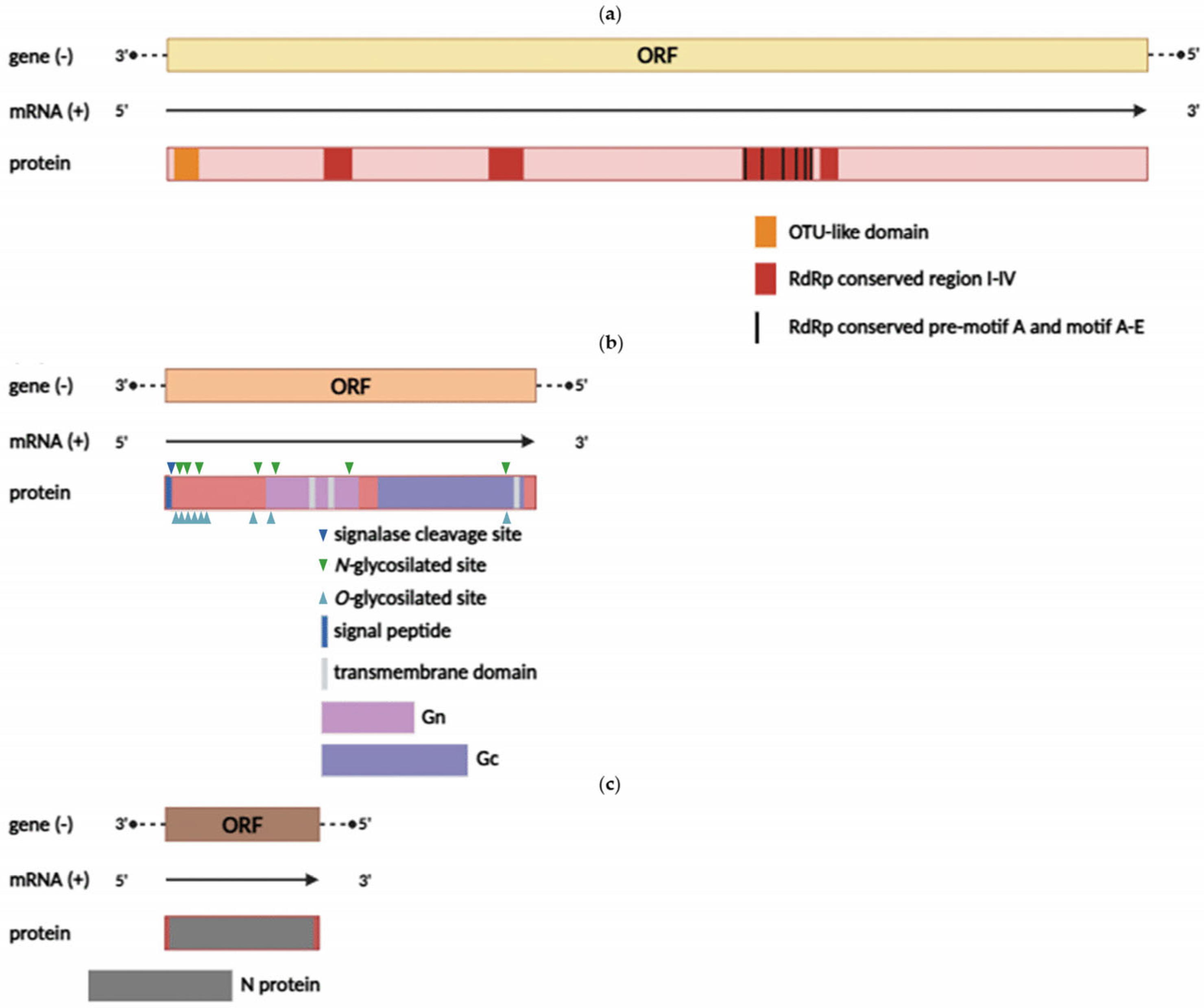
3.2. Genomic Characterization of the Detected Yezo Virus
3.3. Nucleotide and Amino Acid Identity
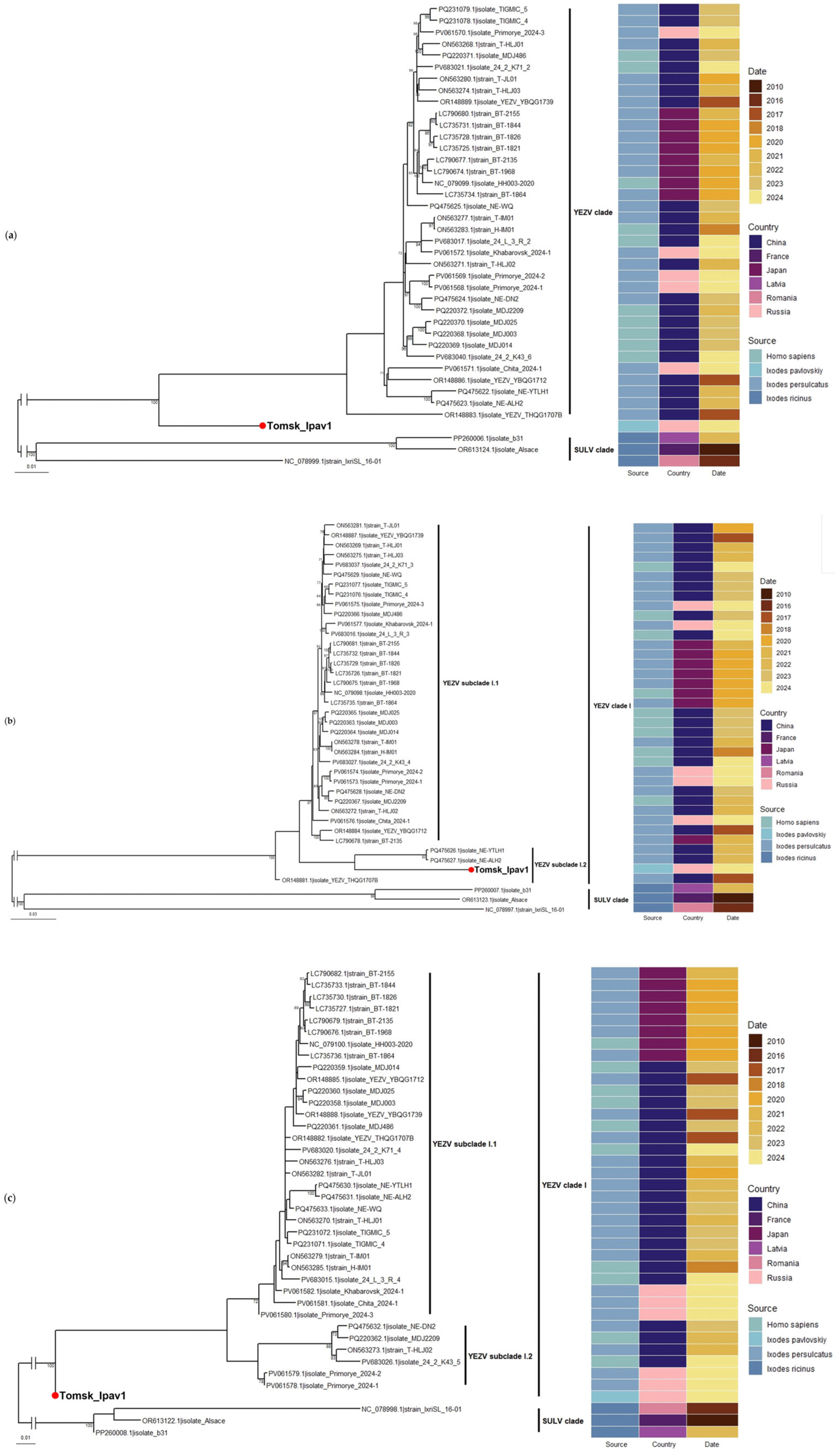
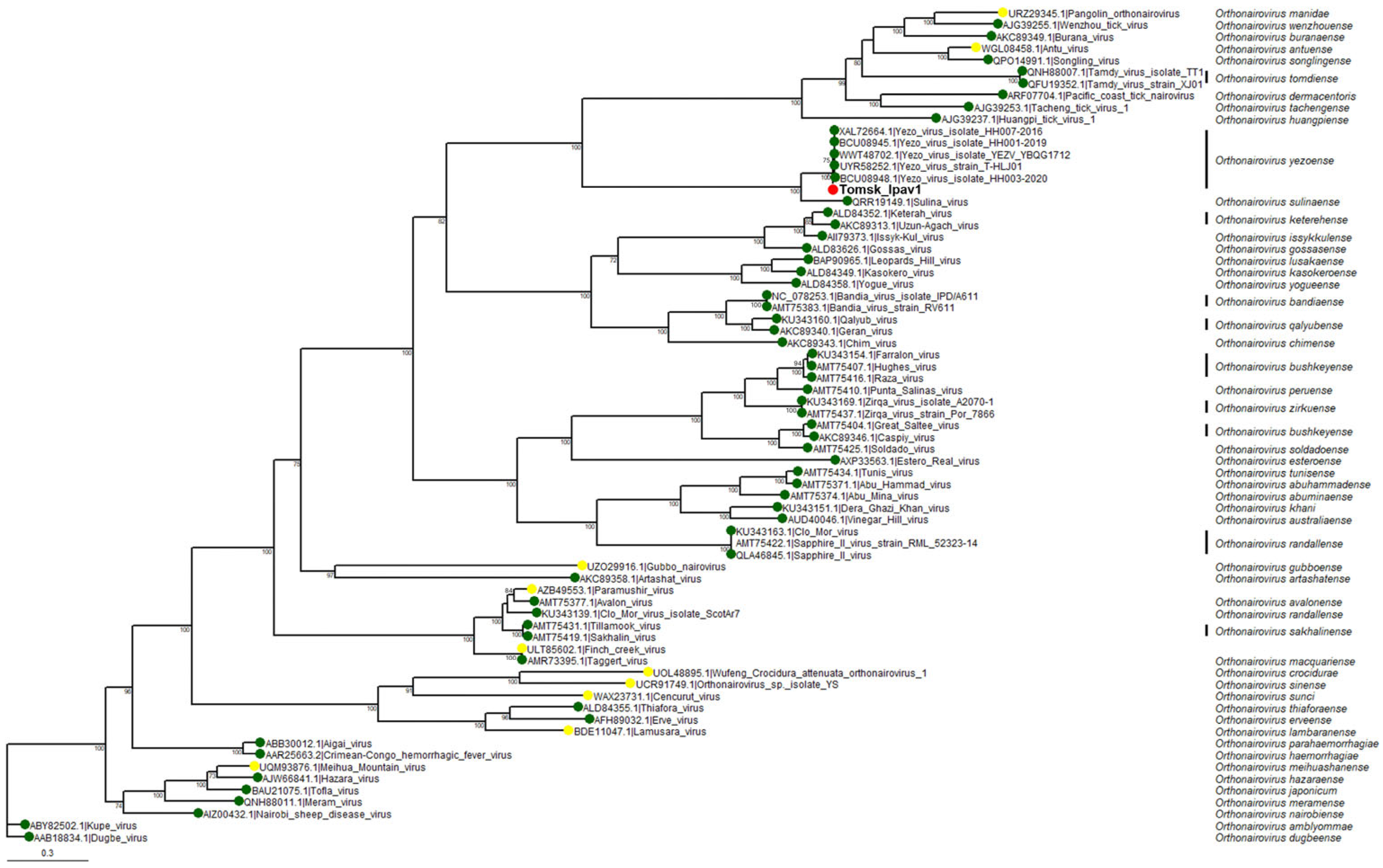
3.4. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| YEZV | Yezo virus |
| TBEV | Tick-borne encephalitis virus |
| ssRNA (–) | Single-stranded negative-sense RNA |
| SULV | Sulina virus |
| RdRp | RNA-dependent RNA polymerase |
| GPC | Glycoprotein precursor complex |
| CCHFV | Crimean-Congo hemorrhagic fever virus |
| NSD | Nairobi sheep disease |
References
- Latif, A.A.; Putterill, J.F.; De Klerk, D.G.; Pienaar, R.; Mans, B.J. Nuttalliella Namaqua (Ixodoidea: Nuttalliellidae): First description of the male, immature stages and re-description of the female. PLoS ONE 2012, 7, e41651. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Maqbool, M.; Sajid, M.S.; Saqib, M.; Anjum, F.R.; Tayyab, M.H.; Rizwan, H.M.; Rashid, M.I.; Rashid, I.; Iqbal, A.; Siddique, R.M.; et al. Potential mechanisms of transmission of tick-borne viruses at the virus-tick interface. Front. Microbiol. 2022, 13, 846884. [Google Scholar] [CrossRef]
- Moskvitina, N.S.; Romanenko, V.N.; Ternovoi, V.A.; Ivanova, N.V.; Protopopova, E.V.; Kravchenko, L.B.; Kononova, Y.V.; Kuranova, V.N.; Chausov, E.A.; Moskvitin, S.S.; et al. Detection of the West Nile virus and its genetic typing in ixodid ticks (Parasitiformes: Ixodidae) in Tomsk city and its suburbs. Parazitologiya 2008, 42, 210–225. [Google Scholar]
- Kuranova, V.N.; Yartsev, V.V.; Kononova, Y.V.; Protopopova, E.V.; Konovalova, S.N.; Ternovoi, V.A.; Tavkina, I.S.; Romanenko, V.N.; Loktev, V.B.; Moskvitina, N.S. Lacertids (Sauria, Lacertidae) in natural foci of infectious human-transmitted ecosystems of the south-east territories of Western Siberia. In Proceedings of the 4th Meeting of the Nikolsky Herpetological Society, Kazan, Russia, 12–17 October 2009; Russian Collection: Saint Petersburg, Russia, 2009; pp. 129–135. [Google Scholar]
- Vilibić-Čavlek, T.; Bogdanić, M.; Savić, V.; Barbić, L.; Stevanović, V.; Kaić, B. Tick-Borne Human Diseases Around the Globe, 7th ed.; Global Health Press: Singapore, 2024. [Google Scholar]
- Romanenko, V.N. The Peculiarities of the biology of ticks inhabiting the environs of Tomsk city. Parazitologiya 2005, 39, 365–370. [Google Scholar]
- Korobitsyn, I.G.; Moskvitina, N.S.; Tyutenkov, O.Y.; Gashkov, S.I.; Kononova, Y.V.; Moskvitin, S.S.; Romanenko, V.N.; Mikryukova, T.P.; Protopopova, E.V.; Kartashov, M.Y.; et al. Detection of tick-borne pathogens in wild birds and their ticks in Western Siberia and high level of their mismatch. Folia Parasitol. 2021, 68, 24. [Google Scholar] [CrossRef]
- Voronkova, O.V.; Romanenko, V.N.; Simakova, A.V.; Esimova, I.E.; D’yakov, D.A.; Motlokhova, E.A.; Chernyshov, N.A.; Yamaletdinova, D.M. Analysis of multiple infection in ixodid ticks Dermacentor reticulatus in a combined natural focus of vector-borne infections in the Tomsk region. Probl. Osob. Opasnykh Infektsii 2023, 2, 106–111. [Google Scholar] [CrossRef]
- Tupota, N.L.; Ternovoi, V.A.; Ponomareva, E.P.; Bayandin, R.B.; Shvalov, A.N.; Malyshev, B.S.; Tregubchak, T.V.; Bauer, T.V.; Protopopova, E.V.; Petrova, N.K.; et al. Detection of the genetic material of the viruses Tacheng uukuvirus and Sara tick phlebovirus in taiga ticks collected in the Sverdlovsk, Tomsk regions and Primorsky territory of Russia and their phylogeny. Probl. Osob. Opasnykh Infektsii 2023, 3, 141–146. [Google Scholar] [CrossRef]
- Kartashov, M.; Svirin, K.; Zheleznova, A.; Yanshin, A.; Radchenko, N.; Kurushina, V.; Tregubchak, T.; Maksimenko, L.; Sivay, M.; Ternovoi, V.; et al. A first report of the Yezo virus isolates detection in Russia. Viruses 2025, 17, 1125. [Google Scholar] [CrossRef] [PubMed]
- Romanenko, V.N. Long-term dynamics of density and diversity of ticks (Ixodidae) on the natural and disturbed territories. Parazitologiya 2011, 45, 384–391. [Google Scholar]
- Moskvitina, N.S.; Korobitsyn, I.G.; Tyuten’kov, O.Y.; Gashkov, S.I.; Kononova, Y.V.; Moskvitin, S.S.; Romanenko, V.N.; Mikryukova, T.P.; Protopopova, E.V.; Kartashov, M.Y.; et al. The role of birds in the maintenance of tick-borne infections in the Tomsk anthropurgic foci. Biol. Bull. Russ. Acad. Sci. 2014, 41, 387–393. [Google Scholar] [CrossRef]
- Kodama, F.; Yamaguchi, H.; Park, E.; Tatemoto, K.; Sashika, M.; Nakao, R.; Terauchi, Y.; Mizuma, K.; Orba, Y.; Kariwa, H.; et al. A novel nairovirus associated with acute febrile illness in Hokkaido, Japan. Nat. Commun. 2021, 12, 5539. [Google Scholar] [CrossRef]
- Lv, X.; Liu, Z.; Li, L.; Xu, W.; Yuan, Y.; Liang, X.; Zhang, L.; Wei, Z.; Sui, L.; Zhao, Y.; et al. Yezo virus infection in tick-bitten patient and ticks, Northeastern China. Emerg. Infect. Dis. 2023, 29, 797–800. [Google Scholar] [CrossRef]
- Ni, X.-B.; Cui, X.-M.; Liu, J.-Y.; Ye, R.-Z.; Wu, Y.-Q.; Jiang, J.-F.; Sun, Y.; Wang, Q.; Shum, M.H.-H.; Chang, Q.-C.; et al. Metavirome of 31 tick species provides a compendium of 1,801 RNA virus genomes. Nat. Microbiol. 2023, 8, 162–173. [Google Scholar] [CrossRef]
- Kumar, P.; Priyanshu, P.; Sharma, R.K.; Sharma, D.; Arora, M.; Gaidhane, A.M.; Zahiruddin, Q.S.; Rustagi, S.; Mawejje, E.; Satapathy, P. Climate change and its role in the emergence of new tick-borne Yezo virus. New Microbes New Infect. 2024, 60–61, 101423. [Google Scholar] [CrossRef]
- Nishino, A.; Tatemoto, K.; Ishijima, K.; Inoue, Y.; Park, E.; Yamamoto, T.; Taira, M.; Kuroda, Y.; Virhuez-Mendoza, M.; Harada, M.; et al. Transboundary movement of Yezo virus via ticks on migratory birds, Japan, 2020–2021. Emerg. Infect. Dis. 2024, 30, 2674–2678. [Google Scholar] [CrossRef]
- Ogata, Y.; Sato, T.; Kato, K.; Kikuchi, K.; Mitsuhashi, K.; Watari, K.; Tamiya, K.; Goto, A.; Yamaguchi, H.; Hisada, R. A Case of tick-borne Yezo virus infection: Concurrent detection in the patient and tick. Int. J. Infect. Dis. 2024, 143, 107038. [Google Scholar] [CrossRef]
- Suzuki, K.; Suzuki, S.; Yamaguchi, H.; Kakinoki, Y. Tick-borne disease with Yezo virus and Borrelia miyamotoi coinfection. Intern. Med. 2024, 63, 2861–2864. [Google Scholar] [CrossRef] [PubMed]
- Matsuno, K. Endemic area of emerging tick-borne Yezo virus infections discovered in China. Lancet Infect. Dis. 2025, 25, 359–360. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, J.H.; Alkhovsky, S.V.; Avšič-Županc, T.; Bergeron, É.; Burt, F.; Ergünay, K.; Garrison, A.R.; Marklewitz, M.; Mirazimi, A.; Papa, A.; et al. ICTV virus taxonomy profile: Nairoviridae 2024: This Article Is Part of the ICTV Virus Taxonomy Profiles Collection. J. Gen. Virol. 2024, 105, 001974. [Google Scholar] [CrossRef]
- Kuhn, J.; Wiley, M.; Rodriguez, S.; Bào, Y.; Prieto, K.; Travassos Da Rosa, A.; Guzman, H.; Savji, N.; Ladner, J.; Tesh, R.; et al. Genomic characterization of the genus Nairovirus (family Bunyaviridae). Viruses 2016, 8, 164. [Google Scholar] [CrossRef] [PubMed]
- Spengler, J.R.; Estrada-Peña, A.; Garrison, A.R.; Schmaljohn, C.; Spiropoulou, C.F.; Bergeron, É.; Bente, D.A. A Chronological review of experimental infection studies of the role of wild animals and livestock in the maintenance and transmission of Crimean-Congo hemorrhagic fever virus. Antivir. Res. 2016, 135, 31–47. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, W.; Zhang, X. Application of metagenomic next-generation sequencing in the diagnosis of infectious diseases. Front. Cell. Infect. Microbiol. 2024, 14, 1458316. [Google Scholar] [CrossRef]
- Vandegrift, K.J.; Kapoor, A. The ecology of new constituents of the tick virome and their relevance to public health. Viruses 2019, 11, 529. [Google Scholar] [CrossRef]
- Filippova, N.A. Ixodid Ticks of the Subfamily Ixodinae, 4th ed.; Strelkov, A.A., Ed.; Nauka: Leningrad, Russia, 1977; 396p. (In Russian) [Google Scholar]
- Conceição-Neto, N.; Yinda, K.C.; Van Ranst, M.; Matthijnssens, J. NetoVIR: Modular Approach to Customize Sample Preparation Procedures for Viral Metagenomics; Moya, A., Pérez Brocal, V., Eds.; Springer: New York, NY, USA, 2018; Volume 1838, pp. 85–95. ISBN 978-1-4939-8682-8. [Google Scholar]
- De Coninck, L.; Faye, L.; Basler, N.; Jansen, D.; Van Espen, L. ViPER, Version 2.3.1; Zenodo: Geneve, Switzerland, 2025. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive metagenomic visualization in a web browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef]
- Vasimuddin, M.D.; Misra, S.; Li, H.; Aluru, S. Efficient architecture-aware acceleration of BWA-MEM for multicore systems. In Proceedings of the 2019 IEEE International Parallel and Distributed Processing Symposium (IPDPS), Rio de Janeiro, Brazil, 20–24 May 2019; IEEE: Rio de Janeiro, Brazil, 2019; pp. 314–324. [Google Scholar] [CrossRef]
- Gasteiger, E. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Almagro Armenteros, J.J.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. Bioinformatics 2022. [Google Scholar] [CrossRef]
- Duckert, P.; Brunak, S.; Blom, N. Prediction of proprotein convertase cleavage sites. Protein Eng. Des. Sel. 2004, 17, 107–112. [Google Scholar] [CrossRef]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 6.0 Predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Gupta, R.; Brunak, S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput. 2002, 7, 310–322. [Google Scholar]
- Steentoft, C.; Vakhrushev, S.Y.; Joshi, H.J.; Kong, Y.; Vester-Christensen, M.B.; Schjoldager, K.T.-B.G.; Lavrsen, K.; Dabelsteen, S.; Pedersen, N.B.; Marcos-Silva, L.; et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013, 32, 1478–1488. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Wong, T.; Ly-Trong, N.; Ren, H.; Baños, H.; Roger, A.; Susko, E.; Bielow, C.; De Maio, N.; Goldman, N.; Hahn, M.; et al. IQ-TREE 3: Phylogenomic inference software using complex evolutionary models. Life Sci. 2025. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T. Ggtree: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Revell, L.J. Phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Waskom, M. Seaborn: Statistical data visualization. JOSS 2021, 6, 3021. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Honig, J.E.; Osborne, J.C.; Nichol, S.T. Crimean–Congo hemorrhagic fever virus genome L RNA segment and encoded protein. Virology 2004, 321, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, H.-Y.; Shao, J.-W.; Pei, M.-C.; Cao, C.; Huang, F.-Q.; Sun, M.-F. Genomic characterization and phylogenetic analysis of a novel Nairobi sheep disease genogroup Orthonairovirus from ticks, Southeastern China. Front. Microbiol. 2022, 13, 977405. [Google Scholar] [CrossRef]
- Estrada, D.F.; De Guzman, R.N. Structural characterization of the Crimean-Congo hemorrhagic fever virus Gn tail provides insight into virus assembly. J. Biol. Chem. 2011, 286, 21678–21686. [Google Scholar] [CrossRef]
- Li, N.; Rao, G.; Li, Z.; Yin, J.; Chong, T.; Tian, K.; Fu, Y.; Cao, S. Cryo-EM structure of glycoprotein C from Crimean-Congo hemorrhagic fever virus. Virol. Sin. 2022, 37, 127–137. [Google Scholar] [CrossRef]
- Wang, W.; Liu, X.; Wang, X.; Dong, H.; Ma, C.; Wang, J.; Liu, B.; Mao, Y.; Wang, Y.; Li, T.; et al. Structural and functional diversity of Nairovirus-encoded nucleoproteins. J. Virol. 2015, 89, 11740–11749. [Google Scholar] [CrossRef]
- Chen, Z.-Y.; Zhang, J.; He, P.-J.; Xiong, T.; Zhu, D.-Y.; Zhu, W.-J.; Ni, X.-B.; Du, L.-F.; Wang, Q.; Zhang, Y.-W.; et al. Characteristics of viral ovarian tumor domain protease from two emerging orthonairoviruses and identification of Yezo virus human infections in northeastern China as early as 2012. J. Virol. 2025, 99, e01727-24. [Google Scholar] [CrossRef]
- Ndiaye, M.; Badji, A.; Dieng, I.; Dolgova, A.S.; Mhamadi, M.; Kirichenko, A.D.; Gladkikh, A.S.; Gaye, A.; Faye, O.; Sall, A.A.; et al. Molecular detection and genetic characterization of two Dugbe orthonairovirus isolates detected from ticks in southern Senegal. Viruses 2024, 16, 964. [Google Scholar] [CrossRef]
- Deyde, V.M.; Khristova, M.L.; Rollin, P.E.; Ksiazek, T.G.; Nichol, S.T. Crimean-Congo hemorrhagic fever virus genomics and global diversity. J. Virol. 2006, 80, 8834–8842. [Google Scholar] [CrossRef] [PubMed]
- Al’khovskiĭ, S.V.; L’vov, D.K.; Shchelkanov, M.I.; Deriabin, P.G.; Shchetinin, A.M.; Samokhvalov, E.I.; Aristova, V.A.; Gitel’man, A.K.; Botikov, A.G. Genetic characterization of the Uzun-Agach virus (UZAV, Bunyaviridae, Nairovirus), isolated from bat Myotis blythii oxygnathus Monticelli, 1885 (Chiroptera; Vespertilionidae) in Kazakhstan. Vopr. Virusol. 2014, 59, 23–26. [Google Scholar]
- Lukashev, A.N.; Klimentov, A.S.; Smirnova, S.E.; Dzagurova, T.K.; Drexler, J.F.; Gmyl, A.P. Phylogeography of Crimean Congo hemorrhagic fever virus. PLoS ONE 2016, 11, e0166744. [Google Scholar] [CrossRef] [PubMed]
- Safonova, M.V.; Shchelkanov, M.Y.; Khafizov, K.; Matsvay, A.D.; Ayginin, A.A.; Dolgova, A.S.; Shchelkanov, E.M.; Pimkina, E.V.; Speranskaya, A.S.; Galkina, I.V.; et al. Sequencing and genetic characterization of two strains Paramushir virus obtained from the Tyuleniy Island in the Okhotsk Sea (2015). Ticks Tick. Borne Dis. 2019, 10, 269–279. [Google Scholar] [CrossRef] [PubMed]
| Taxon (Closest BLASTx Match) | Count of Contigs | Relative Abundance (%) | Coverage of Contigs | Length of Contigs (nt) |
|---|---|---|---|---|
| YEZV (Nairoviridae) (94.91–98.76%) | 4 | 66 | 1, 373, 575 and 5356 | 252, 1748, 4296 and 12,103 |
| Fangzheng tomsu-like virus (Tombusviridae) (94.24–96.11%) | 3 | 28 | 920, 3132 and 6339 | 3211, 1094 and 1479 |
| Great Island virus (Sedoreoviridae) (28.47% and 32.51%) | 2 | 3 | 615 and 904 | 3031 and 2165 |
| Haerbin Reovi tick virus 1 (Unclassified Reiovirales) (99.46%) | 1 | 2 | 690 | 4149 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Apanasevich, M.; Dubovitskiy, N.; Derko, A.; Khozyainova, A.; Tarasov, A.; Kokhanenko, A.; Artemov, G.; Denisov, E.; Shestopalov, A.; Sharshov, K. Genomic Characterization of a Novel Yezo Virus Revealed in Ixodes pavlovskyi Tick Virome in Western Siberia. Viruses 2025, 17, 1362. https://doi.org/10.3390/v17101362
Apanasevich M, Dubovitskiy N, Derko A, Khozyainova A, Tarasov A, Kokhanenko A, Artemov G, Denisov E, Shestopalov A, Sharshov K. Genomic Characterization of a Novel Yezo Virus Revealed in Ixodes pavlovskyi Tick Virome in Western Siberia. Viruses. 2025; 17(10):1362. https://doi.org/10.3390/v17101362
Chicago/Turabian StyleApanasevich, Maxim, Nikita Dubovitskiy, Anastasiya Derko, Anna Khozyainova, Alexander Tarasov, Alina Kokhanenko, Gleb Artemov, Evgeny Denisov, Alexander Shestopalov, and Kirill Sharshov. 2025. "Genomic Characterization of a Novel Yezo Virus Revealed in Ixodes pavlovskyi Tick Virome in Western Siberia" Viruses 17, no. 10: 1362. https://doi.org/10.3390/v17101362
APA StyleApanasevich, M., Dubovitskiy, N., Derko, A., Khozyainova, A., Tarasov, A., Kokhanenko, A., Artemov, G., Denisov, E., Shestopalov, A., & Sharshov, K. (2025). Genomic Characterization of a Novel Yezo Virus Revealed in Ixodes pavlovskyi Tick Virome in Western Siberia. Viruses, 17(10), 1362. https://doi.org/10.3390/v17101362







