Strain-Specific Variability in Viral Kinetics, Cytokine Response, and Cellular Damage in Air–Liquid Cultures of Human Nasal Organoids After Infection with SARS-CoV-2
Abstract
1. Introduction
2. Methods
2.1. HNO-ALI Cell Lines
2.2. Virus Stocks
2.3. Study Design
2.4. Sample Collection
2.5. Viral Infection
2.6. Quantitative PCR Assay
2.7. Cytokine and Chemokine Assay
2.8. Immunohistochemistry (IHC) and Immunofluorescence Staining
2.9. Immunofluorescence Image Quantification and Analysis
2.10. Sex as a Biological Variable
2.11. Statistical Analysis
3. Results
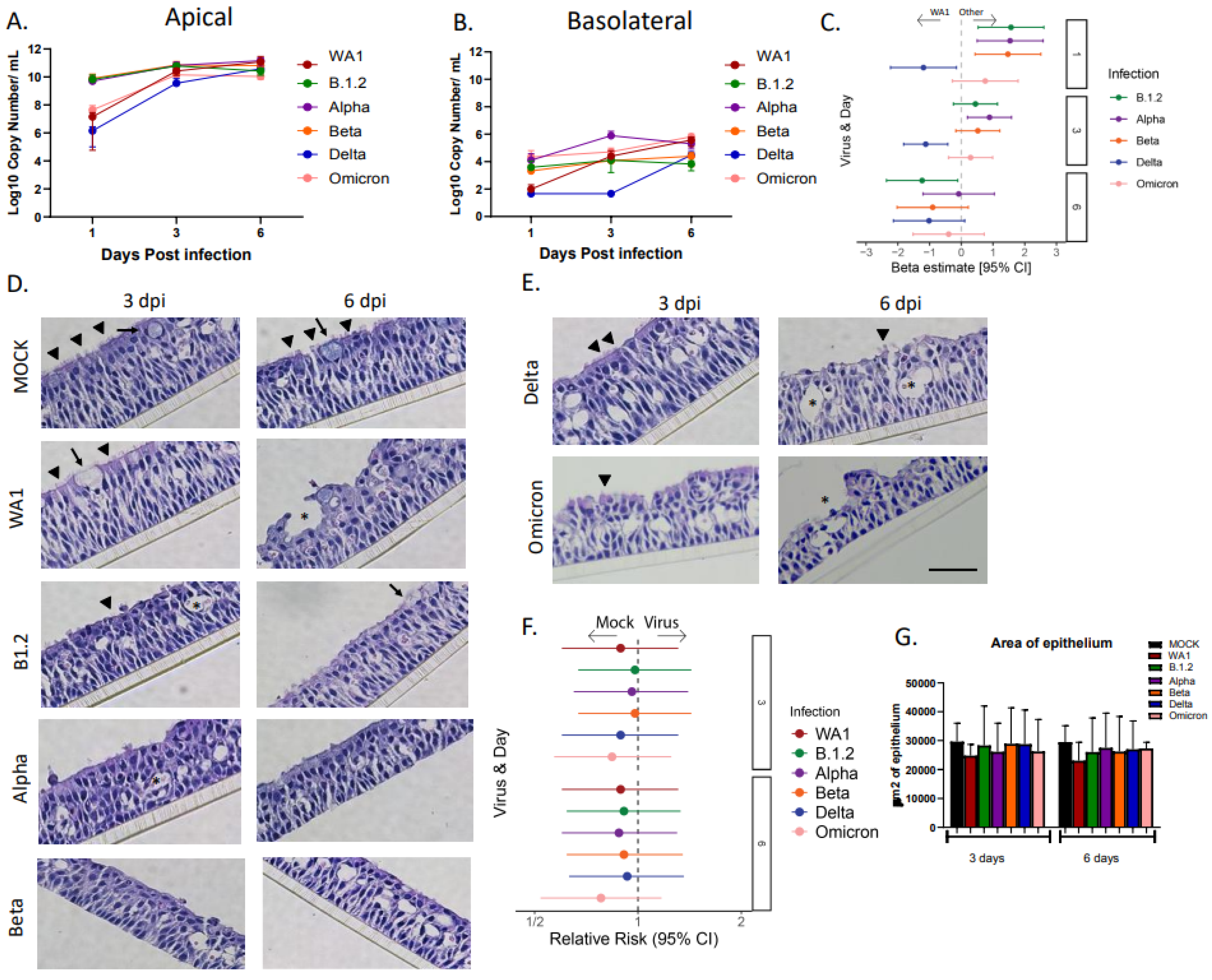
3.1. Delta Reaches a Steady State Later than Other SARS-CoV-2 Variants
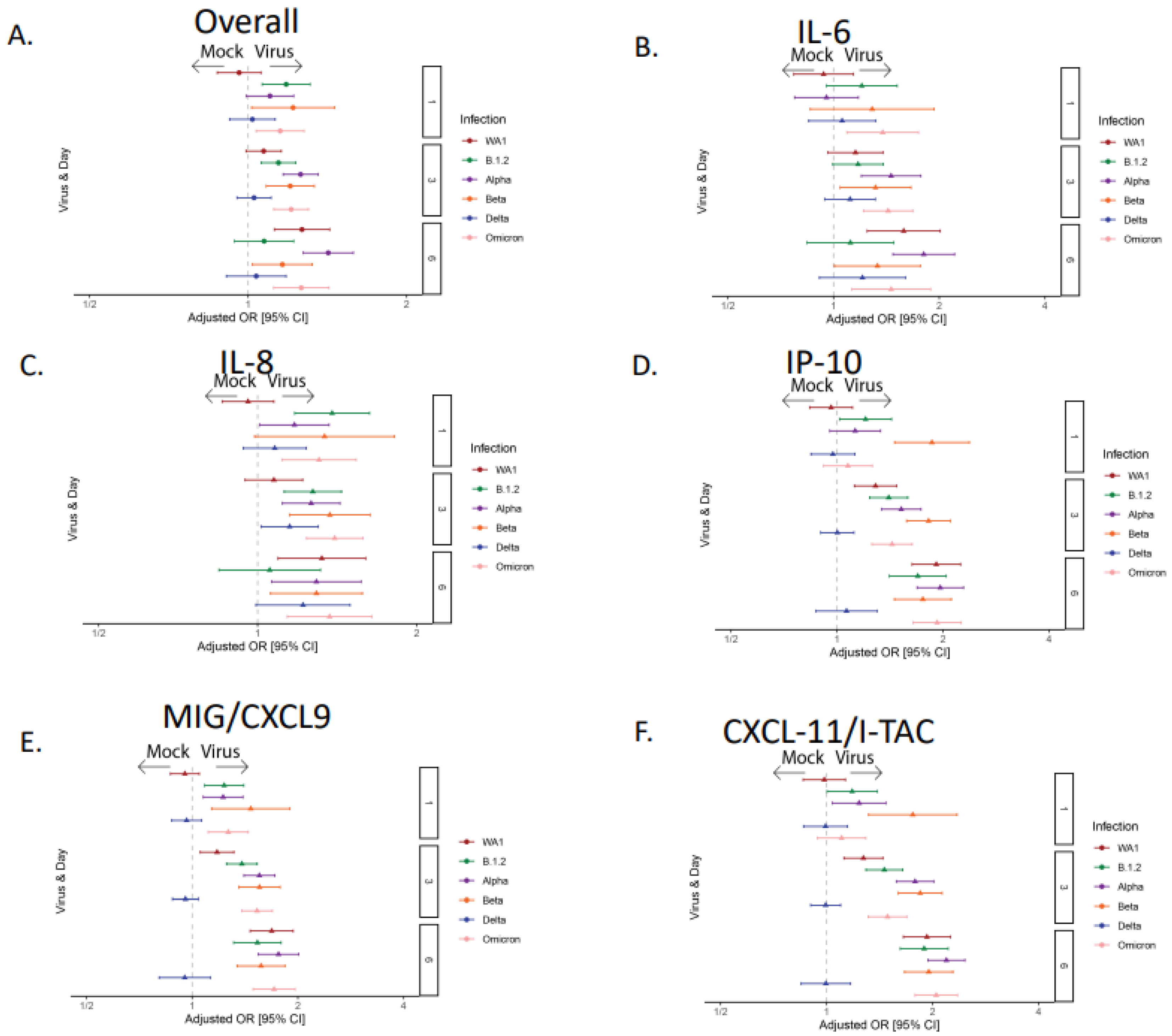
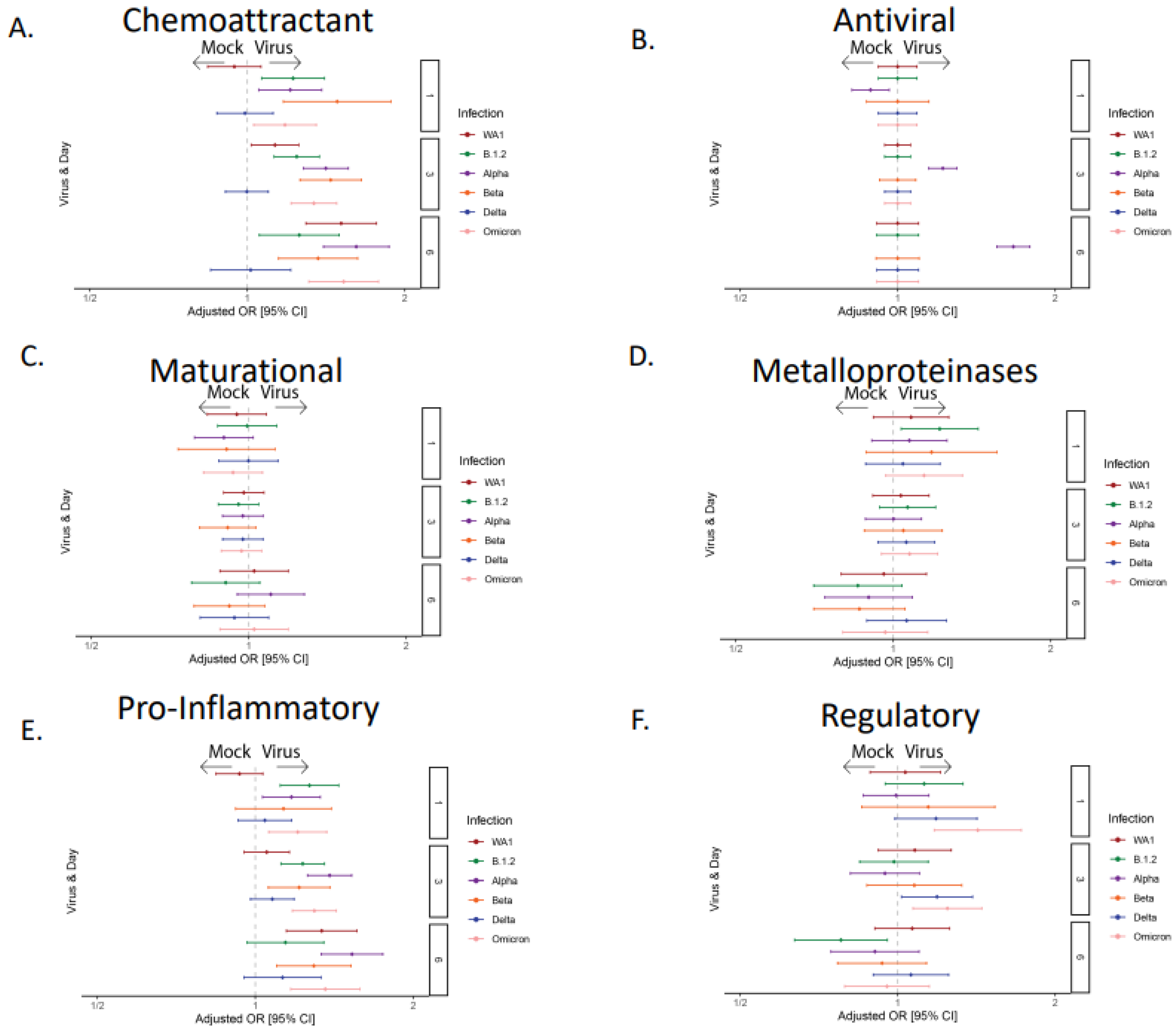
3.2. The Majority of the SARS-CoV-2 Variants Except Delta Induced Robust Cytokine and Chemokine Responses
3.3. Infection with SARS-CoV-2 Variants Causes Significant Ciliary Damage in HNO-ALIs
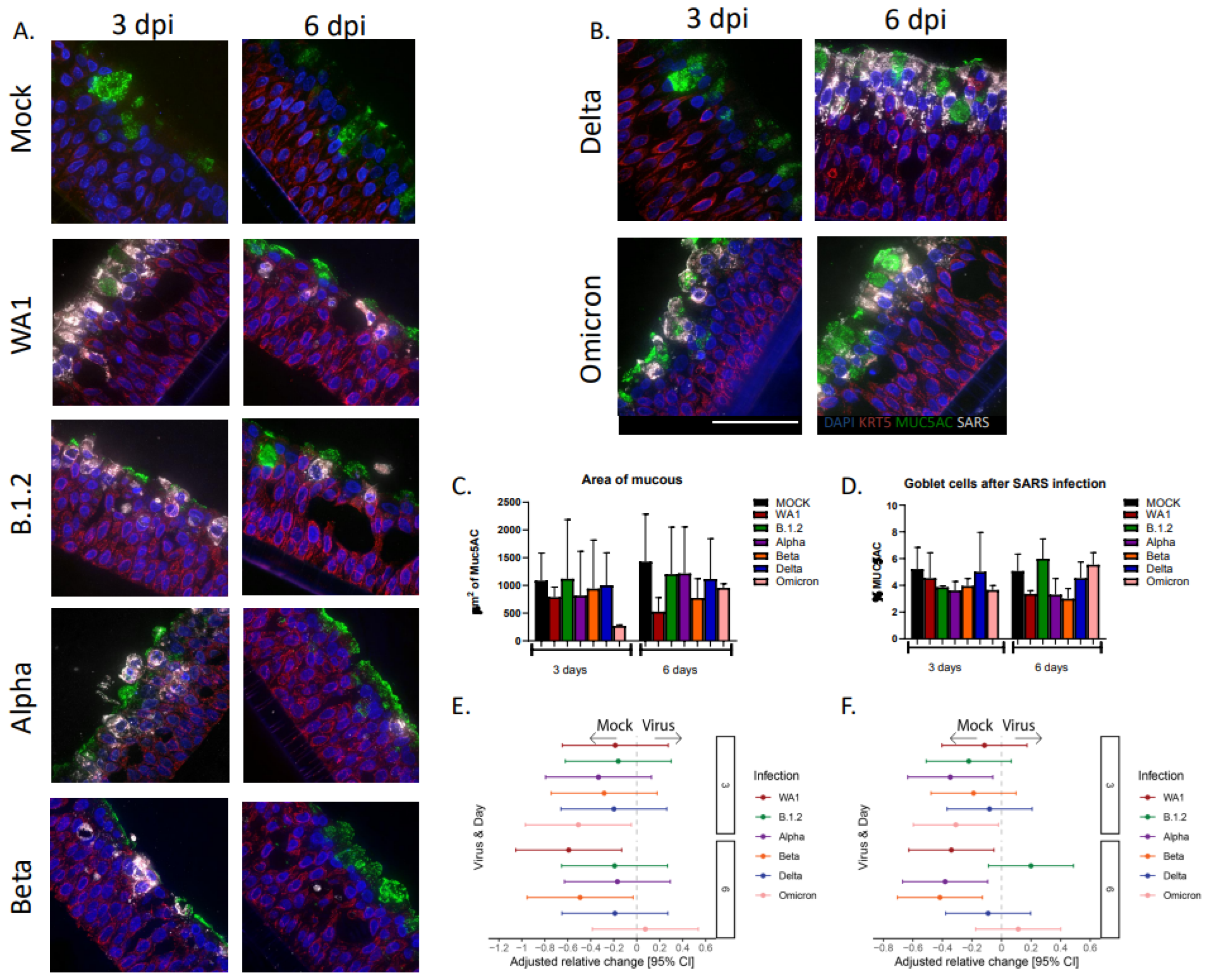
3.4. Goblet Cells and Mucous Decrease After Infections with SARS-CoV-2 Variants
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Mathieu, E.; Ritchie, H.; Rodés-Guirao, L.; Appel, C.; Giattino, C.; Hasell, J.; Macdonald, B.; Dattani, S.; Beltekian, D.; Ortiz-Ospina, E.; et al. Coronavirus Pandemic (COVID-19). Available online: https://ourworldindata.org/ (accessed on 1 October 2025).
- Steiner, S.; Kratzel, A.; Barut, G.T.; Lang, R.M.; Moreira, E.A.; Thomann, L.; Kelly, J.N.; Thiel, V. SARS-CoV-2 biology and host interactions. Nat. Rev. Microbiol. 2024, 22, 206–225. [Google Scholar] [CrossRef]
- Furnon, W.; Cowton, V.M.; De Lorenzo, G.; Orton, R.; Herder, V.; Cantoni, D.; Ilia, G.; Mendonca, D.C.; Kerr, K.; Allan, J.; et al. Phenotypic evolution of SARS-CoV-2 spike during the COVID-19 pandemic. Nat. Microbiol. 2025, 10, 77–93. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization 2023 data.who.int WHO Coronavirus (COVID-19) Dashboard. Available online: https://data.who.int/dashboards/covid19 (accessed on 1 October 2025).
- CDC. CDC Museum COVID-19 Timeline. Available online: https://www.cdc.gov/museum/timeline/covid19.html (accessed on 1 October 2025).
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.B.; Russell, T.W.; Tully, D.C.; Washburne, A.D.; et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 2021, 372, eabg3055. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.Y.; Smith, D.M. SARS-CoV-2 Variants of Concern. Yonsei Med. J. 2021, 62, 961. [Google Scholar] [CrossRef] [PubMed]
- Atherstone, C.J.; Guagliardo, S.A.J.; Hawksworth, A.; O’lAughlin, K.; Wong, K.; Sloan, M.L.; Henao, O.; Rao, C.Y.; McElroy, P.D.; Bennett, S.D. COVID-19 Epidemiology during Delta Variant Dominance Period in 45 High-Income Countries, 2020–2021. Emerg. Infect. Dis. 2023, 29, 1757–1764. [Google Scholar] [CrossRef]
- Bast, E.; Tang, F.; Dahn, J.; Palacio, A. Increased risk of hospitalisation and death with the delta variant in the USA. Lancet Infect. Dis. 2021, 21, 1629–1630. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, J.; Jian, F.; Xiao, T.; Song, W.; Yisimayi, A.; Huang, W.; Li, Q.; Wang, P.; An, R.; et al. Omicron Escapes the Majority of Existing SARS-CoV-2 Neutralizing Antibodies. Nature 2022, 602, 657–663. [Google Scholar] [CrossRef]
- Wang, Y.; Long, Y.; Wang, F.; Li, C.; Liu, W. Characterization of SARS-CoV-2 recombinants and emerging Omicron sublineages. Int. J. Med Sci. 2023, 20, 151–162. [Google Scholar] [CrossRef]
- Yang, W.; Yang, S.; Wang, L.; Zhou, Y.; Xin, Y.; Li, H.; Mu, W.; Wu, Q.; Xu, L.; Zhao, M.; et al. Clinical characteristics of 310 SARS-CoV-2 Omicron variant patients and comparison with Delta and Beta variant patients in China. Virol. Sin. 2022, 37, 704–715. [Google Scholar] [CrossRef]
- Ju, B.; Zheng, Q.; Guo, H.; Fan, Q.; Li, T.; Song, S.; Sun, H.; Shen, S.; Zhou, X.; Xue, W.; et al. Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209. Cell Res. 2022, 32, 491–494. [Google Scholar] [CrossRef]
- Yang, L.; Han, Y.; Nilsson-Payant, B.E.; Gupta, V.; Wang, P.; Duan, X.; Tang, X.; Zhu, J.; Zhao, Z.; Jaffré, F.; et al. A Human Pluripotent Stem Cell-based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids. Cell Stem Cell 2020, 27, 125–136.e7. [Google Scholar] [CrossRef]
- Mlcochova, P.; Kemp, S.A.; Dhar, M.S.; Papa, G.; Meng, B.; Ferreira, I.A.T.M.; Datir, R.; Collier, D.A.; Albecka, A.; Singh, S.; et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021, 599, 114–119. [Google Scholar] [CrossRef]
- Rajan, A.; Weaver, A.M.; Aloisio, G.M.; Jelinski, J.; Johnson, H.L.; Venable, S.F.; McBride, T.; Aideyan, L.; Piedra, F.-A.; Ye, X.; et al. The Human Nose Organoid Respiratory Virus Model: An Ex Vivo Human Challenge Model To Study Respiratory Syncytial Virus (RSV) and Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Pathogenesis and Evaluate Therapeutics. mBio 2022, 13, e0351121. [Google Scholar] [CrossRef] [PubMed]
- Chiu, M.C.; Li, C.; Liu, X.; Song, W.; Wan, Z.; Yu, Y.; Huang, J.; Xiao, D.; Chu, H.; Cai, J.-P.; et al. Human Nasal Organoids Model SARS-CoV-2 Upper Respiratory Infection and Recapitulate the Differential Infectivity of Emerging Variants. mBio 2022, 13, e0194422. [Google Scholar] [CrossRef] [PubMed]
- Tran, B.M.; Grimley, S.L.; McAuley, J.L.; Hachani, A.; Earnest, L.; Wong, S.L.; Caly, L.; Druce, J.; Purcell, D.F.J.; Jackson, D.C.; et al. Air-Liquid-Interface Differentiated Human Nose Epithelium: A Robust Primary Tissue Culture Model of SARS-CoV-2 Infection. Int. J. Mol. Sci. 2022, 23, 835. [Google Scholar] [CrossRef] [PubMed]
- Hui, K.P.Y.; Ho, J.C.W.; Cheung, M.-C.; Ng, K.-C.; Ching, R.H.H.; Lai, K.-L.; Kam, T.T.; Gu, H.; Sit, K.-Y.; Hsin, M.K.Y.; et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature 2022, 603, 715–720. [Google Scholar] [CrossRef]
- Shuai, H.; Chan, J.F.-W.; Hu, B.; Chai, Y.; Yuen, T.T.-T.; Yin, F.; Huang, X.; Yoon, C.; Hu, J.-C.; Liu, H.; et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature 2022, 603, 693–699. [Google Scholar] [CrossRef]
- Suzuki, R.; Yamasoba, D.; Kimura, I.; Wang, L.; Kishimoto, M.; Ito, J.; Morioka, Y.; Nao, N.; Nasser, H.; Uriu, K.; et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature 2022, 603, 700–705. [Google Scholar] [CrossRef]
- Masui, A.; Hashimoto, R.; Matsumura, Y.; Yamamoto, T.; Nagao, M.; Noda, T.; Takayama, K.; Gotoh, S. Micro-patterned culture of iPSC-derived alveolar and airway cells distinguishes SARS-CoV-2 variants. Stem Cell Rep. 2024, 19, 545–561. [Google Scholar] [CrossRef]
- Li, C.; Huang, J.; Yu, Y.; Wan, Z.; Chiu, M.C.; Liu, X.; Zhang, S.; Cai, J.-P.; Chu, H.; Li, G.; et al. Human airway and nasal organoids reveal escalating replicative fitness of SARS-CoV-2 emerging variants. Proc. Natl. Acad. Sci. USA 2023, 120, e2300376120. [Google Scholar] [CrossRef]
- Rajan, A.; Piedra, F.-A.; Aideyan, L.; McBride, T.; Robertson, M.; Johnson, H.L.; Aloisio, G.M.; Henke, D.; Coarfa, C.; Stossi, F.; et al. Multiple Respiratory Syncytial Virus (RSV) Strains Infecting HEp-2 and A549 Cells Reveal Cell Line-Dependent Differences in Resistance to RSV Infection. J. Virol. 2022, 96, e0190421. [Google Scholar] [CrossRef]
- Aloisio, G.M.; Nagaraj, D.; Murray, A.M.; Schultz, E.M.; McBride, T.; Aideyan, L.; Nicholson, E.G.; Henke, D.; Ferlic-Stark, L.; Rajan, A.; et al. Infant-derived human nasal organoids exhibit relatively increased susceptibility, epithelial responses, and cytotoxicity during RSV infection. J. Infect. 2024, 89, 106305. [Google Scholar] [CrossRef] [PubMed]
- Avadhanula, V.; Nicholson, E.G.; Ferlic-Stark, L.; Piedra, F.-A.; Blunck, B.N.; Fragoso, S.; Bond, N.L.; Santarcangelo, P.L.; Ye, X.; McBride, T.J.; et al. Viral Load of Severe Acute Respiratory Syndrome Coronavirus 2 in Adults During the First and Second Wave of Coronavirus Disease 2019 Pandemic in Houston, Texas: The Potential of the Superspreader. J. Infect. Dis. 2021, 223, 1528–1537. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.R.; Chaiwun, B.; Young, L.; Cote, R.J.; Taylor, C.R. Antigen retrieval technique utilizing citrate buffer or urea solution for immunohistochemical demonstration of androgen receptor in formalin-fixed paraffin sections. J. Histochem. Cytochem. 1993, 41, 1599–1604. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Hasegawa, T.; Hato, T.; Okayama, T.; Ikeo, K.; Miyamoto, Y.; Iwanaga, N.; Suzuki, K.; Yoshida, M.; Yamashita, K.; Yamashita, S.; et al. Th1 cytokine endotype discriminates and predicts severe complications in COVID-19. Eur. Cytokine Netw. 2022, 33, 1–12. [Google Scholar] [CrossRef]
- Liao, M.; Liu, Y.; Yuan, J.; Wen, Y.; Xu, G.; Zhao, J.; Cheng, L.; Li, J.; Wang, X.; Wang, F.; et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020, 26, 842–844. [Google Scholar] [CrossRef]
- Chen, H.; Liu, W.; Wang, Y.; Liu, D.; Zhao, L.; Yu, J. SARS-CoV-2 activates lung epithelial cell proinflammatory signaling and leads to immune dysregulation in COVID-19 patients. EBioMedicine 2021, 70, 103500. [Google Scholar] [CrossRef]
- Lowery, S.A.; Sariol, A.; Perlman, S. Innate immune and inflammatory responses to SARS-CoV-2: Implications for COVID-19. Cell Host Microbe 2021, 29, 1052–1062. [Google Scholar] [CrossRef]
- Xiong, Y.; Liu, Y.; Cao, L.; Wang, D.; Guo, M.; Jiang, A.; Guo, D.; Hu, W.; Yang, J.; Tang, Z.; et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020, 9, 761–770. [Google Scholar] [CrossRef]
- Pum, A.; Ennemoser, M.; Adage, T.; Kungl, A.J. Cytokines and Chemokines in SARS-CoV-2 Infections—Therapeutic Strategies Targeting Cytokine Storm. Biomolecules 2021, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, A.J.; Ding, J.; You, Y.; Dong, Z.; Chehade, H.; Alvero, A.; Mor, Y.; Draghici, S.; Mor, G. Identification of key signaling pathways induced by SARS-CoV2 that underlie thrombosis and vascular injury in COVID-19 patients. J. Leukoc. Biol. 2020, 109, 35–47. [Google Scholar] [CrossRef] [PubMed]
- I Bloom, C.; Drake, T.M.; Docherty, A.B.; Lipworth, B.J.; Johnston, S.L.; Nguyen-Van-Tam, J.S.; Carson, G.; Dunning, J.; Harrison, E.M.; Baillie, J.K.; et al. Risk of adverse outcomes in patients with underlying respiratory conditions admitted to hospital with COVID-19: A national, multicentre prospective cohort study using the ISARIC WHO Clinical Characterisation Protocol UK. Lancet Respir. Med. 2021, 9, 699–711. [Google Scholar] [CrossRef] [PubMed]
- Hysenaj, L.; Little, S.; Kulhanek, K.; Magnen, M.; Bahl, K.; Gbenedio, O.M.; Prinz, M.; Rodriguez, L.; Andersen, C.; Rao, A.A.; et al. SARS-CoV-2 infection of airway organoids reveals conserved use of Tetraspanin-8 by Ancestral, Delta, and Omicron variants. Stem Cell Rep. 2023, 18, 636–653. [Google Scholar] [CrossRef]
- Tanneti, N.S.; Patel, A.K.; Tan, L.H.; Marques, A.D.; Perera, R.A.P.M.; Sherrill-Mix, S.; Kelly, B.J.; Renner, D.M.; Collman, R.G.; Rodino, K.; et al. Comparison of SARS-CoV-2 variants of concern in primary human nasal cultures demonstrates Delta as most cytopathic and Omicron as fastest replicating. mBio 2024, 15, e0312923. [Google Scholar] [CrossRef]
- Meganck, R.M.; Edwards, C.E.; Mallory, M.L.; Lee, R.E.; Dang, H.; Bailey, A.B.; Wykoff, J.A.; Gallant, S.C.; Zhu, D.R.; Yount, B.L.; et al. SARS-CoV-2 variant of concern fitness and adaptation in primary human airway epithelia. Cell Rep. 2024, 43, 114076. [Google Scholar] [CrossRef]
- Huang, F.; Liu, X.; Sun, X.; Li, Z. IL-10 Served as an Indicator in Severe COVID-19 Patients. J. Med. Virol. 2021, 93, 1233. [Google Scholar] [CrossRef]
- Yang, Y.; Shen, C.; Li, J.; Yuan, J.; Wei, J.; Huang, F.; Wang, F.; Li, G.; Li, Y.; Xing, L.; et al. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J. Allergy Clin. Immunol. 2020, 146, 119–127.e4. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, C.; Miao, X.; Kong, D.; Zhao, Y.; Gong, W.; Ding, X. Therapeutic targeting of interleukin-6 for the treatment of COVID-19. Eur. Cytokine Netw. 2020, 1. [Google Scholar]
- Qin, Z.; Xiang, K.; Su, D.-F.; Sun, Y.; Liu, X. Activation of the Cholinergic Anti-Inflammatory Pathway as a Novel Therapeutic Strategy for COVID-19. Front. Immunol. 2021, 11, 595342. [Google Scholar] [CrossRef]
- Zhou, Z.; Ren, L.; Zhang, L.; Zhong, J.; Xiao, Y.; Jia, Z.; Guo, L.; Yang, J.; Wang, C.; Jiang, S.; et al. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe 2020, 27, 883–890.e2. [Google Scholar] [CrossRef]
- Anderberg, S.B.; Luther, T.; Berglund, M.; Larsson, R.; Rubertsson, S.; Lipcsey, M.; Larsson, A.; Frithiof, R.; Hultström, M. Increased levels of plasma cytokines and correlations to organ failure and 30-day mortality in critically ill Covid-19 patients. Cytokine 2021, 138, 155389. [Google Scholar] [CrossRef]
- D’rozario, R.; Raychaudhuri, D.; Bandopadhyay, P.; Sarif, J.; Mehta, P.; Liu, C.S.C.; Sinha, B.P.; Roy, J.; Bhaduri, R.; Das, M.; et al. Circulating Interleukin-8 Dynamics Parallels Disease Course and Is Linked to Clinical Outcomes in Severe COVID-19. Viruses 2023, 15, 549. [Google Scholar] [CrossRef]
- Korobova, Z.R.; Arsentieva, N.A.; Liubimova, N.E.; Dedkov, V.G.; Gladkikh, A.S.; Sharova, A.A.; Chernykh, E.I.; Kashchenko, V.A.; Ratnikov, V.A.; Gorelov, V.P.; et al. A Comparative Study of the Plasma Chemokine Profile in COVID-19 Patients Infected with Different SARS-CoV-2 Variants. Int. J. Mol. Sci. 2022, 23, 9058. [Google Scholar] [CrossRef]
- A Twohig, K.; Nyberg, T.; Zaidi, A.; Thelwall, S.; A Sinnathamby, M.; Aliabadi, S.; Seaman, S.R.; Harris, R.J.; Hope, R.; Lopez-Bernal, J.; et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: A cohort study. Lancet Infect. Dis. 2022, 22, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Saito, A.; Irie, T.; Suzuki, R.; Maemura, T.; Nasser, H.; Uriu, K.; Kosugi, Y.; Shirakawa, K.; Sadamasu, K.; Kimura, I.; et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 2021, 602, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Nilsson-Payant, B.E.; Uhl, S.; Grimont, A.; Doane, A.S.; Cohen, P.; Patel, R.S.; Higgins, C.A.; Acklin, J.A.; Bram, Y.; Chandar, V.; et al. The NF-κB Transcriptional Footprint Is Essential for SARS-CoV-2 Replication. J. Virol. 2021, 95, e0125721. [Google Scholar] [CrossRef] [PubMed]
- Neufeldt, C.J.; Cerikan, B.; Cortese, M.; Frankish, J.; Lee, J.-Y.; Plociennikowska, A.; Heigwer, F.; Prasad, V.; Joecks, S.; Burkart, S.S.; et al. SARS-CoV-2 infection induces a pro-inflammatory cytokine response through cGAS-STING and NF-κB. Commun. Biol. 2022, 5, 1–15. [Google Scholar] [CrossRef]
- Saito, A.; Horie, M.; Nagase, T. TGF-β Signaling in Lung Health and Disease. Int. J. Mol. Sci. 2018, 19, 2460. [Google Scholar] [CrossRef]
- Robinot, R.; Hubert, M.; de Melo, G.D.; Lazarini, F.; Bruel, T.; Smith, N.; Levallois, S.; Larrous, F.; Fernandes, J.; Gellenoncourt, S.; et al. SARS-CoV-2 infection induces the dedifferentiation of multiciliated cells and impairs mucociliary clearance. Nat. Commun. 2021, 12, 1–16. [Google Scholar] [CrossRef]
- Ahn, J.H.; Kim, J.M.; Hong, S.P.; Choi, S.Y.; Yang, M.J.; Ju, Y.S.; Kim, Y.T.; Kim, H.M.; Rahman, T.; Chung, M.K.; et al. Nasal ciliated cells are primary targets for SARS-CoV-2 replication in the early stage of COVID-19. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef]
- Wu, C.-T.; Lidsky, P.V.; Xiao, Y.; Cheng, R.; Lee, I.T.; Nakayama, T.; Jiang, S.; He, W.; Demeter, J.; Knight, M.G.; et al. SARS-CoV-2 replication in airway epithelia requires motile cilia and microvillar reprogramming. Cell 2022, 186, 112–130.e20. [Google Scholar] [CrossRef]
- Osan, J.; Talukdar, S.N.; Feldmann, F.; DeMontigny, B.A.; Jerome, K.; Bailey, K.L.; Feldmann, H.; Mehedi, M. Goblet Cell Hyperplasia Increases SARS-CoV-2 Infection in Chronic Obstructive Pulmonary Disease. Microbiol. Spectr. 2022, 10, e0045922. [Google Scholar] [CrossRef]
- Del Valle, D.M.; Kim-Schulze, S.; Huang, H.-H.; Beckmann, N.D.; Nirenberg, S.; Wang, B.; Lavin, Y.; Swartz, T.H.; Madduri, D.; Stock, A.; et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020, 26, 1636–1643. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aloisio, G.M.; McBride, T.J.; Aideyan, L.; Schultz, E.M.; Murray, A.M.; Rajan, A.; Nicholson, E.G.; Henke, D.; Ferlic-Stark, L.; Kambal, A.; et al. Strain-Specific Variability in Viral Kinetics, Cytokine Response, and Cellular Damage in Air–Liquid Cultures of Human Nasal Organoids After Infection with SARS-CoV-2. Viruses 2025, 17, 1343. https://doi.org/10.3390/v17101343
Aloisio GM, McBride TJ, Aideyan L, Schultz EM, Murray AM, Rajan A, Nicholson EG, Henke D, Ferlic-Stark L, Kambal A, et al. Strain-Specific Variability in Viral Kinetics, Cytokine Response, and Cellular Damage in Air–Liquid Cultures of Human Nasal Organoids After Infection with SARS-CoV-2. Viruses. 2025; 17(10):1343. https://doi.org/10.3390/v17101343
Chicago/Turabian StyleAloisio, Gina M., Trevor J. McBride, Letisha Aideyan, Emily M. Schultz, Ashley M. Murray, Anubama Rajan, Erin G. Nicholson, David Henke, Laura Ferlic-Stark, Amal Kambal, and et al. 2025. "Strain-Specific Variability in Viral Kinetics, Cytokine Response, and Cellular Damage in Air–Liquid Cultures of Human Nasal Organoids After Infection with SARS-CoV-2" Viruses 17, no. 10: 1343. https://doi.org/10.3390/v17101343
APA StyleAloisio, G. M., McBride, T. J., Aideyan, L., Schultz, E. M., Murray, A. M., Rajan, A., Nicholson, E. G., Henke, D., Ferlic-Stark, L., Kambal, A., Johnson, H. L., Mosa, E. A., Stossi, F., Blutt, S. E., Piedra, P. A., & Avadhanula, V. (2025). Strain-Specific Variability in Viral Kinetics, Cytokine Response, and Cellular Damage in Air–Liquid Cultures of Human Nasal Organoids After Infection with SARS-CoV-2. Viruses, 17(10), 1343. https://doi.org/10.3390/v17101343





