Molecular and Evolutionary Characteristics of Chicken Parvovirus (ChPV) Genomes Detected in Chickens with Runting–Stunting Syndrome
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples and Viral Metagenomics
2.2. Sequence and Phylogenetic Analysis
2.3. Recombination Analysis
2.4. Selective Pressure Analysis
2.5. Protein Modeling of Viral Capsid and B and T Epitope Prediction
3. Results
3.1. Viral Metagenomics
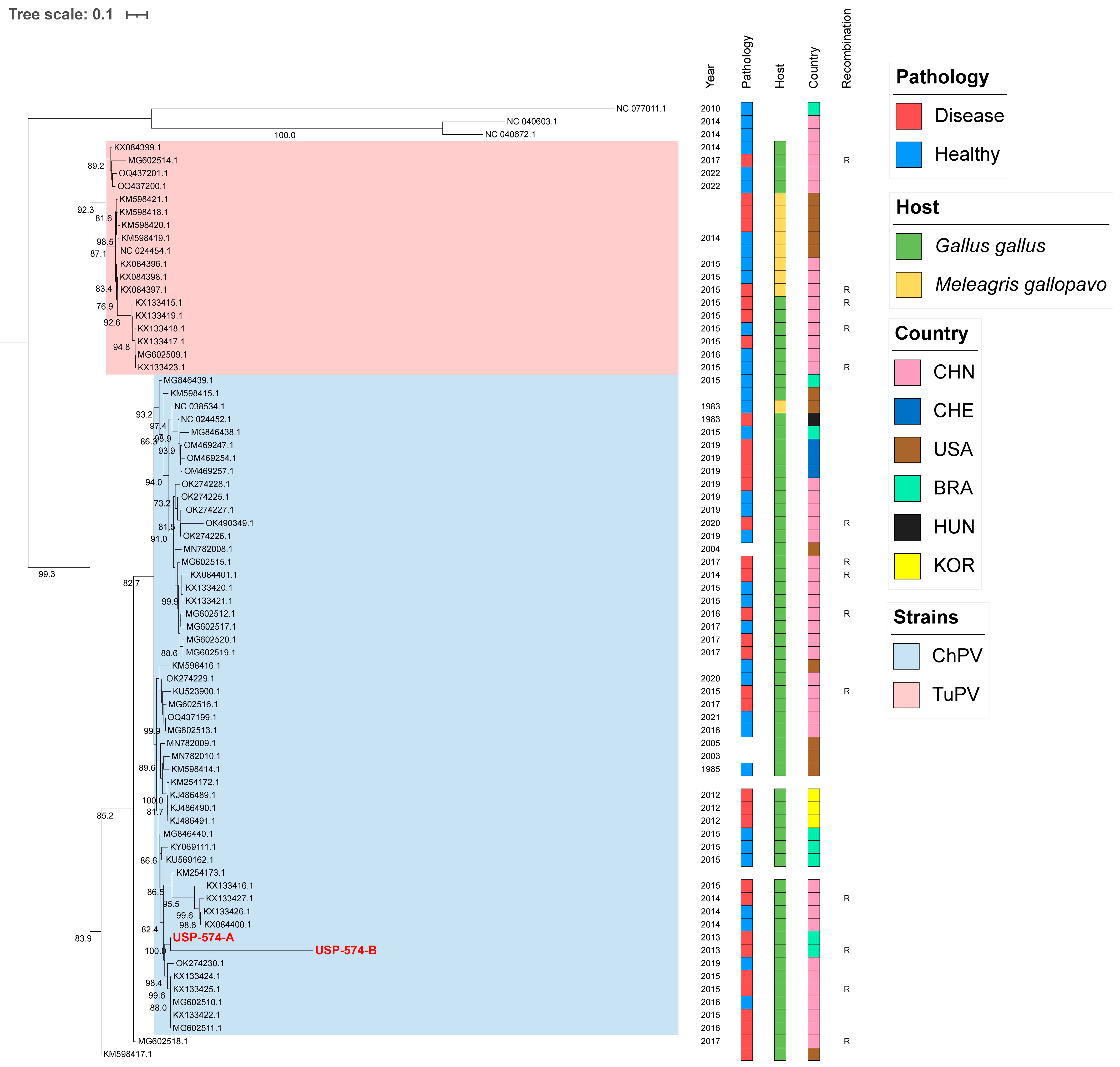
3.2. Sequence and Phylogenetic Analysis
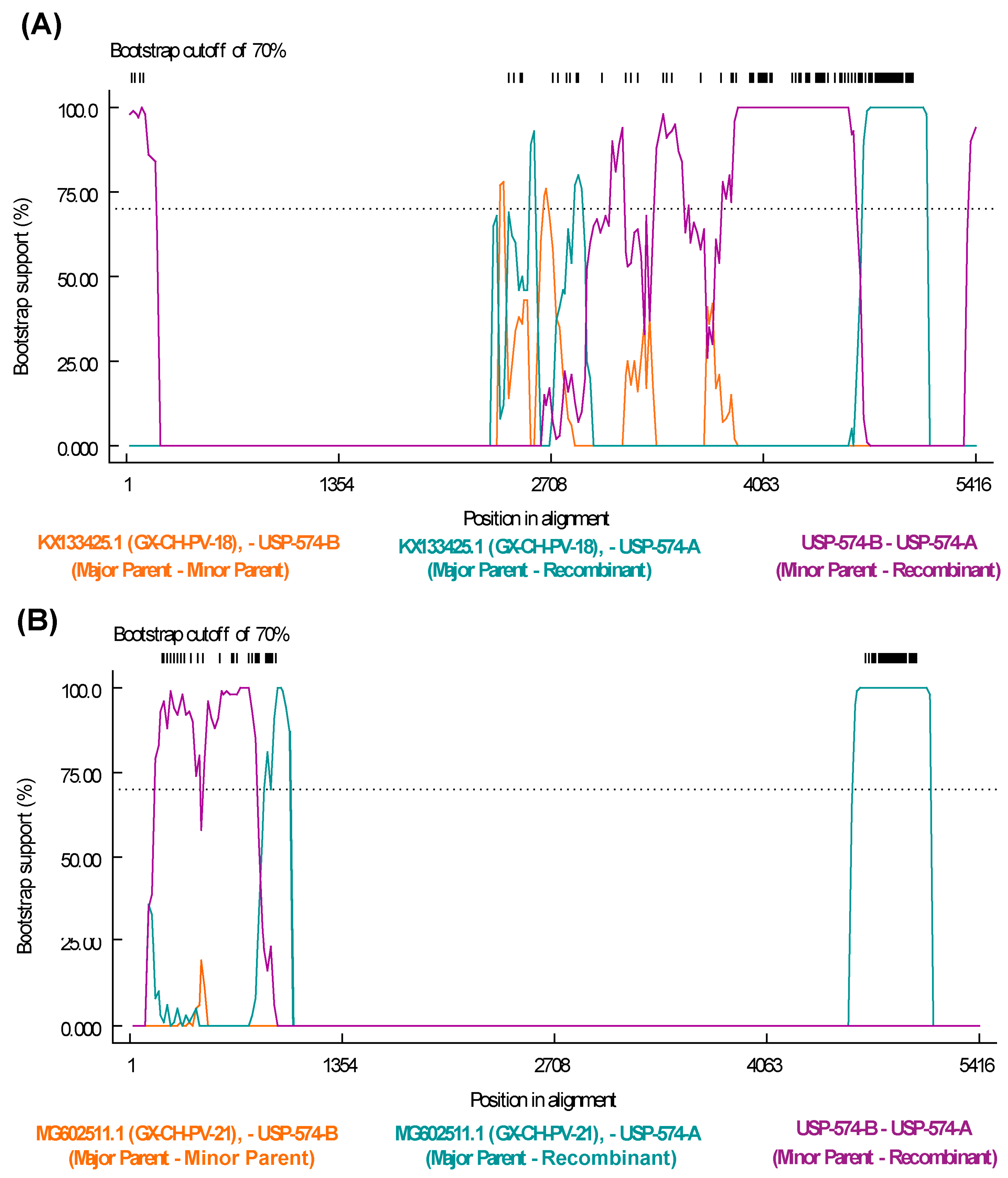
3.3. Recombination Analysis
3.4. Selective Pressure Analysis
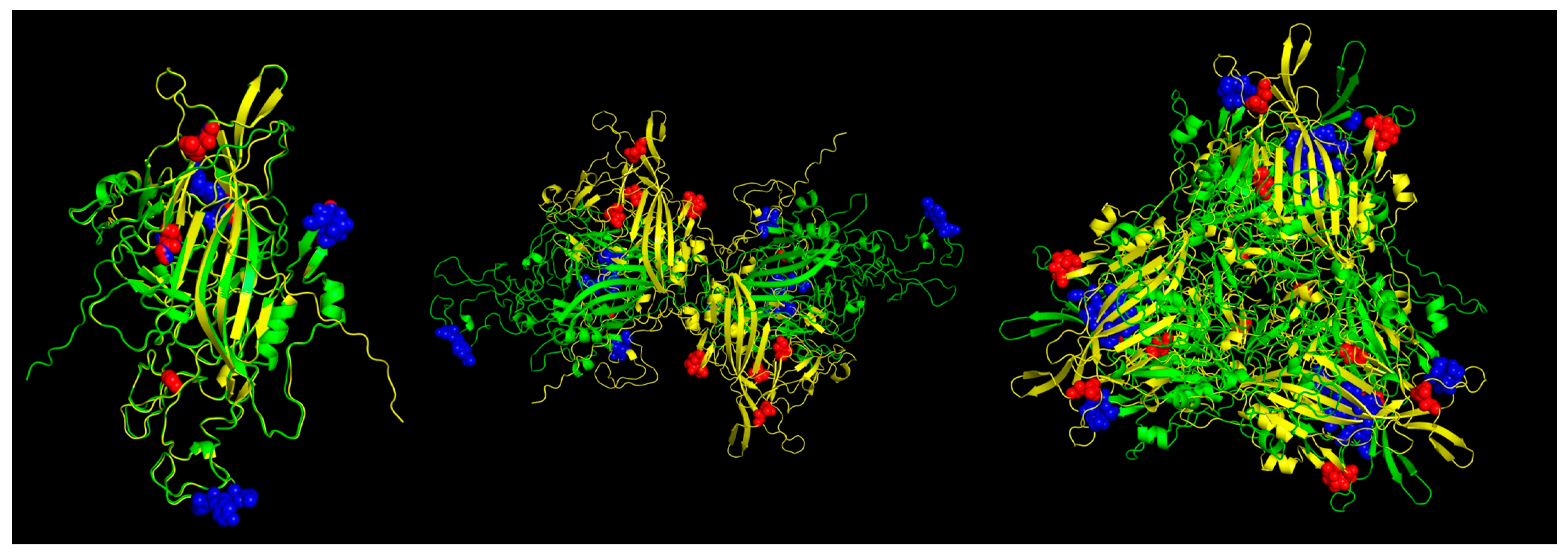
3.5. Protein Modeling of Viral Capsid and T and B Epitope Prediction
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- International Committee on Taxonomy of Viruses Current ICTV Taxonomy Release|ICTV. Available online: https://ictv.global/taxonomy (accessed on 17 October 2023).
- Zsak, L.; Strother, K.O.; Day, J.M. Development of a Polymerase Chain Reaction Procedure for Detection of Chicken and Turkey Parvoviruses. Avian Dis. 2009, 53, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Zsak, L.; Cha, R.M.; Li, F.; Day, J.M. Host Specificity and Phylogenetic Relationships of Chicken and Turkey Parvoviruses. Avian Dis. 2015, 59, 157–161. [Google Scholar] [CrossRef]
- Kapgate, S.S.; Kumanan, K.; Vijayarani, K.; Barbuddhe, S.B. Avian Parvovirus: Classification, Phylogeny, Pathogenesis and Diagnosis. Avian Pathol. 2018, 47, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Nuñez, L.F.; Santander-Parra, S.H.; Chaible, L.; De la Torre, D.I.; Buim, M.R.; Murakami, A.; Zaidan Dagli, M.L.; Astolfi-Ferreira, C.S.; Piantino Ferreira, A.J. Development of a Sensitive Real-Time Fast-qPCR Based on SYBR® Green for Detection and Quantification of Chicken Parvovirus (ChPV). Vet. Sci. 2018, 5, 69. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.; Liu, F.; Chen, S.; Wang, M.; Cheng, A. Role of Capsid Proteins in Parvoviruses Infection. Virol. J. 2015, 12, 114. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, M.A.; Davis, J.F.; McNulty, M.S.; Brown, J.; Player, E.C. Enteritis (so-Called Runting Stunting Syndrome) in Georgia Broiler Chicks. Avian Dis. 1993, 37, 451–458. [Google Scholar] [CrossRef]
- Pantin-Jackwood, M.J.; Spackman, E.; Woolcock, P.R. Molecular Characterization and Typing of Chicken and Turkey Astroviruses Circulating in the United States: Implications for Diagnostics. Avian Dis. 2006, 50, 397–404. [Google Scholar] [CrossRef]
- Mettifogo, E.; Nuñez, L.F.N.; Chacón, J.L.; Santander Parra, S.H.; Astolfi-Ferreira, C.S.; Jerez, J.A.; Jones, R.C.; Piantino Ferreira, A.J. Emergence of Enteric Viruses in Production Chickens Is a Concern for Avian Health. Sci. World J. 2014, 2014, 450423. [Google Scholar] [CrossRef]
- Kang, K.-I.; El-Gazzar, M.; Sellers, H.S.; Dorea, F.; Williams, S.M.; Kim, T.; Collett, S.; Mundt, E. Investigation into the Aetiology of Runting and Stunting Syndrome in Chickens. Avian Pathol. 2012, 41, 41–50. [Google Scholar] [CrossRef]
- Koo, B.S.; Lee, H.R.; Jeon, E.O.; Han, M.S.; Min, K.C.; Lee, S.B.; Mo, I.P. Molecular Survey of Enteric Viruses in Commercial Chicken Farms in Korea with a History of Enteritis. Poult. Sci. 2013, 92, 2876–2885. [Google Scholar] [CrossRef]
- Saif, Y.M.; Guy, J.S.; Day, J.M.; Cattoli, G.; Hayhow, C.S. Viral Enteric Infections. In Diseases of Poultry; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2020; pp. 401–445. ISBN 978-1-119-37119-9. [Google Scholar]
- Kisary, J.; Nagy, B.; Bitay, Z. Presence of Parvoviruses in the Intestine of Chickens Showing Stunting Syndrome. Avian Pathol. 1984, 13, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Zsak, L.; Cha, R.M.; Day, J.M. Chicken Parvovirus-Induced Runting-Stunting Syndrome in Young Broilers. Avian Dis. 2013, 57, 123–127. [Google Scholar] [CrossRef] [PubMed]
- Nuñez, L.F.N.; Sá, L.R.M.; Parra, S.H.S.; Astolfi-Ferreira, C.S.; Carranza, C.; Ferreira, A.J.P. Molecular Detection of Chicken Parvovirus in Broilers with Enteric Disorders Presenting Curving of Duodenal Loop, Pancreatic Atrophy, and Mesenteritis. Poult. Sci. 2016, 95, 802–810. [Google Scholar] [CrossRef] [PubMed]
- Nuñez, L.F.N.; Santander Parra, S.H.; Mettifogo, E.; Astolfi-Ferreira, C.S.; Piantino Ferreira, A.J. Isolation and Molecular Characterisation of Chicken Parvovirus from Brazilian Flocks with Enteric Disorders. Br. Poult. Sci. 2015, 56, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Feng, B.; Xie, Z.; Zhang, M.; Fan, Q.; Deng, X.; Xie, Z.; Li, M.; Zeng, T.; Xie, L.; et al. Molecular Characterization of Emerging Chicken and Turkey Parvovirus Variants and Novel Strains in Guangxi, China. Sci. Rep. 2023, 13, 13083. [Google Scholar] [CrossRef]
- Devaney, R.; Trudgett, J.; Trudgett, A.; Meharg, C.; Smyth, V. A Metagenomic Comparison of Endemic Viruses from Broiler Chickens with Runting-Stunting Syndrome and from Normal Birds. Avian Pathol. 2016, 45, 616–629. [Google Scholar] [CrossRef]
- Lima, D.A.; Cibulski, S.P.; Tochetto, C.; Varela, A.P.M.; Finkler, F.; Teixeira, T.F.; Loiko, M.R.; Cerva, C.; Junqueira, D.M.; Mayer, F.Q.; et al. The Intestinal Virome of Malabsorption Syndrome-Affected and Unaffected Broilers through Shotgun Metagenomics. Virus Res. 2019, 261, 9–20. [Google Scholar] [CrossRef]
- Kim, H.-R.; Kwon, Y.-K.; Jang, I.; Bae, Y.-C. Viral Metagenomic Analysis of Chickens with Runting-Stunting Syndrome in the Republic of Korea. Virol. J. 2020, 17, 53. [Google Scholar] [CrossRef]
- Kubacki, J.; Qi, W.; Fraefel, C. Differential Viral Genome Diversity of Healthy and RSS-Affected Broiler Flocks. Microorganisms 2022, 10, 1092. [Google Scholar] [CrossRef]
- da Costa, C.A.; Thézé, J.; Komninakis, S.C.V.; Sanz-Duro, R.L.; Felinto, M.R.L.; Moura, L.C.C.; Barroso, I.M.d.O.; Santos, L.E.C.; Nunes, M.A.d.L.; Moura, A.A.; et al. Spread of Chikungunya Virus East/Central/South African Genotype in Northeast Brazil. Emerg. Infect. Dis. 2017, 23, 1742–1744. [Google Scholar] [CrossRef]
- Nuñez, L.F.N.; Chacón, R.D.; Charlys da Costa, A.; Parra, S.H.S.; da Costa, R.P.I.; Sánchez-Llatas, C.J.; Cea-Callejo, P.; Valdeiglesias Ichillumpa, S.; Astolfi-Ferreira, C.S.; Sá, L.R.M.; et al. Detection and molecular characterization of Chicken Parvovirus and Chicken Megrivirus in layer breeders affected by Intestinal Dilatation Syndrome. Avian Pathol. 2024, 53, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Naccache, S.N.; Ng, T.; Federman, S.; Li, L.; Chiu, C.Y.; Delwart, E.L. An Ensemble Strategy That Significantly Improves de Novo Assembly of Microbial Genomes from Metagenomic Next-Generation Sequencing Data. Nucleic Acids Res. 2015, 43, e46. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent Updates and New Developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Kosakovsky Pond, S.L.; Posada, D.; Gravenor, M.B.; Woelk, C.H.; Frost, S.D.W. Automated Phylogenetic Detection of Recombination Using a Genetic Algorithm. Mol. Biol. Evol. 2006, 23, 1891–1901. [Google Scholar] [CrossRef]
- Martin, D.P.; Varsani, A.; Roumagnac, P.; Botha, G.; Maslamoney, S.; Schwab, T.; Kelz, Z.; Kumar, V.; Murrell, B. RDP5: A Computer Program for Analyzing Recombination in, and Removing Signals of Recombination from, Nucleotide Sequence Datasets. Virus Evol. 2021, 7, veaa087. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Weaver, S.; Shank, S.D.; Spielman, S.J.; Li, M.; Muse, S.V.; Kosakovsky Pond, S.L. Datamonkey 2.0: A Modern Web Application for Characterizing Selective and Other Evolutionary Processes. Mol. Biol. Evol. 2018, 35, 773–777. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Schrödinger. LLC the PyMOL Molecular Graphics System, Version 1.8. 2015. Available online: http://www.pymol.org/pymol (accessed on 17 October 2023).
- Reynisson, B.; Alvarez, B.; Paul, S.; Peters, B.; Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved Predictions of MHC Antigen Presentation by Concurrent Motif Deconvolution and Integration of MS MHC Eluted Ligand Data. Nucleic Acids Res. 2020, 48, W449–W454. [Google Scholar] [CrossRef]
- Valdivia-Olarte, H.; Requena, D.; Ramirez, M.; Saravia, L.E.; Izquierdo, R.; Falconi-Agapito, F.; Zavaleta, M.; Best, I.; Fernández-Díaz, M.; Zimic, M. Design of a Predicted MHC Restricted Short Peptide Immunodiagnostic and Vaccine Candidate for Fowl Adenovirus C in Chicken Infection. Bioinformation 2015, 11, 460. [Google Scholar] [CrossRef] [PubMed]
- Abergel, C.; Legendre, M.; Claverie, J.-M. The Rapidly Expanding Universe of Giant Viruses: Mimivirus, Pandoravirus, Pithovirus and Mollivirus. FEMS Microbiol. Rev. 2015, 39, 779–796. [Google Scholar] [CrossRef]
- Kim, T.; Mundt, E. Metagenomic Analysis of Intestinal Microbiomes in Chickens. Methods Mol. Biol. 2011, 733, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Domanska-Blicharz, K.; Jacukowicz, A.; Lisowska, A.; Minta, Z. Genetic Characterization of Parvoviruses Circulating in Turkey and Chicken Flocks in Poland. Arch. Virol. 2012, 157, 2425–2430. [Google Scholar] [CrossRef]
- Hueffer, K.; Parrish, C.R. Parvovirus Host Range, Cell Tropism and Evolution. Curr. Opin. Microbiol. 2003, 6, 392–398. [Google Scholar] [CrossRef]
- Tang, Y.; Tang, N.; Zhu, J.; Wang, M.; Liu, Y.; Lyu, Y. Molecular Characteristics and Genetic Evolutionary Analyses of Circulating Parvoviruses Derived from Cats in Beijing. BMC Vet. Res. 2022, 18, 195. [Google Scholar] [CrossRef]
- Chen, L.; Chen, L.; Wang, X.; Huo, S.; Li, Y. Detection and Molecular Characterization of Enteric Viruses in Poultry Flocks in Hebei Province, China. Animals 2022, 12, 2873. [Google Scholar] [CrossRef]
- Stamenković, G.G.; Ćirković, V.S.; Šiljić, M.M.; Blagojević, J.V.; Knežević, A.M.; Joksić, I.D.; Stanojević, M.P. Substitution Rate and Natural Selection in Parvovirus B19. Sci. Rep. 2016, 6, 35759. [Google Scholar] [CrossRef] [PubMed]
- Mira, F.; Canuti, M.; Purpari, G.; Cannella, V.; Di Bella, S.; Occhiogrosso, L.; Schirò, G.; Chiaramonte, G.; Barreca, S.; Pisano, P.; et al. Molecular Characterization and Evolutionary Analyses of Carnivore Protoparvovirus 1 NS1 Gene. Viruses 2019, 11, 308. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xiao, Y.; Qiu, M.; Li, X.; Li, S.; Lin, H.; Li, X.; Zhu, J.; Chen, N. A Systematic Investigation Unveils High Coinfection Status of Porcine Parvovirus Types 1 through 7 in China from 2016 to 2020. Microbiol. Spectr. 2021, 9, e0129421. [Google Scholar] [CrossRef] [PubMed]
- Véliz-Ahumada, A.; Sonia, V.; Daniela, S.; Miguel, G.; Timothy, H.; Valentina, F.; Lisette, L.; Leonardo, S. Molecular Analysis of Full-Length VP2 of Canine Parvovirus Reveals Antigenic Drift in CPV-2b and CPV-2c Variants in Central Chile. Animals 2021, 11, 2387. [Google Scholar] [CrossRef] [PubMed]
- Huo, X.; Chen, Y.; Zhu, J.; Wang, Y. Evolution, Genetic Recombination, and Phylogeography of Goose Parvovirus. Comp. Immunol. Microbiol. Infect. Dis. 2023, 102, 102079. [Google Scholar] [CrossRef] [PubMed]
- Hao, S.; Zhang, J.; Chen, Z.; Xu, H.; Wang, H.; Guan, W. Alternative Polyadenylation of Human Bocavirus at Its 3′ End Is Regulated by Multiple Elements and Affects Capsid Expression. J. Virol. 2017, 91, e02026-16. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Dolja, V.V.; Krupovic, M. Origins and Evolution of Viruses of Eukaryotes: The Ultimate Modularity. Virology 2015, 479–480, 2–25. [Google Scholar] [CrossRef]
- Pénzes, J.J.; de Souza, W.M.; Agbandje-McKenna, M.; Gifford, R.J. An Ancient Lineage of Highly Divergent Parvoviruses Infects Both Vertebrate and Invertebrate Hosts. Viruses 2019, 11, 525. [Google Scholar] [CrossRef] [PubMed]
- Gigler, A.; Dorsch, S.; Hemauer, A.; Williams, C.; Kim, S.; Young, N.S.; Zolla-Pazner, S.; Wolf, H.; Gorny, M.K.; Modrow, S. Generation of Neutralizing Human Monoclonal Antibodies against Parvovirus B19 Proteins. J. Virol. 1999, 73, 1974–1979. [Google Scholar] [CrossRef]
- McKenna, R.; Olson, N.H.; Chipman, P.R.; Baker, T.S.; Booth, T.F.; Christensen, J.; Aasted, B.; Fox, J.M.; Bloom, M.E.; Wolfinbarger, J.B.; et al. Three-Dimensional Structure of Aleutian Mink Disease Parvovirus: Implications for Disease Pathogenicity. J. Virol. 1999, 73, 6882–6891. [Google Scholar] [CrossRef]
- Spatz, S.J.; Volkening, J.D.; Mullis, R.; Li, F.; Mercado, J.; Zsak, L. Expression of Chicken Parvovirus VP2 in Chicken Embryo Fibroblasts Requires Codon Optimization for Production of Naked DNA and Vectored Meleagrid Herpesvirus Type 1 Vaccines. Virus Genes 2013, 47, 259–267. [Google Scholar] [CrossRef]
- Koo, B.-S.; Lee, H.-R.; Jeon, E.-O.; Han, M.-S.; Min, K.-C.; Lee, S.-B.; Bae, Y.-J.; Cho, S.-H.; Mo, J.-S.; Kwon, H.M.; et al. Genetic Characterization of Three Novel Chicken Parvovirus Strains Based on Analysis of Their Coding Sequences. Avian Pathol. 2015, 44, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Chacón, J.L.; Chacón, R.D.; Sánchez-Llatas, C.J.; Morín, J.G.; Astolfi-Ferreira, C.S.; Piantino Ferreira, A.J. Antigenic and Molecular Characterization of Isolates of the Brazilian Genotype BR-I (GI-11) of Infectious Bronchitis Virus Supports Its Recognition as BR-I Serotype. Avian Pathol. 2023, 52, 323–338. [Google Scholar] [CrossRef] [PubMed]
- Chacón, R.D.; Sánchez-Llatas, C.J.; Diaz Forero, A.J.; Guimarães, M.B.; Pajuelo, S.L.; Astolfi-Ferreira, C.S.; Ferreira, A.J.P. Evolutionary Analysis of a Parrot Bornavirus 2 Detected in a Sulphur-Crested Cockatoo (Cacatua Galerita) Suggests a South American Ancestor. Animals 2023, 14, 47. [Google Scholar] [CrossRef] [PubMed]
- Chacón, R.D.; Sánchez-Llatas, C.J.; L Pajuelo, S.; Diaz Forero, A.J.; Jimenez-Vasquez, V.; Médico, J.A.; Soto-Ugaldi, L.F.; Astolfi-Ferreira, C.S.; Piantino Ferreira, A.J. Molecular Characterization of the Meq Oncogene of Marek’s Disease Virus in Vaccinated Brazilian Poultry Farms Reveals Selective Pressure on Prevalent Strains. Vet. Q. 2024, 44, 1–13. [Google Scholar] [CrossRef]
- Truyen, U.; Parrish, C.R. The Evolution and Control of Parvovirus Host Ranges. Semin. Virol. 1995, 6, 311–317. [Google Scholar] [CrossRef]
- Govindasamy, L.; Hueffer, K.; Parrish, C.R.; Agbandje-McKenna, M. Structures of Host Range-Controlling Regions of the Capsids of Canine and Feline Parvoviruses and Mutants. J. Virol. 2003, 77, 12211–12221. [Google Scholar] [CrossRef]
- López-Astacio, R.A.; Adu, O.F.; Lee, H.; Hafenstein, S.L.; Parrish, C.R. The Structures and Functions of Parvovirus Capsids and Missing Pieces: The Viral DNA and Its Packaging, Asymmetrical Features, Nonprotein Components, and Receptor or Antibody Binding and Interactions. J. Virol. 2023, 97, e0016123. [Google Scholar] [CrossRef]



| Gene | Codon Position | FEL p-Value 2 | FUBAR Probability α < β Positive 2 | SLAC P-[dN/dS > 1] 2 |
|---|---|---|---|---|
| NS1 | 131 | — | 0.006 | |
| 331 | — | 0.042 | — | |
| 349 | — | 0.020 | — | |
| 534 | 0.0819 | 0.015 | — | |
| 615 | 0.0182 | 0.006 | 0.0879 | |
| 617 1 | 0.0940 | — | — | |
| 637 | 0.0098 | 0.000 | 0.0223 | |
| 640 | 0.0018 | 0.000 | 0.0061 | |
| 645 | 0.0044 | 0.000 | 0.0303 | |
| 647 | 0.0003 | 0.000 | 0.0018 | |
| 654 | 0.0133 | 0.006 | 0.0403 | |
| NP1 | 2 | 0.0872 | 0.934 | — |
| 6 | 0.0855 | 0.946 | — | |
| NP | 6 | 0.0133 | 0.972 | — |
| 15 | 0.0737 | 0.956 | — | |
| 16 | — | 0.948 | — | |
| 31 | — | 0.904 | — | |
| 35 | 0.0751 | — | — | |
| 45 | — | 0.917 | — | |
| 58 | 0.0835 | 0.916 | — | |
| 66 | — | 0.930 | — | |
| 67 | 0.0822 | — | — | |
| 68 | 0.0887 | — | — | |
| 69 | 0.0045 | 0.995 | 0.0427 | |
| VP1 | 8 | 0.0360 | 0.918 | 0.0702 |
| 85 | 0.0777 | — | — | |
| 107 | 0.0342 | 0.904 | 0.0517 | |
| 134 | 0.0724 | 0.918 | — | |
| 187 | 0.0042 | 0.983 | — | |
| 188 | 0.0416 | 0.916 | 0.0677 | |
| 192 | 0.0261 | 0.940 | 0.0618 | |
| 210 | 0.0657 | — | — | |
| 215 | 0.0056 | 0.983 | 0.0214 |
| VP1 Position 1 | Peptide | Chicken MHC-I Substitute Alleles 2 | |||
|---|---|---|---|---|---|
| ChPV | TuPV | HLA-B*40:06 | HLA-B*41:03 | HLA-B*41:04 | |
| 47 | ARKELTPQQKA | — | C | — | — |
| 48 | RKELTPQQKA | KKELTAQQKA | C + T | — | — |
| 49 | KELTPQQKA | KELTAQQKA | C + T | C + T | C |
| 84 | KEFFKNHQGA | KEFFKNHQGA | C + T | — | — |
| 128 | EEHPFNQEEL | EEAPFNEQEL | — | C + T | — |
| 135 | — | NEQELEEAM | — | T | — |
| 202 | RDMDQYKAI | RDFDKYQAI | T | C + T | C + T |
| 221 | SENQTQYF | AENETQYF | — | C + T | C + T |
| 221 | — | AENETQYFGF | — | T | T |
| 301 | QEGKYPRL | — | — | — | C |
| 301 | QEGKYPRLL | QEGRYPRIL | — | C + T | C + T |
| 353 | RESAFYCL | — | — | C | C |
| 372 | NEWETTFVF | NEWQTSYEF | C + T | C + T | C + T |
| 378 | — | YEFPDSTP | T | — | — |
| 446 | LENLANVAV | — | C | C | C |
| 469 | RPESDKDEYL | RPETDKDEYL | — | C + T | — |
| 581 | KESPGHIF | KESPGHVF | — | C + T | C + T |
| 581 | KESPGHIFV | KESPGHVFV | C + T | C + T | C + T |
| 613 | VEIEWELEP | — | C | — | — |
| 615 | — | IEWELEHFT | T | — | — |
| Total peptides | 16 | 22 | 17 | ||
| VP1 Position 1 | Peptide | Chicken MHC-II Substitute Alleles 2 | ||||
|---|---|---|---|---|---|---|
| ChPV | TuPV | DRB1:1310 | DRB1:1366 | DRB1:1445 | DRB1:1482 | |
| 5 | APKGYVPSLPTTDEE | PPKGYVPSLPTTDEE | C + T | — | — | — |
| 6 | PKGYVPSLPTTDEEA | — | C | C | — | — |
| 58 | ERKRFFITQAQKNKK | DRKRFFITQAQKNKK | C + T | — | — | — |
| 59 | RKRFFITQAQKNKKP | RKRFFITQAQKNKKP | C + T | C + T | — | — |
| 293 | GTIQIFADQEGKYPR | GTIQIFADQEGRYPR | — | — | — | C + T |
| 295 | — | TIQIFADQEGRYPRI | — | — | — | T |
| 403 | LYDTWNVNGRGDDAK | — | C | C | — | — |
| 404 | YDTWNVNGRGDDAKR | — | C | C | — | — |
| 405 | DTWNVNGRGDDAKRG | — | C | C | — | — |
| 431 | — | GPYIYLSDTTAAGQQ | — | T | — | — |
| 494 | VRNSQIQVSTANKVQ | — | — | — | C | C |
| 495 | RNSQIQVSTANKVQV | — | — | — | C | C |
| 496 | NSQIQVSTANKVQVD | — | — | — | C | C |
| 497 | SQIQVSTANKVQVDT | — | — | — | C | C |
| 498 | QIQVSTANKVQVDTS | — | — | — | C | C |
| 585 | GHIFVKVTPKPTGAA | GHVFVKVTPKPTGAA | — | — | C + T | — |
| 586 | HIFVKVTPKPTGAAN | HVFVKVTPKPTGAAN | — | — | C + T | — |
| 648 | DENGQYQVNVNSGDI | DENGQYQINTTSADL | C + T | T | — | — |
| 649 | ENGQYQVNVNSGDIT | ENGQYQINTTSADLA | C + T | C + T | — | — |
| 650 | NGQYQVNVNSGDITR | NGQYQINTTSADLAR | C + T | C + T | — | — |
| 651 | — | GQYQINTTSADLARL | T | T | — | — |
| 661 | DITRLYMTKRAPRTN | — | — | — | C | — |
| Total peptides | 17 | 13 | 10 | 8 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chacón, R.D.; Sánchez-Llatas, C.J.; da Costa, A.C.; Valdeiglesias Ichillumpa, S.; Cea-Callejo, P.; Marín-Sánchez, O.; Astolfi-Ferreira, C.S.; Santander-Parra, S.; Nuñez, L.F.N.; Piantino Ferreira, A.J. Molecular and Evolutionary Characteristics of Chicken Parvovirus (ChPV) Genomes Detected in Chickens with Runting–Stunting Syndrome. Viruses 2024, 16, 1389. https://doi.org/10.3390/v16091389
Chacón RD, Sánchez-Llatas CJ, da Costa AC, Valdeiglesias Ichillumpa S, Cea-Callejo P, Marín-Sánchez O, Astolfi-Ferreira CS, Santander-Parra S, Nuñez LFN, Piantino Ferreira AJ. Molecular and Evolutionary Characteristics of Chicken Parvovirus (ChPV) Genomes Detected in Chickens with Runting–Stunting Syndrome. Viruses. 2024; 16(9):1389. https://doi.org/10.3390/v16091389
Chicago/Turabian StyleChacón, Ruy D., Christian J. Sánchez-Llatas, Antonio Charlys da Costa, Stefhany Valdeiglesias Ichillumpa, Pablo Cea-Callejo, Obert Marín-Sánchez, Claudete S. Astolfi-Ferreira, Silvana Santander-Parra, Luis F. N. Nuñez, and Antonio J. Piantino Ferreira. 2024. "Molecular and Evolutionary Characteristics of Chicken Parvovirus (ChPV) Genomes Detected in Chickens with Runting–Stunting Syndrome" Viruses 16, no. 9: 1389. https://doi.org/10.3390/v16091389
APA StyleChacón, R. D., Sánchez-Llatas, C. J., da Costa, A. C., Valdeiglesias Ichillumpa, S., Cea-Callejo, P., Marín-Sánchez, O., Astolfi-Ferreira, C. S., Santander-Parra, S., Nuñez, L. F. N., & Piantino Ferreira, A. J. (2024). Molecular and Evolutionary Characteristics of Chicken Parvovirus (ChPV) Genomes Detected in Chickens with Runting–Stunting Syndrome. Viruses, 16(9), 1389. https://doi.org/10.3390/v16091389







