mRNA Splicing of UL44 and Secretion of Alphaherpesvirinae Glycoprotein C (gC) Is Conserved among the Mardiviruses
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Cultures
2.2. Viruses
2.3. Western Blot Analysis of Secreted gC Proteins
2.4. RNA Extraction and RT-PCR Analysis
2.5. DNA Sequencing
2.6. Generation of Recombinant (r)HVT
2.7. Generation of Recombinant 301B/1 with Mutations in the Donor, BP, and Acceptor Sites
3. Results
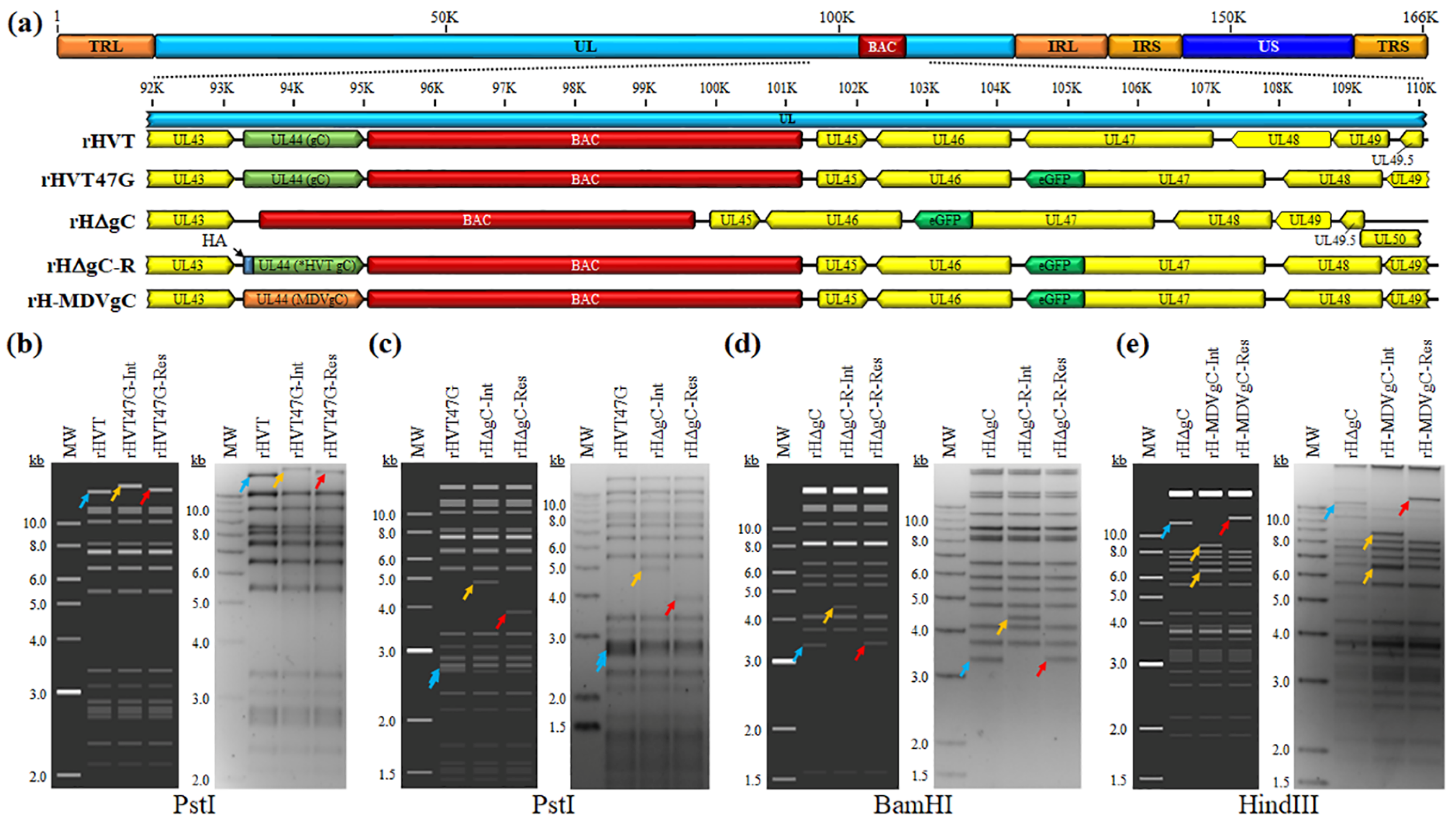
3.1. Generation of rHVT
3.1.1. Generation of rHVT47G
3.1.2. Generation of rHVT Lacking gC or Expressing HA-Tagged HVT gC or MDV gC
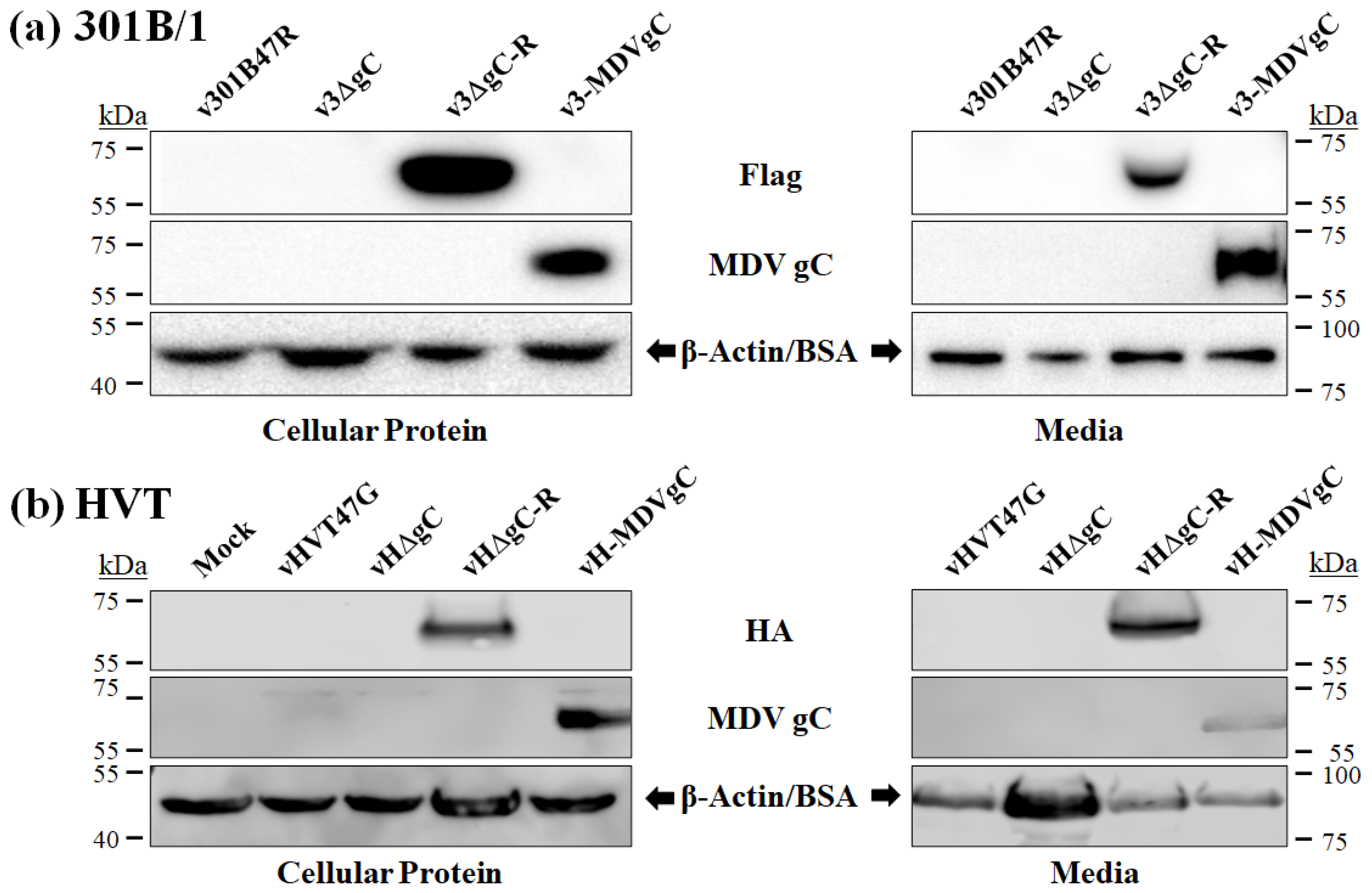
3.2. Secretion of HVT gC Protein
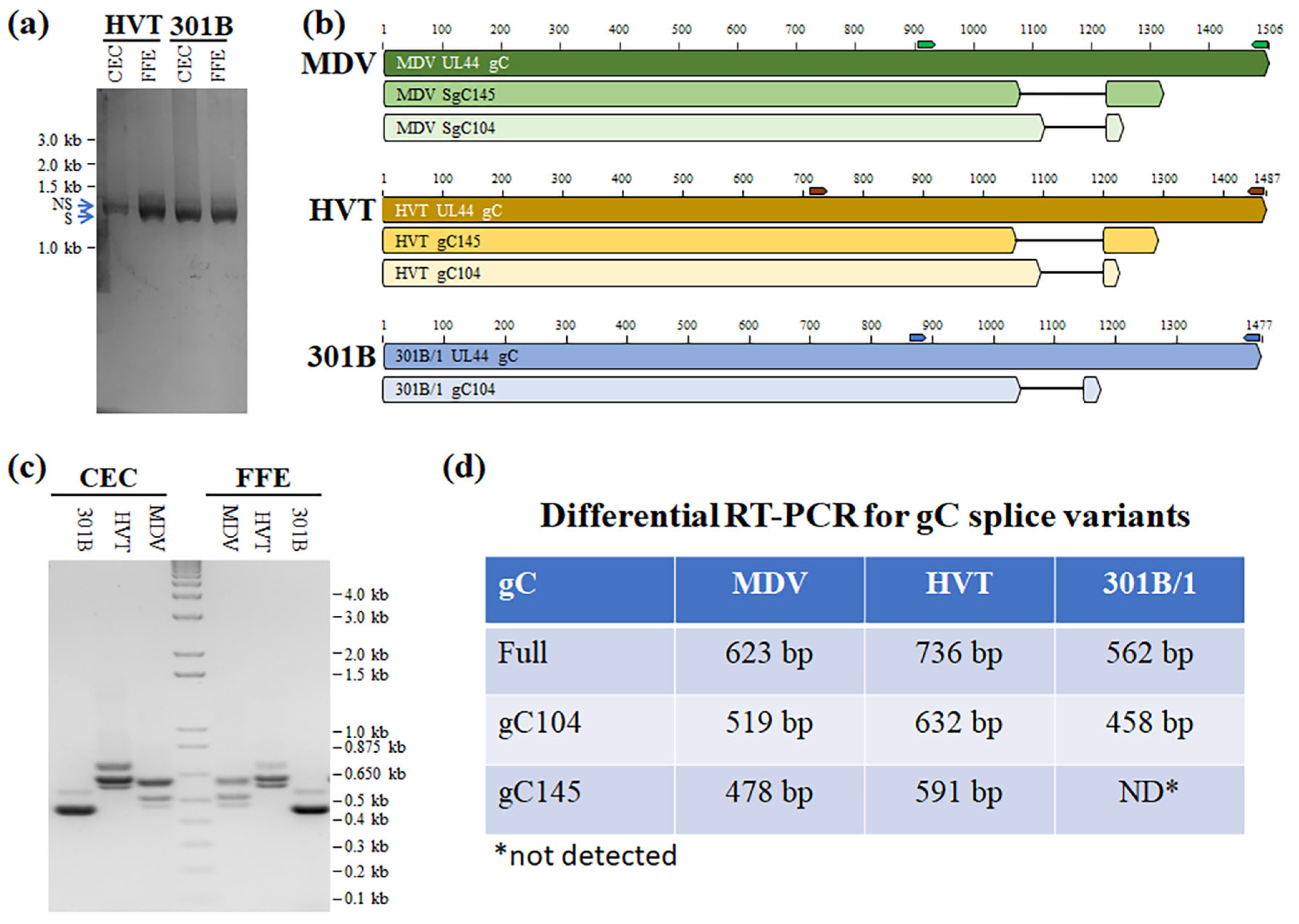
3.3. Identification of HVT and 301B/1 UL44 (gC) mRNA Splice Variants
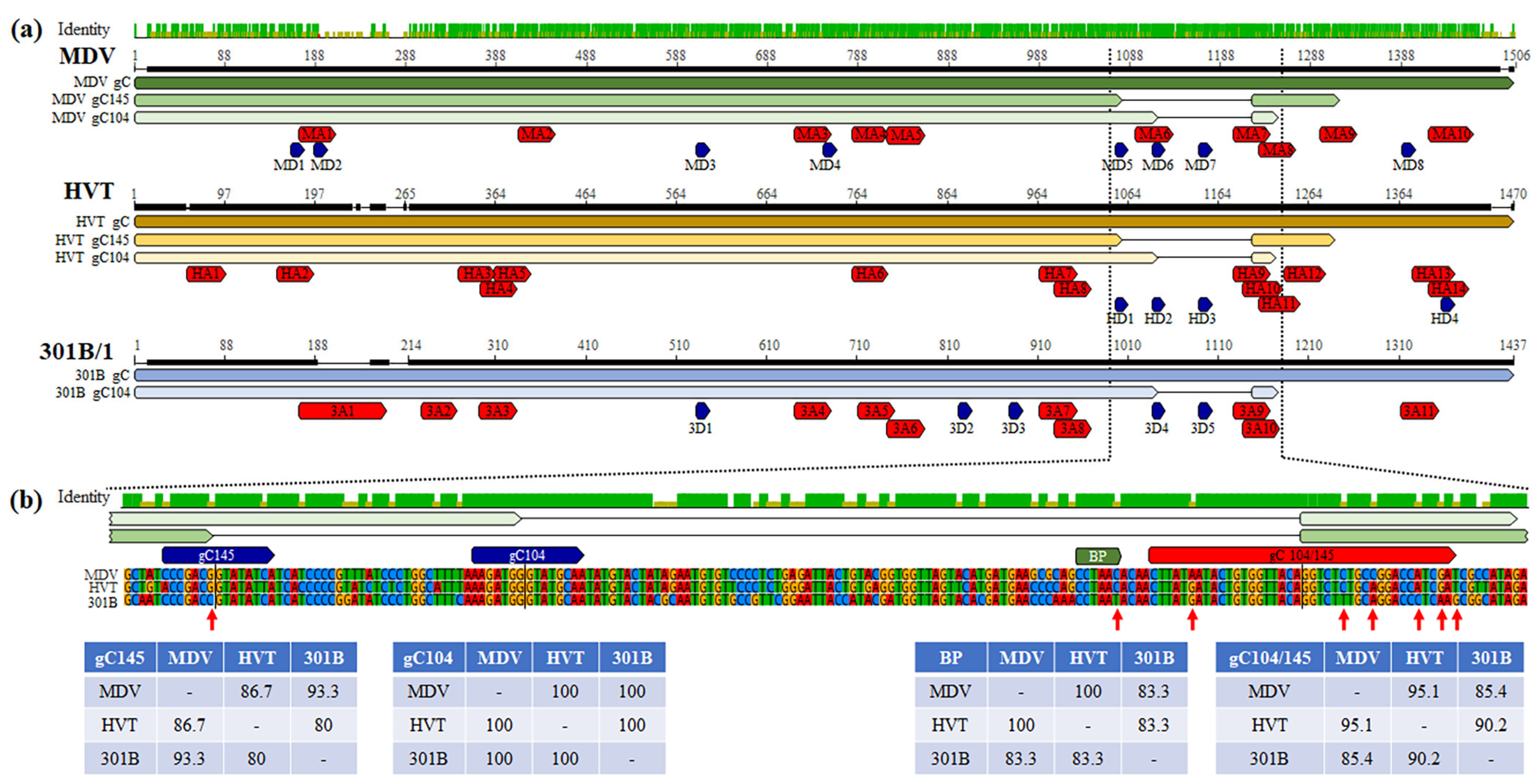
3.4. Conserved mRNA Splicing Donor and Acceptor Sequences
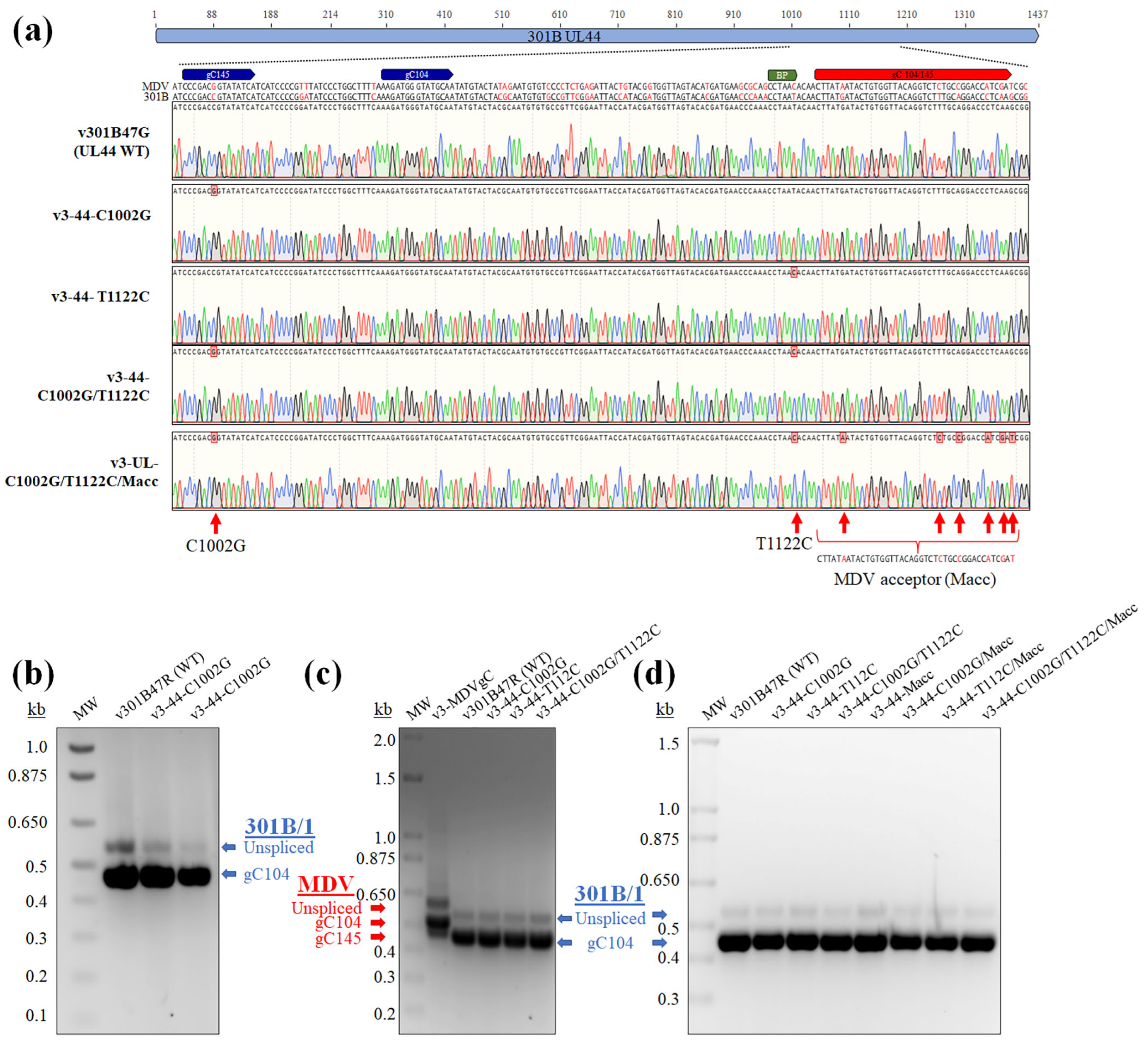
3.5. Modification of 301B/1 Donor, BP, and Acceptor Sequences Does Not Rescue gC145 Splicing
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gatherer, D.; Depledge, D.P.; Hartley, C.A.; Szpara, M.L.; Vaz, P.K.; Benko, M.; Brandt, C.R.; Bryant, N.A.; Dastjerdi, A.; Doszpoly, A.; et al. ICTV Virus Taxonomy Profile: Herpesviridae 2021. J. Gen. Virol. 2021, 102, 001673. [Google Scholar] [CrossRef] [PubMed]
- Calnek, B.W. Pathogenesis of Marek’s disease virus infection. Curr. Top. Microbiol. Immunol. 2001, 255, 25–55. [Google Scholar] [CrossRef] [PubMed]
- Baaten, B.J.; Staines, K.A.; Smith, L.P.; Skinner, H.; Davison, T.F.; Butter, C. Early replication in pulmonary B cells after infection with Marek’s disease herpesvirus by the respiratory route. Viral Immunol. 2009, 22, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Witter, R.L. Increased virulence of Marek’s disease virus field isolates. Avian Dis. 1997, 41, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Banfield, B.W.; Leduc, Y.; Esford, L.; Visalli, R.J.; Brandt, C.R.; Tufaro, F. Evidence for an interaction of herpes simplex virus with chondroitin sulfate proteoglycans during infection. Virology 1995, 208, 531–539. [Google Scholar] [CrossRef]
- Mettenleiter, T.C. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology 1989, 171, 623–625. [Google Scholar] [CrossRef] [PubMed]
- Rue, C.A.; Ryan, P. Characterization of pseudorabies virus glycoprotein C attachment to heparan sulfate proteoglycans. J. Gen. Virol. 2002, 83, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Osterrieder, N. Construction and characterization of an equine herpesvirus 1 glycoprotein C negative mutant. Virus Res. 1999, 59, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Allen, G.P.; Coogle, L.D. Characterization of an equine herpesvirus type 1 gene encoding a glycoprotein (gp13) with homology to herpes simplex virus glycoprotein C. J. Virol. 1988, 62, 2850–2858. [Google Scholar] [CrossRef]
- Eisenberg, R.J.; Ponce de Leon, M.; Friedman, H.M.; Fries, L.F.; Frank, M.M.; Hastings, J.C.; Cohen, G.H. Complement component C3b binds directly to purified glycoprotein C of herpes simplex virus types 1 and 2. Microb. Pathog. 1987, 3, 423–435. [Google Scholar] [CrossRef]
- Huemer, H.P.; Larcher, C.; van Drunen Littel-van den Hurk, S.; Babiuk, L.A. Species selective interaction of Alphaherpesvirinae with the “unspecific” immune system of the host. Arch. Virol. 1993, 130, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Huemer, H.P.; Nowotny, N.; Crabb, B.S.; Meyer, H.; Hubert, P.H. gp13 (EHV-gC): A complement receptor induced by equine herpesviruses. Virus Res. 1995, 37, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Smiley, L.; Seidel, C.; Friedman, H.M. Glycoprotein-C of Herpes-Simplex Virus Type-I Functions as a C3b Receptor. Clin. Res. 1984, 32, A384. [Google Scholar]
- Jarosinski, K.W.; Margulis, N.G.; Kamil, J.P.; Spatz, S.J.; Nair, V.K.; Osterrieder, N. Horizontal transmission of Marek’s disease virus requires US2, the UL13 protein kinase, and gC. J. Virol. 2007, 81, 10575–10587. [Google Scholar] [CrossRef] [PubMed]
- Vega-Rodriguez, W.; Xu, H.; Ponnuraj, N.; Akbar, H.; Kim, T.; Jarosinski, K.W. The requirement of glycoprotein C (gC) for interindividual spread is a conserved function of gC for avian herpesviruses. Sci. Rep. 2021, 11, 7753. [Google Scholar] [CrossRef] [PubMed]
- Moffat, J.F.; Zerboni, L.; Kinchington, P.R.; Grose, C.; Kaneshima, H.; Arvin, A.M. Attenuation of the vaccine Oka strain of varicella-zoster virus and role of glycoprotein C in alphaherpesvirus virulence demonstrated in the SCID-hu mouse. J. Virol. 1998, 72, 965–974. [Google Scholar] [CrossRef] [PubMed]
- Isfort, R.J.; Stringer, R.A.; Kung, H.J.; Velicer, L.F. Synthesis, processing, and secretion of the Marek’s disease herpesvirus A antigen glycoprotein. J. Virol. 1986, 57, 464–474. [Google Scholar] [CrossRef] [PubMed]
- Jarosinski, K.W.; Osterrieder, N. Marek’s disease virus expresses multiple UL44 (gC) variants through mRNA splicing that are all required for efficient horizontal transmission. J. Virol. 2012, 86, 7896–7906. [Google Scholar] [CrossRef] [PubMed]
- Volkening, J.D.; Spatz, S.J.; Ponnuraj, N.; Akbar, H.; Arrington, J.V.; Vega-Rodriguez, W.; Jarosinski, K.W. Viral proteogenomic and expression profiling during productive replication of a skin-tropic herpesvirus in the natural host. PLoS Pathog. 2023, 19, e1011204. [Google Scholar] [CrossRef] [PubMed]
- Sedlackova, L.; Perkins, K.D.; Lengyel, J.; Strain, A.K.; van Santen, V.L.; Rice, S.A. Herpes simplex virus type 1 ICP27 regulates expression of a variant, secreted form of glycoprotein C by an intron retention mechanism. J. Virol. 2008, 82, 7443–7455. [Google Scholar] [CrossRef]
- Sedlackova, L.; Perkins, K.D.; Meyer, J.; Strain, A.K.; Goldman, O.; Rice, S.A. Identification of an ICP27-responsive element in the coding region of a herpes simplex virus type 1 late gene. J. Virol. 2010, 84, 2707–2718. [Google Scholar] [CrossRef] [PubMed]
- Ponnuraj, N.; Tien, Y.T.; Vega-Rodriguez, W.; Krieter, A.; Jarosinski, K.W. The Herpesviridae Conserved Multifunctional Infected-Cell Protein 27 (ICP27) Is Important but Not Required for Replication and Oncogenicity of Marek’s Disease Alphaherpesvirus. J. Virol. 2019, 93, e01903–e01918. [Google Scholar] [CrossRef] [PubMed]
- Chuard, A.; Courvoisier-Guyader, K.; Remy, S.; Spatz, S.; Denesvre, C.; Pasdeloup, D. The Tegument Protein pUL47 of Marek’s Disease Virus Is Necessary for Horizontal Transmission and Is Important for Expression of Glycoprotein gC. J. Virol. 2020, 95, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Schat, K.A.; Sellers, H.S. Cell-culture methods. In A Laboratory Manual for the Identification and Characterization of Avian Pathogens, 5th ed.; Dufour-Zavala, L., Swayne, D.E., Glisson, J.R., Pearson, J.E., Reed, W.M., Jackwood, M.W., Woolcock, P.R., Eds.; American Association of Avian Pathologists: Jacksonville, FL, USA, 2008; pp. 195–203. [Google Scholar]
- Niikura, M.; Kim, T.; Silva, R.F.; Dodgson, J.; Cheng, H.H. Virulent Marek’s disease virus generated from infectious bacterial artificial chromosome clones with complete DNA sequence and the implication of viral genetic homogeneity in pathogenesis. J. Gen. Virol. 2011, 92, 598–607. [Google Scholar] [CrossRef]
- Tischer, B.K.; Schumacher, D.; Chabanne-Vautherot, D.; Zelnik, V.; Vautherot, J.F.; Osterrieder, N. High-level expression of Marek’s disease virus glycoprotein C is detrimental to virus growth in vitro. J. Virol. 2005, 79, 5889–5899. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tischer, B.K.; Smith, G.A.; Osterrieder, N. En passant mutagenesis: A two step markerless red recombination system. Methods Mol. Biol. 2010, 634, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Jarosinski, K.W.; O’Connell, P.H.; Schat, K.A. Impact of deletions within the Bam HI-L fragment of attenuated Marek’s disease virus on vIL-8 expression and the newly identified transcript of open reading frame LORF4. Virus Genes 2003, 26, 255–269. [Google Scholar] [CrossRef] [PubMed]
- Jarosinski, K.W.; Schat, K.A. Multiple alternative splicing to exons II and III of viral interleukin-8 (vIL-8) in the Marek’s disease virus genome: The importance of vIL-8 exon I. Virus Genes 2007, 34, 9–22. [Google Scholar] [CrossRef]
- Jarosinski, K.W. Dual infection and superinfection inhibition of epithelial skin cells by two alphaherpesviruses co-occur in the natural host. PLoS ONE 2012, 7, e37428. [Google Scholar] [CrossRef][Green Version]
- Jarosinski, K.W.; Arndt, S.; Kaufer, B.B.; Osterrieder, N. Fluorescently tagged pUL47 of Marek’s disease virus reveals differential tissue expression of the tegument protein in vivo. J. Virol. 2012, 86, 2428–2436. [Google Scholar] [CrossRef][Green Version]
- Jarosinski, K.W.; Carpenter, J.E.; Buckingham, E.M.; Jackson, W.; Knudtson, K.; Moffat, J.F.; Kita, H.; Grose, C. Cellular Stress Response to Varicella-Zoster Virus Infection of Human Skin Includes Highly Elevated Interleukin-6 Expression. Open Forum Infect. Dis. 2018, 5, ofy118. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, M.; Elliott, G. Fluorescent tagging of herpes simplex virus tegument protein VP13/14 in virus infection. J. Virol. 2001, 75, 2575–2583. [Google Scholar] [CrossRef] [PubMed]
- Verhagen, J.; Hutchinson, I.; Elliott, G. Nucleocytoplasmic shuttling of bovine herpesvirus 1 UL47 protein in infected cells. J. Virol. 2006, 80, 1059–1063. [Google Scholar] [CrossRef][Green Version]
- Reese, M.G.; Eeckman, F.H.; Kulp, D.; Haussler, D. Improved splice site detection in Genie. J. Comput. Biol. 1997, 4, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Majerciak, V.; Yamanegi, K.; Allemand, E.; Kruhlak, M.; Krainer, A.R.; Zheng, Z.M. Kaposi’s sarcoma-associated herpesvirus ORF57 functions as a viral splicing factor and promotes expression of intron-containing viral lytic genes in spliceosome-mediated RNA splicing. J. Virol. 2008, 82, 2792–2801. [Google Scholar] [CrossRef]
- Nojima, T.; Oshiro-Ideue, T.; Nakanoya, H.; Kawamura, H.; Morimoto, T.; Kawaguchi, Y.; Kataoka, N.; Hagiwara, M. Herpesvirus protein ICP27 switches PML isoform by altering mRNA splicing. Nucleic Acids Res. 2009, 37, 6515–6527. [Google Scholar] [CrossRef]
| Virus 1 | Gene 2 | Direction | Sequence (5′→3′) 3 | Purpose 4 |
|---|---|---|---|---|
| HVT | UL47 | Forward | CGTGGCAAGTCTGCGGTTGG | Sequencing |
| Reverse | CAACCACATTGCTTCCGC | |||
| HVT | UL44 | Forward | CGTAAGCTTTGTGTTTTATTGAGCGGTCG | Cloning |
| Reverse | CGTTCTAGATTTGGCCGCTGCGTGATACC | |||
| HVT | UL44 | Forward | CGACGGGATCCCCAGGGTTCTTTCTGGACTAGTCCTACACCCCGTGGAAATAGGGATAACAGGGTAATCGATTT | Shuttle Vector |
| Reverse | TTAACGGATCCGCCAGTGTTACAACCAATTAACC | |||
| MDV | UL44 | Forward | TATCTTCCCTCATGCTCACG | RT-PCR (full) |
| Forward | CTCGGAAAGATCGCGATGGA | RT-PCR (diff) | ||
| Reverse | CACAACTACATAACAATGAGATTATAATCG | RT-PCR (full/diff) | ||
| 301B | UL44 | Forward | CCCATGCACGCGTCACG | RT-PCR (full) |
| Forward | GGGAGCCACCCTGGTATCTA | RT-PCR (diff) | ||
| Reverse | GACGGACGAAAGTAATATGTATTTTTTCCCG | RT-PCR (full/diff) | ||
| HVT | UL44 | Forward | CCGCAAGTATATGGTTTCCAACATGC | RT-PCR (full) |
| Forward | GTGTGCCAACGACCCGCATC | RT-PCR (diff) | ||
| Reverse | CTTAATTCCGCCCCGGTAGG | RT-PCR (full/diff) |
| Modification 1 | Direction | Sequence (5′→3′) 2 |
|---|---|---|
| eGFP at C-term of HVT UL47 | Forward | gatgccaaagatgtaatatttgaaccgcgcccttttaaaATGgtgagcaagggcgaggag |
| Reverse | aacacccctccgcgtcgggcaagagtttcggaagcacgCTActtgtacagctcgtccatgccg | |
| Removal of HVT gC (ΔgC) | Forward | tttataccatattacgcatctatcgaaacttgttcgagaaccgcaagtatTAAgattaaccatcgtattagggataacagggtaatcgattt |
| Reverse | ggttataacacttaataatttttatatcacatacgatggttaatcTTAatacttgcggttctcgaacagccagtgttacaaccaattaacc | |
| HVT gC into rHΔgC | Forward | tttataccatattacgcatctatcgaaacttgtgttttattgagcggtcg |
| Reverse | ggttataacacttaataatttttatatcacctgatcagcgggtttaaacg | |
| HA at N-term of HVT gC | Forward | agttctgtctttacagcaaacctcttgtgccggattgccctatccgtatgatgtgccggattatgcgtagggataacagggtaatcgattt |
| Reverse | ttgaaagttaggatatgatgggtatcgacgttatgcgcataatccggcacatcatacggatagggcaatccggcagccagtgttacaaccaattaacc | |
| MDV gC into rHΔgC | Forward | tttataccatattacgcatctatcgaaacttgttcgagaatatcttccctcatgctcacg |
| Reverse | ggttataacacttaataatttttatatcacatacgatggtcataacaatgagattataat |
| Modification 1 | Direction | Sequence (5′→3′) 2 |
|---|---|---|
| UL44 C1002G (gC145 donor) | Forward | agcagggaaatatgatgagtactacgaacgccactgcaatcccgacGgtatatcatcatccccggtagggataacagggtaatcgattt |
| Reverse | ttgcatacccatctttgaaagccagggatatccggggatgatgatataCcgtcgggattgcagtgggccagtgttacaaccaattaacc | |
| UL44 T1122C (BP) | Forward | ggaattaccatacgatggttagtacacgatgaacccaaacctaaCacaacttatgatacttagggataacagggtaatcgattt |
| Reverse | cttgagggtcctgcaaagacctgtaaccacagtatcataagttgtGttaggtttgggttcgccagtgttacaaccaattaacc | |
| UL44 MDV acceptor (Macc) | Forward | tagtacacgatgaacccaaacctaatacaacttatAatactgtggttacaggtctCtgcCggaccAtcGaTtagggataacagggtaatcgattt |
| Forward * | tagtacacgatgaacccaaacctaaCacaacttatAatactgtggttacaggtctCtgcCggaccAtcGaTtagggataacagggtaatcgattt | |
| Reverse | aatattcggctgatgatatttctatgccgAtCgaTggtccGgcaGagacctgtaaccacagtatTgccagtgttacaaccaattaacc |
| Virus | Name 1 | Start 2 | End 3 | Score 4 | Exon/Intron (5′-3′) |
|---|---|---|---|---|---|
| MDV | MD1 | 162 | 176 | 0.90 | aactgag/gtacctca |
| MD2 | 187 | 201 | 0.29 | acagaaa/gtgtgtca | |
| MD3 | 611 | 625 | 0.25 | gaaacaa/gtacttca | |
| MD4 | 751 | 765 | 0.18 | tacatac/gtgtgtgt | |
| MD5 (gC145) | 1074 | 1088 | 0.24 | cccgacg/gtatatca | |
| MD6 (gC104) | 1115 | 1129 | 0.69 | aagatgg/gtatgcaa | |
| MD7 | 1166 | 1180 | 0.95 | tacggtg/gttagtac | |
| MD8 | 1392 | 1406 | 0.16 | acccatg/gttattac | |
| HVT | HD1 (gC145) | 1050 | 1064 | 0.15 | accgacg/gtatatta |
| HD2 (gC104) | 1091 | 1105 | 0.69 | aagatgg/gtatgcaa | |
| HD3 | 1142 | 1156 | 0.54 | tgaggtg/gttagttc | |
| HD4 | 1411 | 1425 | 0.12 | tgaggtg/gttagttc | |
| 301B/1 | 3D1 | 533 | 547 | 0.18 | gagacaa/gtacttca |
| 3D2 | 823 | 837 | 0.99 | acaccac/gtaaggac | |
| 3D3 | 879 | 893 | 0.14 | caccctg/gtatctac | |
| 3D4 (gC104) | 1037 | 1051 | 0.69 | aagatgg/gtatgcaa | |
| 3D5 | 1088 | 1102 | 0.98 | tacgatg/gttagtac |
| Virus | Name 1 | Start 2 | End 3 | Score 4 | Intron/Exon (5′-3′) |
|---|---|---|---|---|---|
| MDV | MA1 | 170 | 210 | 0.70 | tacctcatgcaccttccaca/gaaagtgtgtcaacaaattcg |
| MA2 | 413 | 453 | 0.89 | aatggtccaactttgttcta/gatctgatctttaacccaatt | |
| MA3 | 719 | 759 | 0.19 | attttaatcggcctttaata/gataaacatatttacatacgt | |
| MA4 | 782 | 822 | 0.20 | tggatgtactggcccctcca/gtcctcagcggagaaaattac | |
| MA5 | 822 | 862 | 0.11 | caaggcatcttgtatcgtta/gacacttttatccccctggat | |
| MA6 | 1097 | 1137 | 0.23 | gtttatccctggcttttaaa/gatgggtatgcaatatgtact | |
| MA7 (gC104/145) | 1205 | 1245 | 0.73 | cttataatactgtggttaca/ggtctctgccggaccatcgat | |
| MA8 | 1233 | 1273 | 0.42 | ccggaccatcgatcgccata/gaaatctcctcagccgcattc | |
| MA9 | 1301 | 1341 | 0.31 | aatatacgtgcagactcata/ggctaccccttcgatgaagat | |
| MA10 | 1421 | 1461 | 0.48 | tgggattggctgtaatttta/gggatggggataatcatgact | |
| HVT | HA1 | 58 | 98 | 0.96 | tttctagttctgtctttaca/gcaaacctcttgtgccggatt |
| HA2 | 155 | 195 | 0.47 | ccgatggcgttcctttgtca/gaggtgcccaattcgcctacg | |
| HA3 | 323 | 363 | 0.13 | tcgtattccttaacaataca/ggaagaattttgtgtgacctt | |
| HA4 | 347 | 387 | 0.33 | gaattttgtgtgaccttata/gtcgaccccccttcagacgat | |
| HA5 | 362 | 402 | 0.38 | ttatagtcgaccccccttca/gacgatgaatggtccaacttc | |
| HA6 | 758 | 798 | 0.69 | tggatgtattggcccctcca/gttctcagcggagaaaactac | |
| HA7 | 967 | 1007 | 0.84 | ggactcgaatctcctccaaa/ggtttcctgcttggtagcgtg | |
| HA8 | 983 | 1023 | 0.28 | caaaggtttcctgcttggta/gcgtggaggcaaggcgatatg | |
| HA9 (gC104/145) | 1181 | 1221 | 0.51 | cttatgatactgtggttaca/ggtctctgcaggaccatcgat | |
| HA10 | 1191 | 1231 | 0.81 | tgtggttacaggtctctgca/ggaccatcgatcgttatagaa | |
| HA11 | 1209 | 1249 | 0.58 | caggaccatcgatcgttata/gaaatctcgccagtcggattc | |
| HA12 | 1237 | 1277 | 0.37 | gccagtcggattccagtcca/ggacaactgggcgaaaacgaa | |
| HA13 | 1379 | 1419 | 0.29 | taacaattacggccgttcta/ggactggccttgtttttaggt | |
| HA14 | 1397 | 1437 | 0.95 | taggactggccttgttttta/ggtattggtatcattatcaca | |
| 301B/1 | 3A1 | 170 | 210 | 0.17 | ttcctgcatccccgccggca/ggggagaaagaggagagccac |
| 3A2 | 227 | 267 | 0.13 | cgcgtaggatgcctagtata/gtttgcgataaagaagaagtt | |
| 3A3 | 292 | 332 | 0.56 | cgtttcgtgtgcactcttaa/gatcgcccctccctccgacaa | |
| 3A4 | 641 | 681 | 0.45 | ctttcaatcggcctttacta/gataagcacgtgtacatccgc | |
| 3A5 | 711 | 751 | 0.43 | tctagccccccccgtcctca/gtggcgataagtacaaggctt | |
| 3A6 | 744 | 784 | 0.26 | caaggcttcatgcatcgtta/ggcatttttatccaccgggct | |
| 3A7 | 913 | 953 | 0.36 | gccatcgaccctcctcccaa/gatttcatgtctggtagcctg | |
| 3A8 | 929 | 969 | 0.17 | ccaagatttcatgtctggta/gcctggaagcagggaaatatg | |
| 3A9 (gC104/145) | 1127 | 1167 | 0.56 | cttatgatactgtggttaca/ggtctttgcaggaccctcaag | |
| 3A10 | 1137 | 1177 | 0.85 | tgtggttacaggtctttgca/ggaccctcaagcggcatagaa | |
| 3A11 | 1313 | 1353 | 0.10 | cccccatggttctcgcgata/gcggctgttgtgggactagct |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, H.; Vega-Rodriguez, W.; Campos, V.; Jarosinski, K.W. mRNA Splicing of UL44 and Secretion of Alphaherpesvirinae Glycoprotein C (gC) Is Conserved among the Mardiviruses. Viruses 2024, 16, 782. https://doi.org/10.3390/v16050782
Xu H, Vega-Rodriguez W, Campos V, Jarosinski KW. mRNA Splicing of UL44 and Secretion of Alphaherpesvirinae Glycoprotein C (gC) Is Conserved among the Mardiviruses. Viruses. 2024; 16(5):782. https://doi.org/10.3390/v16050782
Chicago/Turabian StyleXu, Huai, Widaliz Vega-Rodriguez, Valeria Campos, and Keith W. Jarosinski. 2024. "mRNA Splicing of UL44 and Secretion of Alphaherpesvirinae Glycoprotein C (gC) Is Conserved among the Mardiviruses" Viruses 16, no. 5: 782. https://doi.org/10.3390/v16050782
APA StyleXu, H., Vega-Rodriguez, W., Campos, V., & Jarosinski, K. W. (2024). mRNA Splicing of UL44 and Secretion of Alphaherpesvirinae Glycoprotein C (gC) Is Conserved among the Mardiviruses. Viruses, 16(5), 782. https://doi.org/10.3390/v16050782





