Exploring Viral Genome Profile in Mpox Patients during the 2022 Outbreak, in a North-Eastern Centre of Italy
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples
2.2. DNA Extraction, Library Preparation and NGS
2.3. Bioinformatics Analysis
- -
- Quality control (QC), performed using FastQC v.0.11.9;
- -
- Adapter removal, performed using Fastp v.0.23.2;
- -
- Contamination control, performed using Kraken2 v. 2.1.2;
- -
- Alignment of the reads to the MPXV reference genome (NC_063383.1), performed using BWA-mem2 v. 2.2.1;
- -
- Genome assembly, performed using iVar consensus v.1.3.1 tool (options “-aa -A -d o -Q 0”).
2.4. GISAID Data Download
2.5. Sanger Sequencing
2.6. Ethics
3. Results
3.1. QC Analyses, Read Alignment and Consensus Generation
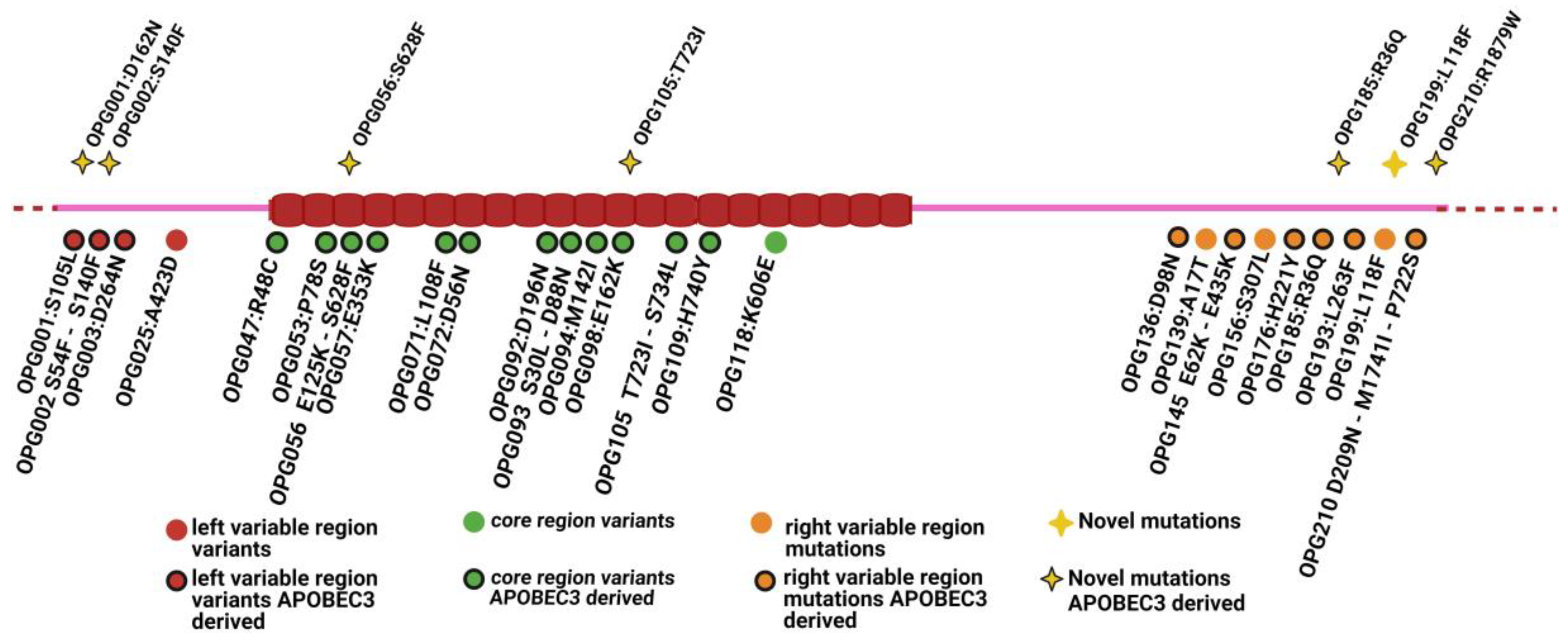
3.2. Variant Calling and Annotation
3.3. APOBEC3-Driven Mutations
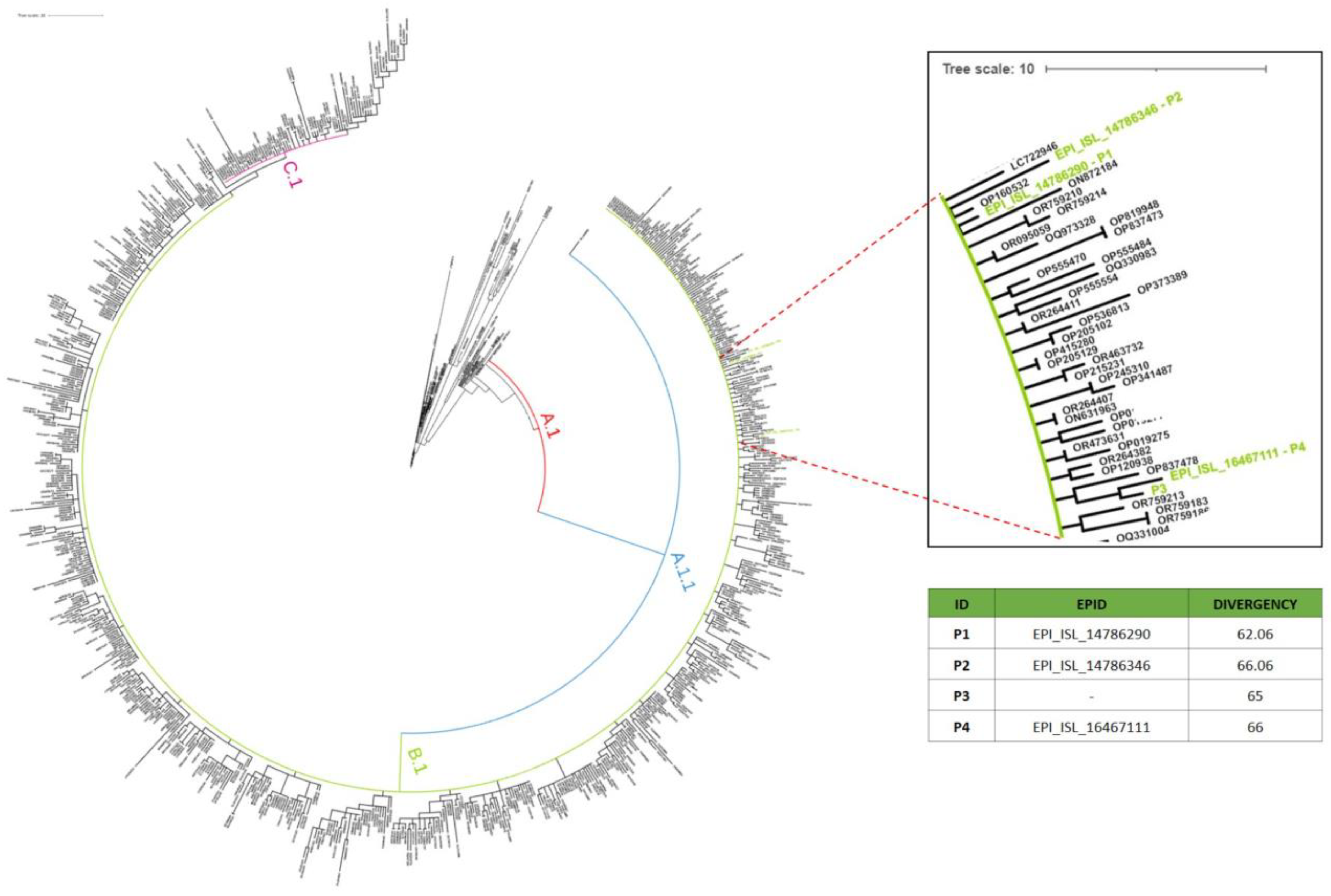
3.4. Phylogenetic Analysis
3.5. APOBEC3-Driven Mutations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Petersen, E.; Kantele, A.; Koopmans, M.; Asogun, D.; Yinka-Ogunleye, A.; Ihekweazu, C.; Zumla, A. Human Monkeypox. Infect. Dis. Clin. N. Am. 2019, 33, 1027–1043. [Google Scholar] [CrossRef]
- Shchelkunova, G.A.; Shchelkunov, S.N. Smallpox, Monkeypox and Other Human Orthopoxvirus Infections. Viruses 2022, 15, 103. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Director-General’s Statement at the Press Conference Following IHR Emergency Committee Regarding the Multi-Country Outbreak of Monkeypox—23 July 2022. Available online: https://www.who.int/director-general/speeches/detail/who-director-general-s-statement-on-the-press-conference-following-IHR-emergency-committee-regarding-the-multi--country-outbreak-of-monkeypox--23-july-2022 (accessed on 27 April 2024).
- Centers for Disease Control and Prevention. Monkeypox and Orthopoxvirus Outbreak Global Map. 2022. Available online: https://www.cdc.gov/poxvirus/mpox/response/2022/world-map.html (accessed on 23 April 2024).
- Shchelkunov, S.N.; Totmenin, A.V.; Safronov, P.F.; Mikheev, M.V.; Gutorov, V.V.; Ryazankina, O.I.; Petrov, N.A.; Babkin, I.V.; Uvarova, E.A.; Sandakhchiev, L.S.; et al. Analysis of the Monkeypox Virus Genome. Virology 2002, 297, 172–194. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Shang, J.; Weng, S.; Aliyari, S.R.; Ji, C.; Cheng, G.; Wu, A. Genomic Annotation and Molecular Evolution of Monkeypox Virus Outbreak in 2022. J. Med. Virol. 2023, 95, e28036. [Google Scholar] [CrossRef] [PubMed]
- Bunge, E.M.; Hoet, B.; Chen, L.; Lienert, F.; Weidenthaler, H.; Baer, L.R.; Steffen, R. The Changing Epidemiology of Human Monkeypox—A Potential Threat? A Systematic Review. PLoS Negl. Trop. Dis. 2022, 16, e0010141. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Bhattacharya, M.; Sharma, A.R.; Dhama, K. Evolution, Epidemiology, Geographical Distribution, and Mutational Landscape of Newly Emerging Monkeypox Virus. GeroScience 2022, 44, 2895–2911. [Google Scholar] [CrossRef] [PubMed]
- Desingu, P.A.; Rubeni, T.P.; Sundaresan, N.R. Evolution of Monkeypox Virus from 2017 to 2022: In the Light of Point Mutations. Front. Microbiol. 2022, 13, 1037598. [Google Scholar] [CrossRef]
- Forni, D.; Moltrasio, C.; Sironi, M.; Mozzi, A.; Quattri, E.; Venegoni, L.; Zamprogno, M.; Citterio, A.; Clerici, M.; Marzano, A.V.; et al. Whole-genome Sequencing of hMPXV1 in Five Italian Cases Confirms the Occurrence of the Predominant Epidemic Lineage. J. Med. Virol. 2023, 95, e28493. [Google Scholar] [CrossRef] [PubMed]
- Luna, N.; Muñoz, M.; Bonilla-Aldana, D.K.; Patiño, L.H.; Kasminskaya, Y.; Paniz-Mondolfi, A.; Ramírez, J.D. Monkeypox Virus (MPXV) Genomics: A Mutational and Phylogenomic Analyses of B.1 Lineages. Travel Med. Infect. Dis. 2023, 52, 102551. [Google Scholar] [CrossRef] [PubMed]
- Lum, F.-M.; Torres-Ruesta, A.; Tay, M.Z.; Lin, R.T.P.; Lye, D.C.; Rénia, L.; Ng, L.F.P. Monkeypox: Disease Epidemiology, Host Immunity and Clinical Interventions. Nat. Rev. Immunol. 2022, 22, 597–613. [Google Scholar] [CrossRef] [PubMed]
- Firth, C.; Kitchen, A.; Shapiro, B.; Suchard, M.A.; Holmes, E.C.; Rambaut, A. Using Time-Structured Data to Estimate Evolutionary Rates of Double-Stranded DNA Viruses. Mol. Biol. Evol. 2010, 27, 2038–2051. [Google Scholar] [CrossRef] [PubMed]
- Terajima, M.; Cruz, J.; Leporati, A.M.; Demkowicz, W.E.; Kennedy, J.S.; Ennis, F.A. Identification of Vaccinia CD8+ T-Cell Epitopes Conserved among Vaccinia and Variola Viruses Restricted by Common MHC Class I Molecules, HLA-A2 or HLA-B7. Hum. Immunol. 2006, 67, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Hammarlund, E.; Lewis, M.W.; Carter, S.V.; Amanna, I.; Hansen, S.G.; Strelow, L.I.; Wong, S.W.; Yoshihara, P.; Hanifin, J.M.; Slifka, M.K. Multiple Diagnostic Techniques Identify Previously Vaccinated Individuals with Protective Immunity against Monkeypox. Nat. Med. 2005, 11, 1005–1011. [Google Scholar] [CrossRef] [PubMed]
- Dobrovolná, M.; Brázda, V.; Warner, E.F.; Bidula, S. Inverted Repeats in the Monkeypox Virus Genome Are Hot Spots for Mutation. J. Med. Virol. 2023, 95, e28322. [Google Scholar] [CrossRef] [PubMed]
- Alcamí, A.; Symons, J.A.; Collins, P.D.; Williams, T.J.; Smith, G.L. Blockade of Chemokine Activity by a Soluble Chemokine Binding Protein from Vaccinia Virus. J. Immunol. 1998, 160, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Huddleston, J.; Hadfield, J.; Sibley, T.R.; Lee, J.; Fay, K.; Ilcisin, M.; Harkins, E.; Bedford, T.; Neher, R.A.; Hodcroft, E.B. Augur: A Bioinformatics Toolkit for Phylogenetic Analyses of Human Pathogens. J. Open Source Softw. 2021, 6, 2906. [Google Scholar] [CrossRef] [PubMed]
- Elbe, S.; Buckland-Merrett, G. Data, Disease and Diplomacy: GISAID’s Innovative Contribution to Global Health. Glob. Chall. 2017, 1, 33–46. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, Á.; Rambaut, A. Initial Observations about Putative APOBEC3 Deaminase Editing Driving Short-Term Evolution of MPXV Since 2017; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, UK, 2022. [Google Scholar] [CrossRef]
- Isidro, J.; Borges, V.; Pinto, M.; Sobral, D.; Santos, J.D.; Nunes, A.; Mixão, V.; Ferreira, R.; Santos, D.; Duarte, S.; et al. Phylogenomic Characterization and Signs of Microevolution in the 2022 Multi-Country Outbreak of Monkeypox Virus. Nat. Med. 2022, 28, 1569–1572. [Google Scholar] [CrossRef] [PubMed]
- Vauhkonen, H.; Kallio-Kokko, H.; Hiltunen-Back, E.; Lönnqvist, L.; Leppäaho-Lakka, J.; Mannonen, L.; Kant, R.; Sironen, T.; Kurkela, S.; Lappalainen, M.; et al. Intrahost Monkeypox Virus Genome Variation in Patient with Early Infection, Finland, 2022. Emerg. Infect. Dis. 2023, 29, 649–652. [Google Scholar] [CrossRef] [PubMed]
- Alzhanova, D.; Hammarlund, E.; Reed, J.; Meermeier, E.; Rawlings, S.; Ray, C.A.; Edwards, D.M.; Bimber, B.; Legasse, A.; Planer, S.; et al. T Cell Inactivation by Poxviral B22 Family Proteins Increases Viral Virulence. PLoS Pathog. 2014, 10, e1004123. [Google Scholar] [CrossRef]
| (A) | |||||||||||
| ID | EPID | DoC(X) | SNV | GENE/AA SUBs | GENE FUNCTION | READ FREQ | |||||
| P4 | EPI_ISL_16467111 | 91 | C1092T | OPG001:D162N | Chemokine binding protein | 78% mutated 22% wild-type | |||||
| P2 | EPI_ISL_14786346 | 102 | G2333A | OPG002:S140F | Crm-B-secreted TNF-alpha-receptor-like protein | 72% mutated 28% wild-type | |||||
| P4 | EPI_ISL_16467111 | 107 | G37152A | OPG056:S628F | EEV maturation protein | 100% mutated | |||||
| P3 | - | 23 | 100% mutated | ||||||||
| P1 | EPI_ISL_14786290 | 45 | C83293T | OPG105:T723I | DNA-dependent RNA polymerase subunit rpo147 | 100% mutated | |||||
| P2 | EPI_ISL_14786346 | 70 | G159023A | OPG185:R36Q | Hemagglutinin | 77% mutated 23% wild-type | |||||
| P4 | EPI_ISL_16467111 | 110 | G171341T | OPG199:L118F | Serpin | 80% mutated 20% wild-type | |||||
| P3 | - | 11 | 27% mutated 73% wild-type | ||||||||
| P4 | EPI_ISL_16467111 | 92 | C186990T | OPG210:R1879W | B22R family protein | 80% mutated 20% wild-type | |||||
| P3 | - | 13 | 54% mutated 46% wild-type | ||||||||
| (B) | |||||||||||
| NGS | SANGER | ||||||||||
| P2 | P3 | P4 | P2 | P3 | P4 | ||||||
| OPG185:G159023A | 77% mutated 23% wild-type | wild-type and mutated | - | - | |||||||
| OPG199:C171341T | 27% mutated 73% wild-type | 80% mutated 20% wild-type | - | wild-type | mutated | ||||||
| OPG210:C186990T | 54% mutated 46% wild-type | 80% mutated 20% wild-type | - | wild-type | mutated | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deiana, M.; Lavezzari, D.; Mori, A.; Accordini, S.; Pomari, E.; Piubelli, C.; Malagò, S.; Cordioli, M.; Ronzoni, N.; Angheben, A.; et al. Exploring Viral Genome Profile in Mpox Patients during the 2022 Outbreak, in a North-Eastern Centre of Italy. Viruses 2024, 16, 726. https://doi.org/10.3390/v16050726
Deiana M, Lavezzari D, Mori A, Accordini S, Pomari E, Piubelli C, Malagò S, Cordioli M, Ronzoni N, Angheben A, et al. Exploring Viral Genome Profile in Mpox Patients during the 2022 Outbreak, in a North-Eastern Centre of Italy. Viruses. 2024; 16(5):726. https://doi.org/10.3390/v16050726
Chicago/Turabian StyleDeiana, Michela, Denise Lavezzari, Antonio Mori, Silvia Accordini, Elena Pomari, Chiara Piubelli, Simone Malagò, Maddalena Cordioli, Niccolò Ronzoni, Andrea Angheben, and et al. 2024. "Exploring Viral Genome Profile in Mpox Patients during the 2022 Outbreak, in a North-Eastern Centre of Italy" Viruses 16, no. 5: 726. https://doi.org/10.3390/v16050726
APA StyleDeiana, M., Lavezzari, D., Mori, A., Accordini, S., Pomari, E., Piubelli, C., Malagò, S., Cordioli, M., Ronzoni, N., Angheben, A., Tacconelli, E., Capobianchi, M. R., Gobbi, F. G., & Castilletti, C. (2024). Exploring Viral Genome Profile in Mpox Patients during the 2022 Outbreak, in a North-Eastern Centre of Italy. Viruses, 16(5), 726. https://doi.org/10.3390/v16050726








