Avian H6 Influenza Viruses in Vietnamese Live Bird Markets during 2018–2021
Abstract
1. Introduction
2. Materials and Methods
2.1. Virus Isolation and Identification
2.2. Viral Genomic Sequence Analysis
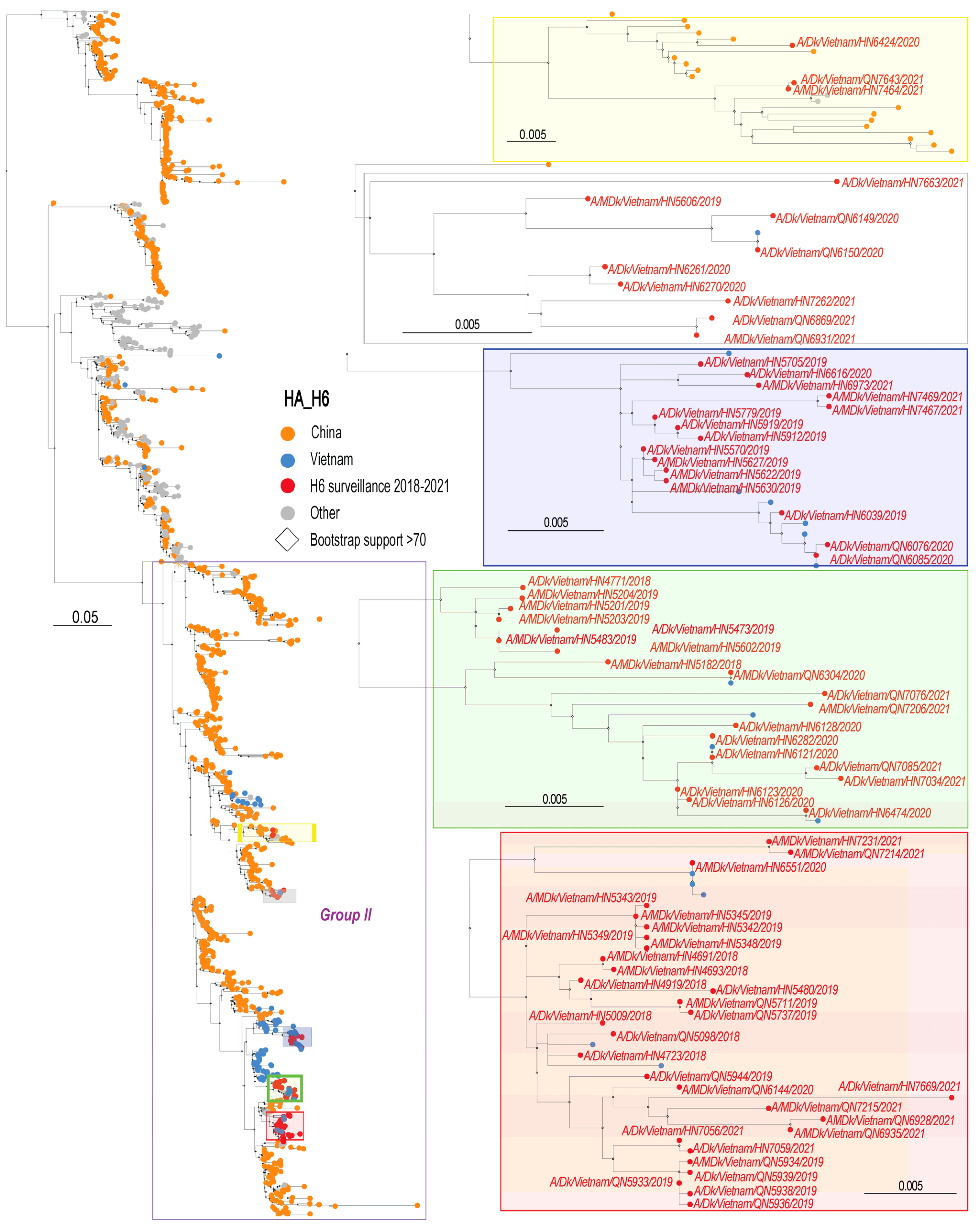
2.3. Phylogenetic Analysis
2.4. Biosafety Statement
3. Results
3.1. Viruses of the H6 Subtype Isolated from Vietnamese Live Bird Markets from 2018 to 2021
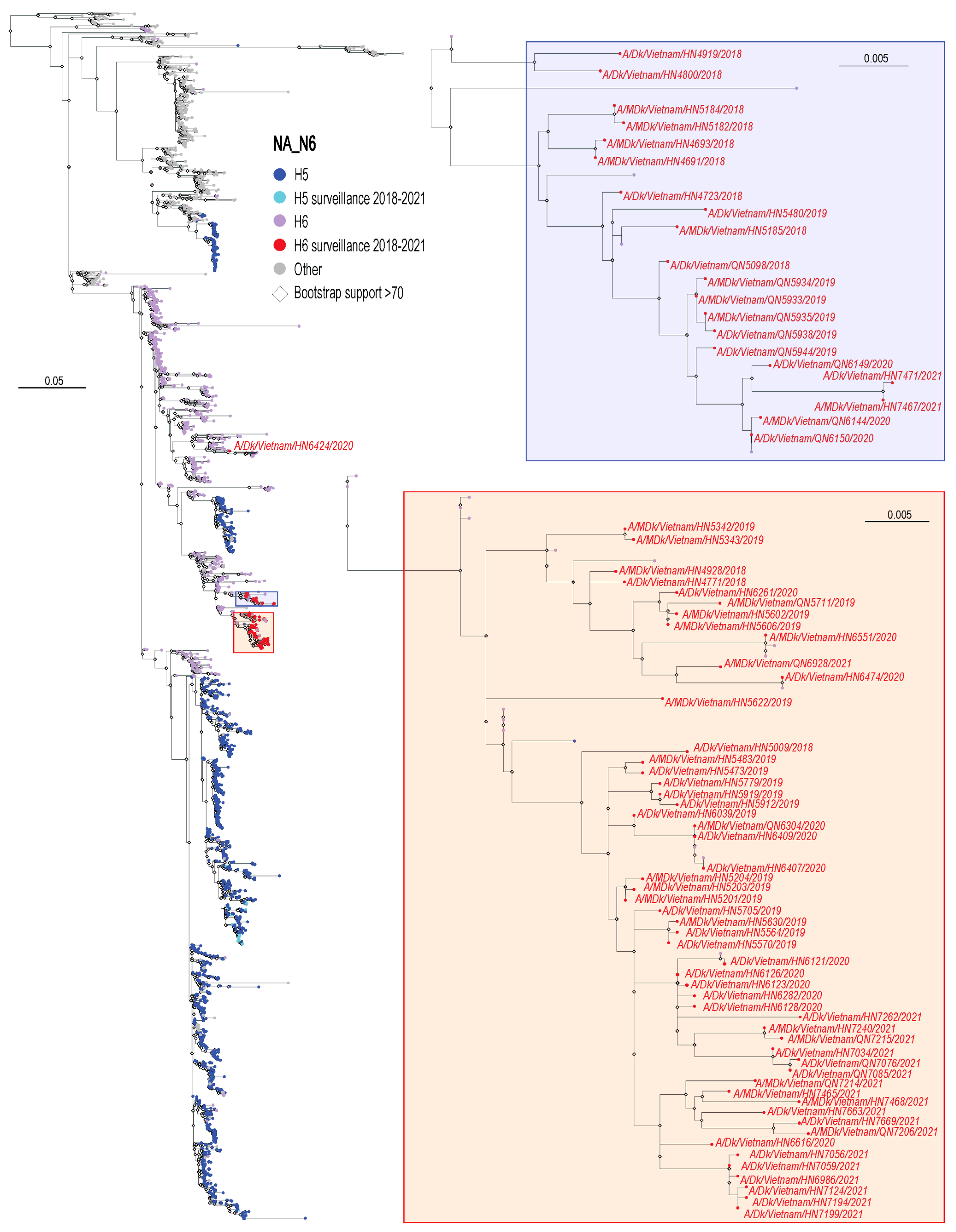
3.2. Phylogenetic Analysis of H6 Viruses
3.3. Analysis of H6 Virus Sequences
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yan, Z.; Li, Y.; Huang, S.; Wen, F. Global distribution, receptor binding, and cross-species transmission of H6 influenza viruses: Risks and implications for humans. J. Virol. 2023, 97, e0137023. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, E.; Stech, J.; Leneva, I.; Krauss, S.; Scholtissek, C.; Chin, P.S.; Peiris, M.; Shortridge, K.F.; Webster, R.G. Characterization of the influenza A virus gene pool in avian species in southern China: Was H6N1 a derivative or a precursor of H5N1? J. Virol. 2000, 74, 6309–6315. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Ren, H.; Teng, Y.; Hu, M.; Peng, X.; Yue, J.; Liang, L. Novel reassortment of avian influenza A(H7N9) virus with subtype H6N6 and H5N6 viruses circulating in Guangdong Province, China. J. Infect. 2017, 75, 179–182. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Zhang, L.; Kan, X.; Jiang, L.; Yang, J.; Guo, Z.; Ren, Q. Origin and molecular characteristics of a novel 2013 avian influenza A(H6N1) virus causing human infection in Taiwan. Clin. Infect. Dis. 2013, 57, 1367–1368. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Chen, C.; Huang, J.; Wang, Y.; Peng, D.; Liu, X. Characterisation of one H6N6 influenza virus isolated from swine in China. Res. Vet. Sci. 2013, 95, 434–436. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Kong, W.; Qi, W.; Long, L.-P.; Cao, Z.; Huang, L.; Qi, H.; Cao, N.; Wang, W.; Zhao, F.; et al. Identification of an H6N6 swine influenza virus in southern China. Infect. Genet. Evol. 2011, 11, 1174–1177. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.T.; Wang, C.H.; Chueh, L.L.; Su, B.L.; Wang, L.C. Influenza A(H6N1) Virus in Dogs, Taiwan. Emerg. Infect. Dis. 2015, 21, 2154–2157. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, H.; Guan, Y.; Peiris, M.; Webster, R.; Webby, R. Changing patterns of h6 influenza viruses in Hong Kong poultry markets. Influenza Res. Treat. 2011, 2011, 702092. [Google Scholar] [CrossRef]
- Huang, K.; Bahl, J.; Fan, X.H.; Vijaykrishna, D.; Cheung, C.L.; Webby, R.J.; Webster, R.G.; Chen, H.; Smith, G.J.D.; Peiris, J.S.M.; et al. Establishment of an H6N2 influenza virus lineage in domestic ducks in southern China. J. Virol. 2010, 84, 6978–6986. [Google Scholar] [CrossRef]
- Huang, K.; Zhu, H.; Fan, X.; Wang, J.; Cheung, C.-L.; Duan, L.; Hong, W.; Liu, Y.; Li, L.; Smith, D.K.; et al. Establishment and lineage replacement of H6 influenza viruses in domestic ducks in southern China. J. Virol. 2012, 86, 6075–6083. [Google Scholar] [CrossRef]
- Zhao, G.; Lu, X.; Gu, X.; Zhao, K.; Song, Q.; Pan, J.; Xu, Q.; Duan, Z.; Peng, D.; Hu, S.; et al. Molecular evolution of the H6 subtype influenza A viruses from poultry in eastern China from 2002 to 2010. Virol. J. 2011, 8, 470. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Zhou, T.; Zhang, J.; Zeng, Q.; Jiang, D.; Wei, M.; Li, X. The Genomic Evolution and the Transmission Dynamics of H6N2 Avian Influenza A Viruses in Southern China. Viruses 2022, 14, 1154. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Quan, C.; Xie, Y.; Ke, C.; Nie, Y.; Chen, Q.; Hu, T.; Chen, J.; Wong, G.; Wang, Q.; et al. Continued reassortment of avian H6 influenza viruses from Southern China, 2014–2016. Transbound Emerg. Dis. 2019, 66, 592–598. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Cui, H.; Teng, Q.; Li, L.; Shi, Y.; Li, X.; Yang, J.; Liu, Q.; Deng, J.; Li, Z. Evolution and pathogenicity of H6 avian influenza viruses isolated from Southern China during 2011 to 2017 in mice and chickens. Sci. Rep. 2020, 10, 20583. [Google Scholar] [CrossRef]
- Cui, J.; Cui, P.; Shi, J.; Fan, W.; Xing, X.; Gu, W.; Zhang, Y.; Zhang, Y.; Zeng, X.; Jiang, Y.; et al. Continued evolution of H6 avian influenza viruses isolated from farms in China between 2014 and 2018. Transbound Emerg. Dis. 2022, 69, 2156–2172. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Zheng, H.; Xiong, J.; Ma, L.; Gui, R.; Zhu, G.; Li, Y.; Yang, G.; Chen, G.; Zhang, J.; et al. Genetic and Pathogenic Characterization of Avian Influenza Virus in Migratory Birds between 2015 and 2019 in Central China. Microbiol. Spectr. 2022, 10, e0165222. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, Q.; Tan, M.; Liu, J.; Li, X.; Yang, L.; Shu, Y.; Wang, D.; Zhu, W. Epidemiology, evolution, and biological characteristics of H6 avian influenza viruses in China. Emerg. Microbes Infect. 2023, 12, 2151380. [Google Scholar] [CrossRef]
- Wu, H.; Yang, F.; Liu, F.; Lu, R.; Peng, X.; Chen, B.; Yao, H.; Wu, N. Isolation and characterization of novel reassortant H6N1 avian influenza viruses from chickens in Eastern China. Virol. J. 2018, 15, 164. [Google Scholar] [CrossRef]
- Hu, C.; Li, X.; Zhu, C.; Zhou, F.; Tang, W.; Wu, D.; Li, Z.; Zhou, L.; Liu, J.; Wei, X.; et al. Co-circulation of multiple reassortant H6 subtype avian influenza viruses in wild birds in eastern China, 2016–2017. Virol. J. 2020, 17, 62. [Google Scholar] [CrossRef]
- Wan, Z.; Kan, Q.; Zhao, Z.; Shao, H.; Deliberto, T.J.; Wan, X.F.; Qin, A.; Ye, J. Characterization of Subtype H6 Avian Influenza A Viruses Isolated from Wild Birds in Poyang Lake, China. Front. Vet. Sci. 2021, 8, 685399. [Google Scholar] [CrossRef]
- Wang, G.; Deng, G.; Shi, J.; Luo, W.; Zhang, G.; Zhang, Q.; Liu, L.; Jiang, Y.; Li, C.; Sriwilaijaroen, N.; et al. H6 influenza viruses pose a potential threat to human health. J. Virol. 2014, 88, 3953–3964. [Google Scholar] [CrossRef]
- Ge, Y.; Chai, H.; Fan, Z.; Wang, X.; Yao, Q.; Ma, J.; Chen, S.; Hua, Y.; Deng, G.; Chen, H. New H6 influenza virus reassortment strains isolated from Anser fabalis in Anhui Province, China. Virol. J. 2017, 14, 36. [Google Scholar] [CrossRef]
- Li, Y.; Li, M.; Tian, J.; Bai, X.; Li, Y. Genetic characteristics and pathogenicity of novel reassortant H6 viruses isolated from wild birds in China. Vet. Microbiol. 2021, 254, 108978. [Google Scholar] [CrossRef]
- Yang, H.; Carney, P.J.; Chang, J.C.; Villanueva, J.M.; Stevens, J. Structure and receptor binding preferences of recombinant hemagglutinins from avian and human H6 and H10 influenza A virus subtypes. J. Virol. 2015, 89, 4612–4623. [Google Scholar] [CrossRef]
- Qu, Z.; Ma, S.; Kong, H.; Deng, G.; Shi, J.; Liu, L.; Suzuki, Y.; Chen, H. Identification of a key amino acid in hemagglutinin that increases human-type receptor binding and transmission of an H6N2 avian influenza virus. Microbes Infect. 2017, 19, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Gao, R.; Zhang, Y.; Li, X.; Chen, W.; Bai, T.; Dong, L.; Wang, D.; Shu, Y. Molecular characterization of H6 subtype influenza viruses in southern China from 2009 to 2011. Emerg. Microbes Infect. 2016, 5, e73. [Google Scholar] [CrossRef] [PubMed]
- Ni, F.; Kondrashkina, E.; Wang, Q. Structural and Functional Studies of Influenza Virus A/H6 Hemagglutinin. PLoS ONE 2015, 10, e0134576. [Google Scholar] [CrossRef] [PubMed][Green Version]
- de Vries, R.P.; Tzarum, N.; Peng, W.; Thompson, A.J.; Wickramasinghe, I.N.A.; de la Pena, A.T.T.; van Breemen, M.J.; Bouwman, K.M.; Zhu, X.; McBride, R.; et al. A single mutation in Taiwanese H6N1 influenza hemagglutinin switches binding to human-type receptors. EMBO Mol. Med. 2017, 9, 1314–1325. [Google Scholar] [CrossRef] [PubMed]
- Le, K.T.; Nguyen, L.T.; Huynh, L.T.; Chu, D.H.; Nguyen, L.V.; Nguyen, T.N.; Tien, T.N.; Matsuno, K.; Okamatsu, M.; Hiono, T.; et al. Genetic, Antigenic, and Pathobiological Characterization of H9 and H6 Low Pathogenicity Avian Influenza Viruses Isolated in Vietnam from 2014 to 2018. Microorganisms 2023, 11, 244. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.-H.; Okamatsu, M.; Matsuno, K.; Hiono, T.; Ogasawara, K.; Nguyen, L.T.; Van Nguyen, L.; Nguyen, T.N.; Nguyen, T.T.; Van Pham, D.; et al. Genetic and antigenic characterization of H5, H6 and H9 avian influenza viruses circulating in live bird markets with intervention in the center part of Vietnam. Vet. Microbiol. 2016, 192, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.; Babujee, L.; Browning, V.L.; Presler, R.; Pattinson, D.; Nguyen, H.L.K.; Hoang, V.M.P.; Le, M.Q.; van Bakel, H.; Neumann, G.; et al. Continued Circulation of Highly Pathogenic H5 Influenza Viruses in Vietnamese Live Bird Markets in 2018–2021. Viruses 2023, 15, 1596. [Google Scholar] [CrossRef]
- Shepard, S.S.; Meno, S.; Bahl, J.; Wilson, M.M.; Barnes, J.; Neuhaus, E. Erratum to: Viral deep sequencing needs an adaptive approach: IRMA, the iterative refinement meta-assembler. BMC Genom. 2016, 17, 801. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Pattengale, N.D.; Alipour, M.; Bininda-Emonds, O.R.; Moret, B.M.; Stamatakis, A. How many bootstrap replicates are necessary? J. Comput. Biol. 2010, 17, 337–354. [Google Scholar] [CrossRef]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T. ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evolution 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Colman, P.M.; Varghese, J.N.; Laver, W.G. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature 1983, 303, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Varghese, J.N.; Colman, P.M.; van Donkelaar, A.; Blick, T.J.; Sahasrabudhe, A.; McKimm-Breschkin, J.L. Structural evidence for a second sialic acid binding site in avian influenza virus neuraminidases. Proc. Natl. Acad. Sci. USA 1997, 94, 11808–11812. [Google Scholar] [CrossRef]
- Neumann, G.; Treanor, J.; Kawaoka, Y. Orthomyxoviruses. In Fields Virology. 1, 7th ed.; Knipe, D.M., Howley, P.M., Whelan, S.P.J., Cohen, J.I., Damania, B., Enquist, L., Freed, E.O., Eds.; Wolters Kluwer: Philadelphia, PA, USA, 2021; pp. 596–668. [Google Scholar]
- Hatta, M.; Gao, P.; Halfmann, P.; Kawaoka, Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 2001, 293, 1840–1842. [Google Scholar] [CrossRef]
- Subbarao, E.K.; London, W.; Murphy, B.R. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 1993, 67, 1761–1764. [Google Scholar] [CrossRef]
- Li, Z.; Chen, H.; Jiao, P.; Deng, G.; Tian, G.; Li, Y.; Hoffmann, E.; Webster, R.G.; Matsuoka, Y.; Yu, K. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J. Virol. 2005, 79, 12058–12064. [Google Scholar] [CrossRef]
- Fan, S.; Hatta, M.; Kim, J.H.; Halfmann, P.; Imai, M.; Macken, C.A.; Le, M.Q.; Nguyen, T.; Neumann, G.; Kawaoka, Y. Novel residues in avian influenza virus PB2 protein affect virulence in mammalian hosts. Nat. Commun. 2014, 5, 5021. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Li, Y.; Halpin, R.; Hine, E.; Spiro, D.J.; Wentworth, D.E. PB2 residue 158 is a pathogenic determinant of pandemic H1N1 and H5 influenza A viruses in mice. J. Virol. 2011, 85, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Bussey, K.A.; Bousse, T.L.; Desmet, E.A.; Kim, B.; Takimoto, T. PB2 residue 271 plays a key role in enhanced polymerase activity of influenza A viruses in mammalian host cells. J. Virol. 2010, 84, 4395–4406. [Google Scholar] [CrossRef] [PubMed]
- Mehle, A.; Doudna, J.A. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc. Natl. Acad. Sci. USA 2009, 106, 21312–21316. [Google Scholar] [CrossRef] [PubMed]
- Czudai-Matwich, V.; Otte, A.; Matrosovich, M.; Gabriel, G.; Klenk, H.D. PB2 mutations D701N and S714R promote adaptation of an influenza H5N1 virus to a mammalian host. J. Virol. 2014, 88, 8735–8742. [Google Scholar] [CrossRef] [PubMed]
- Mänz, B.; de Graaf, M.; Mögling, R.; Richard, M.; Bestebroer, T.M.; Rimmelzwaan, G.F.; Fouchier, R.A.M. Multiple Natural Substitutions in Avian Influenza A Virus PB2 Facilitate Efficient Replication in Human Cells. J. Virol. 2016, 90, 5928–5938. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, G.; Dauber, B.; Wolff, T.; Planz, O.; Klenk, H.D.; Stech, J. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc. Natl. Acad. Sci. USA 2005, 102, 18590–18595. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, Q.; Wang, J.; Mizumoto, K.; Toyoda, T. Two mutations in the C-terminal domain of influenza virus RNA polymerase PB2 enhance transcription by enhancing cap-1 RNA binding activity. Biochim. Biophys. Acta 2012, 1819, 78–83. [Google Scholar] [CrossRef]
- Kong, H.; Ma, S.; Wang, J.; Gu, C.; Wang, Z.; Shi, J.; Deng, G.; Guan, Y.; Chen, H. Identification of Key Amino Acids in the PB2 and M1 Proteins of H7N9 Influenza Virus That Affect Its Transmission in Guinea Pigs. J. Virol. 2019, 94, e01180-19. [Google Scholar] [CrossRef]
- Gao, W.; Zu, Z.; Liu, J.; Song, J.; Wang, X.; Wang, C.; Liu, L.; Tong, Q.; Wang, M.; Sun, H.; et al. Prevailing I292V PB2 mutation in avian influenza H9N2 virus increases viral polymerase function and attenuates IFN-beta induction in human cells. J. Gen. Virol. 2019, 100, 1273–1281. [Google Scholar] [CrossRef]
- Zhou, Y.; Wu, X.; Yan, D.; Chen, C.; Liu, X.; Huang, C.; Fu, X.; Tian, G.; Ding, C.; Wu, J.; et al. V292I mutation in PB2 polymerase induces increased effects of E627K on influenza H7N9 virus replication in cells. Virus Res. 2021, 291, 198186. [Google Scholar] [CrossRef]
- Youk, S.S.; Leyson, C.M.; Seibert, B.A.; Jadhao, S.; Perez, D.R.; Suarez, D.L.; Pantin-Jackwood, M.J. Mutations in PB1, NP, HA, and NA Contribute to Increased Virus Fitness of H5N2 Highly Pathogenic Avian Influenza Virus Clade 2.3.4.4 in Chickens. J. Virol. 2021, 95, e01675. [Google Scholar] [CrossRef]
- Yu, Z.; Cheng, K.; Sun, W.; Zhang, X.; Li, Y.; Wang, T.; Wang, H.; Zhang, Q.; Xin, Y.; Xue, L.; et al. A PB1 T296R substitution enhance polymerase activity and confer a virulent phenotype to a 2009 pandemic H1N1 influenza virus in mice. Virology 2015, 486, 180–186. [Google Scholar] [CrossRef]
- Xu, C.; Hu, W.B.; Xu, K.; He, Y.X.; Wang, T.Y.; Chen, Z.; Li, T.X.; Liu, J.H.; Buchy, P.; Sun, B. Amino acids 473V and 598P of PB1 from an avian-origin influenza A virus contribute to polymerase activity, especially in mammalian cells. J. Gen. Virol. 2012, 93 Pt 3, 531–540. [Google Scholar] [CrossRef]
- Chu, C.; Fan, S.; Li, C.; Macken, C.; Kim, J.H.; Hatta, M.; Neumann, G.; Kawaoka, Y. Functional analysis of conserved motifs in influenza virus PB1 protein. PLoS ONE. 2012, 7, e36113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, Y.; Jin, S.; Zhang, Y.; Sun, L.; Hu, X.; Zhao, M.; Li, F.; Wang, T.; Sun, W.; et al. PB1 S524G mutation of wild bird-origin H3N8 influenza A virus enhances virulence and fitness for transmission in mammals. Emerg. Microbes Infect. 2021, 10, 1038–1051. [Google Scholar] [CrossRef]
- Kamiki, H.; Matsugo, H.; Kobayashi, T.; Ishida, H.; Takenaka-Uema, A.; Murakami, S.; Horimoto, T. A PB1-K577E Mutation in H9N2 Influenza Virus Increases Polymerase Activity and Pathogenicity in Mice. Viruses 2018, 10, 653. [Google Scholar] [CrossRef] [PubMed]
- Hayden, F.G.; Sugaya, N.; Hirotsu, N.; Lee, N.; De Jong, M.D.; Hurt, A.C.; Ishida, T.; Sekino, H.; Yamada, K.; Portsmouth, S.; et al. Baloxavir Marboxil for Uncomplicated Influenza in Adults and Adolescents. N. Engl. J. Med. 2018, 379, 913–923. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.W.; Chang, S.C.; Mok, C.K.; Lo, Y.L.; Kung, Y.N.; Huang, J.H.; Shih, Y.H.; Wang, J.Y.; Chiang, C.; Chen, C.J.; et al. Genomic signatures of human versus avian influenza A viruses. Emerg. Infect. Dis. 2006, 12, 1353–1360. [Google Scholar] [CrossRef]
- Allen, J.E.; Gardner, S.N.; Vitalis, E.A.; Slezak, T.R. Conserved amino acid markers from past influenza pandemic strains. BMC Microbiol. 2009, 9, 77. [Google Scholar] [CrossRef]
- Shaw, M.; Cooper, L.; Xu, X.; Thompson, W.; Krauss, S.; Guan, Y.; Zhou, N.; Klimov, A.; Cox, N.; Webster, R.; et al. Molecular changes associated with the transmission of avian influenza a H5N1 and H9N2 viruses to humans. J. Med. Virol. 2002, 66, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Miotto, O.; Heiny, A.T.; Albrecht, R.; García-Sastre, A.; Tan, T.W.; August, J.T.; Brusic, V. Complete-proteome mapping of human influenza A adaptive mutations: Implications for human transmissibility of zoonotic strains. PLoS ONE 2010, 5, e9025. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, D.B.; Mukatira, S.; Mehta, P.K.; Obenauer, J.C.; Su, X.; Webster, R.G.; Naeve, C.W. Persistent host markers in pandemic and H5N1 influenza viruses. J. Virol. 2007, 81, 10292–10299. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Hatta, M.; Watanabe, S.; Neumann, G.; Watanabe, T.; Kawaoka, Y. Role of host-specific amino acids in the pathogenicity of avian H5N1 influenza viruses in mice. J. Gen. Virol. 2010, 91 Pt 5, 1284–1289. [Google Scholar] [CrossRef] [PubMed]
- Rekstin, A.R.; Kiseleva, I.V.; Desheva Iu, A.; Kats, D.; Aleksandrova, G.I.; Klimov, A.I.; Rudenko, L.G. Growth characteristics of single gene mutants of A/Leningrad/134/57(H2N2) influenza virus and cold-adapted mutant A/Leningrad/134/17/57(H2N2). Vopr. Virusol. 2007, 52, 13–16. [Google Scholar] [PubMed]
- Bialas, K.M.; Desmet, E.A.; Takimoto, T. Specific residues in the 2009 H1N1 swine-origin influenza matrix protein influence virion morphology and efficiency of viral spread in vitro. PLoS ONE 2012, 7, e50595. [Google Scholar] [CrossRef] [PubMed]
- Kainov, D.E.; Muller, K.H.; Theisen, L.L.; Anastasina, M.; Kaloinen, M.; Muller, C.P. Differential effects of NS1 proteins of human pandemic H1N1/2009, avian highly pathogenic H5N1, and low pathogenic H5N2 influenza A viruses on cellular pre-mRNA polyadenylation and mRNA translation. J. Biol. Chem. 2011, 286, 7239–7247. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Calvo, P.A.; Malide, D.; Gibbs, J.; Schubert, U.; Bacik, I.; Basta, S.; O’Neill, R.; Schickli, J.; Palese, P.; et al. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 2001, 7, 1306–1312. [Google Scholar] [CrossRef]
- Hassan, K.E.; El-Kady, M.F.; El-Sawah, A.A.A.; Luttermann, C.; Parvin, R.; Shany, S.; Beer, M.; Harder, T. Respiratory disease due to mixed viral infections in poultry flocks in Egypt between 2017 and 2018: Upsurge of highly pathogenic avian influenza virus subtype H5N8 since 2018. Transbound Emerg. Dis. 2021, 68, 21–36. [Google Scholar] [CrossRef]
- Shehata, A.A.; Sedeik, M.E.; Elbestawy, A.R.; El-Abideen, M.A.Z.; Ibrahim, H.H.; Kilany, W.H.; Ali, A. Co-infections, genetic, and antigenic relatedness of avian influenza H5N8 and H5N1 viruses in domestic and wild birds in Egypt. Poult. Sci. 2019, 98, 2371–2379. [Google Scholar] [CrossRef] [PubMed]
- Young, S.G.; Carrel, M.; Malanson, G.P.; Ali, M.A.; Kayali, G. Predicting Avian Influenza Co-Infection with H5N1 and H9N2 in Northern Egypt. Int. J. Environ. Res. Public. Health 2016, 13, 886. [Google Scholar] [CrossRef] [PubMed]
- Pepin, K.M.; Wang, J.; Webb, C.T.; Smith, G.J.D.; Poss, M.; Hudson, P.J.; Hong, W.; Zhu, H.; Riley, S.; Guan, Y. Multiannual patterns of influenza A transmission in Chinese live bird market systems. Influenza Other Respir. Viruses 2013, 7, 97–107. [Google Scholar] [CrossRef]
- Horwood, P.F.; Horm, S.V.; Suttie, A.; Thet, S.; Phalla, Y.; Rith, S.; Sorn, S.; Holl, D.; Tum, S.; Ly, S.; et al. Co-circulation of Influenza A H5, H7, and H9 Viruses and Co-infected Poultry in Live Bird Markets, Cambodia. Emerg. Infect. Dis. 2018, 24, 352–355. [Google Scholar] [CrossRef] [PubMed]
- Myers, K.P.; Setterquist, S.F.; Capuano, A.W.; Gray, G.C. Infection due to 3 avian influenza subtypes in United States veterinarians. Clin. Infect. Dis. 2007, 45, 4–9. [Google Scholar] [CrossRef]
- Xin, L.; Bai, T.; Zhou, J.F.; Chen, Y.K.; Li, X.D.; Zhu, W.F.; Li, Y.; Tang, J.; Chen, T.; Qin, K.; et al. Seropositivity for Avian Influenza H6 Virus among Humans, China. Emerg. Infect. Dis. 2015, 21, 1267–1269. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, L.; Babujee, L.; Presler, R.; Pattinson, D.; Nguyen, H.L.K.; Hoang, V.M.P.; Le, M.Q.; Bakel, H.v.; Kawaoka, Y.; Neumann, G. Avian H6 Influenza Viruses in Vietnamese Live Bird Markets during 2018–2021. Viruses 2024, 16, 367. https://doi.org/10.3390/v16030367
Guan L, Babujee L, Presler R, Pattinson D, Nguyen HLK, Hoang VMP, Le MQ, Bakel Hv, Kawaoka Y, Neumann G. Avian H6 Influenza Viruses in Vietnamese Live Bird Markets during 2018–2021. Viruses. 2024; 16(3):367. https://doi.org/10.3390/v16030367
Chicago/Turabian StyleGuan, Lizheng, Lavanya Babujee, Robert Presler, David Pattinson, Hang Le Khanh Nguyen, Vu Mai Phuong Hoang, Mai Quynh Le, Harm van Bakel, Yoshihiro Kawaoka, and Gabriele Neumann. 2024. "Avian H6 Influenza Viruses in Vietnamese Live Bird Markets during 2018–2021" Viruses 16, no. 3: 367. https://doi.org/10.3390/v16030367
APA StyleGuan, L., Babujee, L., Presler, R., Pattinson, D., Nguyen, H. L. K., Hoang, V. M. P., Le, M. Q., Bakel, H. v., Kawaoka, Y., & Neumann, G. (2024). Avian H6 Influenza Viruses in Vietnamese Live Bird Markets during 2018–2021. Viruses, 16(3), 367. https://doi.org/10.3390/v16030367




