Porcine Sapovirus Protease Controls the Innate Immune Response and Targets TBK1
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Virus
2.2. Expression Vectors
2.3. Transfection of Cell Lines
2.4. Reporter Gene Assays
2.5. 5BR Assay
2.6. Poly(I:C) Stimulation and RT-qPCR
2.7. CRISPR/Cas9-Mediated Genome Editing
2.8. Immunoprecipitation (IP)
2.9. SDS-PAGE and Immunoblotting
2.10. PSaV Infection
2.11. TCID50 Assay
2.12. Statistical Analysis and Software
3. Results
3.1. PSaV NS6 Is a Type I IFN Antagonist
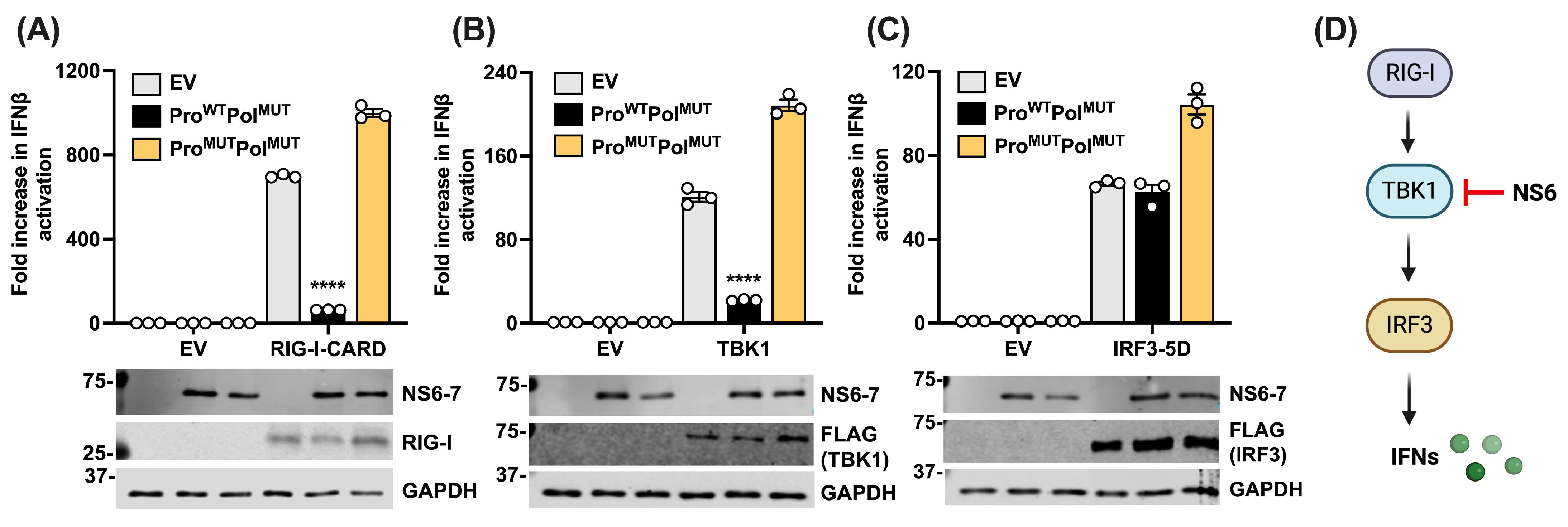
3.2. NS6 Restricts the Type I IFN Pathway at the Level of TBK1
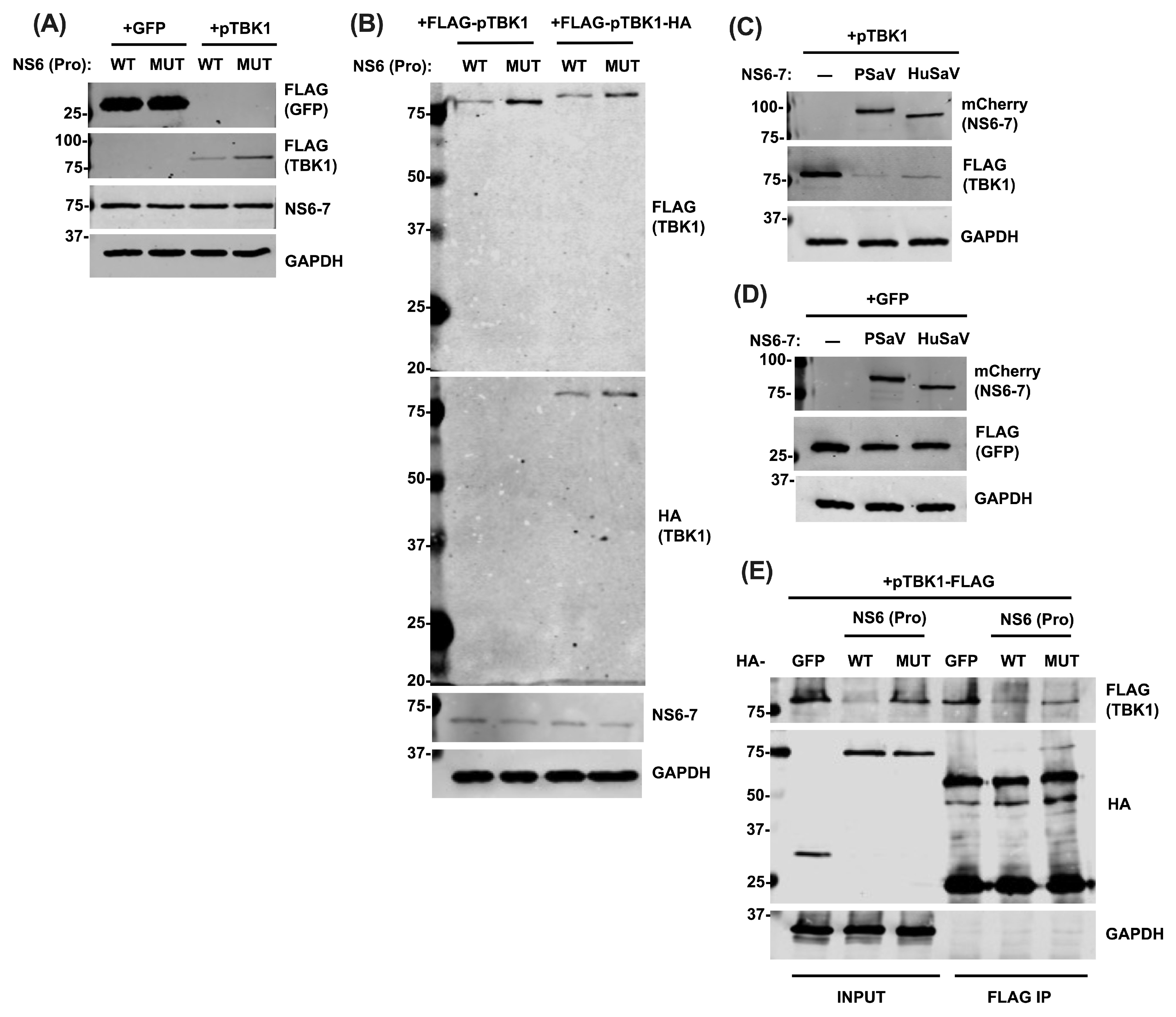
3.3. NS6 Interacts with and Modulates the Protein Levels of TBK1
3.4. TBK1 Protein Levels Are Reduced during PSaV Infection
3.5. NS6-Mediated Reduction in TBK1 Levels Is Proteasomal-Dependent
3.6. PSaV Infection in TBK1-Deficient Cells Results in Higher Viral Titres
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, J.; Zhang, W.; Cui, L.; Shen, Q.; Hua, X. Metagenomic identification, genetic characterization and genotyping of porcine sapoviruses. Infect. Genet. Evol. 2018, 62, 244–252. [Google Scholar] [CrossRef]
- Yinda, C.K.; Conceição-Neto, N.; Zeller, M.; Heylen, E.; Maes, P.; Ghogomu, S.M.; Van Ranst, M.; Matthijnssens, J. Novel highly divergent sapoviruses detected by metagenomics analysis in straw-colored fruit bats in Cameroon. Emerg. Microbes Infect. 2017, 6, e38. [Google Scholar] [CrossRef]
- Oka, T.; Wang, Q.; Katayama, K.; Saif Linda, J. Comprehensive Review of Human Sapoviruses. Clin. Microbiol. Rev. 2015, 28, 32–53. [Google Scholar] [CrossRef] [PubMed]
- Oka, T.; Katayama, K.; Ogawa, S.; Hansman, G.S.; Kageyama, T.; Ushijima, H.; Miyamura, T.; Takeda, N. Proteolytic Processing of Sapovirus ORF1 Polyprotein. J. Virol. 2005, 79, 7283–7290. [Google Scholar] [CrossRef] [PubMed]
- Li, T.-C.; Kataoka, M.; Doan, Y.H.; Saito, H.; Takagi, H.; Muramatsu, M.; Oka, T. Characterization of a Human Sapovirus Genotype GII.3 Strain Generated by a Reverse Genetics System: VP2 Is a Minor Structural Protein of the Virion. Viruses 2022, 14, 1649. [Google Scholar] [CrossRef] [PubMed]
- Sosnovtseva, S.A.; Sosnovtsev, S.V.; Green, K.Y. Mapping of the feline calicivirus proteinase responsible for autocatalytic processing of the nonstructural polyprotein and identification of a stable proteinase-polymerase precursor protein. J. Virol. 1999, 73, 6626–6633. [Google Scholar] [CrossRef] [PubMed]
- Chiba, S.; Sakuma, Y.; Kogasaka, R.; Akihara, M.; Horino, K.; Nakao, T.; Fukui, S. An outbreak of gastroenteritis associated with calicivirus in an infant home. J. Med. Virol. 1979, 4, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Madeley, C.R.; Cosgrove, B.P. CALICIVIRUSES IN MAN. Lancet 1976, 307, 199–200. [Google Scholar] [CrossRef] [PubMed]
- Euller-Nicolas, G.; Le Mennec, C.; Schaeffer, J.; Zeng, X.-L.; Ettayebi, K.; Atmar Robert, L.; Le Guyader Françoise, S.; Estes Mary, K.; Desdouits, M. Human Sapovirus Replication in Human Intestinal Enteroids. J. Virol. 2023, 97, e00383-23. [Google Scholar] [CrossRef]
- Matsumoto, N.; Kurokawa, S.; Tamiya, S.; Nakamura, Y.; Sakon, N.; Okitsu, S.; Ushijima, H.; Yuki, Y.; Kiyono, H.; Sato, S. Replication of Human Sapovirus in Human-Induced Pluripotent Stem Cell-Derived Intestinal Epithelial Cells. Viruses 2023, 15, 1929. [Google Scholar] [CrossRef]
- Takagi, H.; Oka, T.; Shimoike, T.; Saito, H.; Kobayashi, T.; Takahashi, T.; Tatsumi, C.; Kataoka, M.; Wang, Q.; Saif, L.J.; et al. Human sapovirus propagation in human cell lines supplemented with bile acids. Proc. Natl. Acad. Sci. USA 2020, 117, 32078–32085. [Google Scholar] [CrossRef]
- Chang, K.-O.; Kim, Y.; Green, K.Y.; Saif, L.J. Cell-Culture Propagation of Porcine Enteric Calicivirus Mediated by Intestinal Contents Is Dependent on the Cyclic AMP Signaling Pathway. Virology 2002, 304, 302–310. [Google Scholar] [CrossRef][Green Version]
- Chang, K.O.; Sosnovtsev, S.V.; Belliot, G.; Kim, Y.; Saif, L.J.; Green, K.Y. Bile acids are essential for porcine enteric calicivirus replication in association with down-regulation of signal transducer and activator of transcription 1. Proc. Natl. Acad. Sci. USA 2004, 101, 8733–8738. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.O.; Sosnovtsev, S.V.; Belliot, G.; Wang, Q.; Saif, L.J.; Green, K.Y. Reverse genetics system for porcine enteric calicivirus, a prototype sapovirus in the Caliciviridae. J. Virol. 2005, 79, 1409–1416. [Google Scholar] [CrossRef] [PubMed]
- Nagai, M.; Wang, Q.; Oka, T.; Saif, L.J. Porcine sapoviruses: Pathogenesis, epidemiology, genetic diversity, and diagnosis. Virus Res. 2020, 286, 198025. [Google Scholar] [CrossRef] [PubMed]
- Martella, V.; Lorusso, E.; Banyai, K.; Decaro, N.; Corrente, M.; Elia, G.; Cavalli, A.; Radogna, A.; Costantini, V.; Saif, L.J.; et al. Identification of a porcine calicivirus related genetically to human sapoviruses. J. Clin. Microbiol. 2008, 46, 1907–1913. [Google Scholar] [CrossRef] [PubMed]
- Jahun, A.S.; Goodfellow, I.G. Interferon responses to norovirus infections: Current and future perspectives. J. Gen. Virol. 2021, 102, 001660. [Google Scholar] [CrossRef] [PubMed]
- Hosmillo, M.; Sorgeloos, F.; Hiraide, R.; Lu, J.; Goodfellow, I.; Cho, K.O. Porcine sapovirus replication is restricted by the type I interferon response in cell culture. J. Gen. Virol. 2015, 96, 74–84. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rehwinkel, J.; Gack, M.U. RIG-I-like receptors: Their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020, 20, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Carty, M.; Guy, C.; Bowie, A.G. Detection of Viral Infections by Innate Immunity. Biochem. Pharmacol. 2021, 183, 114316. [Google Scholar] [CrossRef]
- Fitzgerald, K.A.; McWhirter, S.M.; Faia, K.L.; Rowe, D.C.; Latz, E.; Golenbock, D.T.; Coyle, A.J.; Liao, S.-M.; Maniatis, T. IKKε and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 2003, 4, 491–496. [Google Scholar] [CrossRef]
- Smale, S.T. Selective Transcription in Response to an Inflammatory Stimulus. Cell 2010, 140, 833–844. [Google Scholar] [CrossRef]
- Berschneider, H. Development of normal cultured small intestinal epithelial cell lines which transport Na and Cl. (Abstract). Gasteroenterology 1989, 96, A41. [Google Scholar]
- Geens, M.M.; Niewold, T.A. Optimizing culture conditions of a porcine epithelial cell line IPEC-J2 through a histological and physiological characterization. Cytotechnology 2011, 63, 415–423. [Google Scholar] [CrossRef]
- Odon, V.; Georgana, I.; Holley, J.; Morata, J.; Maluquer de Motes, C. Novel Class of Viral Ankyrin Proteins Targeting the Host E3 Ubiquitin Ligase Cullin-2. J. Virol. 2018, 92, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Heylbroeck, C.; Pitha, P.M.; Hiscott, J. Virus-Dependent Phosphorylation of the IRF-3 Transcription Factor Regulates Nuclear Translocation, Transactivation Potential, and Proteasome-Mediated Degradation. Mol. Cell. Biol. 1998, 18, 2986–2996. [Google Scholar] [CrossRef] [PubMed]
- Ranjith-Kumar, C.T.; Wen, Y.; Baxter, N.; Bhardwaj, K.; Cheng Kao, C. A Cell-Based Assay for RNA Synthesis by the HCV Polymerase Reveals New Insights on Mechanism of Polymerase Inhibitors and Modulation by NS5A. PLoS ONE 2011, 6, e22575. [Google Scholar] [CrossRef][Green Version]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- REED, L.J.; Muench, H. A Simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Lei, J.; Hilgenfeld, R. RNA-virus proteases counteracting host innate immunity. FEBS Lett. 2017, 591, 3190–3210. [Google Scholar] [CrossRef]
- Zhang, H.; Jiang, P.; Chen, Z.; Wang, D.; Zhou, Y.; Zhu, X.; Xiao, S.; Fang, L. Norovirus 3C-Like protease antagonizes interferon-β production by cleaving NEMO. Virology 2022, 571, 12–20. [Google Scholar] [CrossRef]
- Hornung, V.; Ellegast, J.; Kim, S.; Brzózka, K.; Jung, A.; Kato, H.; Poeck, H.; Akira, S.; Conzelmann, K.-K.; Schlee, M.; et al. 5′-Triphosphate RNA Is the Ligand for RIG-I. Science 2006, 314, 994–997. [Google Scholar] [CrossRef]
- Pichlmair, A.; Schulz, O.; Tan, C.P.; Näslund, T.I.; Liljeström, P.; Weber, F.; Reis e Sousa, C. RIG-I-Mediated Antiviral Responses to Single-Stranded RNA Bearing 5′-Phosphates. Science 2006, 314, 997–1001. [Google Scholar] [CrossRef]
- Oka, T.; Yamamoto, M.; Katayama, K.; Hansman, G.S.; Ogawa, S.; Miyamura, T.; Takeda, N. Identification of the cleavage sites of sapovirus open reading frame 1 polyprotein. J. Gen. Virol. 2006, 87, 3329–3338. [Google Scholar] [CrossRef]
- Yang, Y.; Liang, Y.; Qu, L.; Chen, Z.; Yi, M.; Li, K.; Lemon, S.M. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc. Natl. Acad. Sci. USA 2007, 104, 7253–7258. [Google Scholar] [CrossRef]
- Mukherjee, A.; Morosky, S.A.; Delorme-Axford, E.; Dybdahl-Sissoko, N.; Oberste, M.S.; Wang, T.; Coyne, C.B. The Coxsackievirus B 3Cpro Protease Cleaves MAVS and TRIF to Attenuate Host Type I Interferon and Apoptotic Signaling. PLoS Pathog. 2011, 7, e1001311. [Google Scholar] [CrossRef]
- Wang, D.; Fang, L.; Li, K.; Zhong, H.; Fan, J.; Ouyang, C.; Zhang, H.; Duan, E.; Luo, R.; Zhang, Z.; et al. Foot-and-Mouth Disease Virus 3C Protease Cleaves NEMO To Impair Innate Immune Signaling. J. Virol. 2012, 86, 9311–9322. [Google Scholar] [CrossRef] [PubMed]
- Men, Y.; Wang, Y.; Wang, H.; Zhang, M.; Liu, J.; Chen, Y.; Han, X.; Chen, R.; Chen, Q.; Hu, A. RHDV 3C protein antagonizes type I interferon signaling by cleaving interferon promoter stimulated 1 protein. Virus Genes 2023, 59, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Weidman, M.K.; Yalamanchili, P.; Ng, B.; Tsai, W.; Dasgupta, A. Poliovirus 3C Protease-Mediated Degradation of Transcriptional Activator p53 Requires a Cellular Activity. Virology 2001, 291, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Feng, H.; Zhang, X.; Li, K.; Yang, F.; Cao, W.; Liu, H.; Gao, L.; Xue, Z.; Liu, X.; et al. Porcine Picornavirus 3C Protease Degrades PRDX6 to Impair PRDX6-mediated Antiviral Function. Virol. Sin. 2021, 36, 948–957. [Google Scholar] [CrossRef] [PubMed]
- Jeremiah, S.S.; Miyakawa, K.; Matsunaga, S.; Nishi, M.; Kudoh, A.; Takaoka, A.; Sawasaki, T.; Ryo, A. Cleavage of TANK-Binding Kinase 1 by HIV-1 Protease Triggers Viral Innate Immune Evasion. Front. Microbiol. 2021, 12, 643407. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Lin, X.-T.; Zhao, S.; Zheng, X.-Q.; Zhou, Y.-Q.; Xiao, L.-L.; Chen, H.; Zhang, Z.-Y.; Zhang, L.-J.; Wu, X.-X. Tripartite motif-containing protein 46 accelerates influenza A H7N9 virus infection by promoting K48-linked ubiquitination of TBK1. Virol. J. 2022, 19, 176. [Google Scholar] [CrossRef]
- Sawaged, S.; Mota, T.; Piplani, H.; Thakur, R.; Lall, D.; McCabe, E.; Seo, S.; Sutterwala, F.S.; Feuer, R.; Gottlieb, R.A.; et al. TBK1 and GABARAP family members suppress Coxsackievirus B infection by limiting viral production and promoting autophagic degradation of viral extracellular vesicles. PLoS Pathog. 2022, 18, e1010350. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, I.; Volety, I.; Shukla, D. OPTN-TBK1 axis and a role for PLK1 in HSV-1 infection. mBio 2023, 14, e02715-23. [Google Scholar] [CrossRef] [PubMed]
- Jahun, A.S.; Sorgeloos, F.; Chaudhry, Y.; Arthur, S.E.; Hosmillo, M.; Georgana, I.; Izuagbe, R.; Goodfellow, I.G. Leaked genomic and mitochondrial DNA contribute to the host response to noroviruses in a STING-dependent manner. Cell Rep. 2023, 42, 112179. [Google Scholar] [CrossRef] [PubMed]
- Larabi, A.; Devos, J.M.; Ng, S.-L.; Nanao, M.H.; Round, A.; Maniatis, T.; Panne, D. Crystal Structure and Mechanism of Activation of TANK-Binding Kinase 1. Cell Rep. 2013, 3, 734–746. [Google Scholar] [CrossRef]
- Sharif, M.; Baek, Y.-B.; Nguyen, T.H.; Soliman, M.; Cho, K.-O. Porcine sapovirus-induced RIPK1-dependent necroptosis is proviral in LLC-PK cells. PLoS ONE 2023, 18, e0279843. [Google Scholar] [CrossRef]
- Delanghe, T.; Dondelinger, Y.; Bertrand, M.J.M. RIPK1 Kinase-Dependent Death: A Symphony of Phosphorylation Events. Trends Cell Biol. 2020, 30, 189–200. [Google Scholar] [CrossRef]
- Schierack, P.; Nordhoff, M.; Pollmann, M.; Weyrauch, K.D.; Amasheh, S.; Lodemann, U.; Jores, J.; Tachu, B.; Kleta, S.; Blikslager, A.; et al. Characterization of a porcine intestinal epithelial cell line for in vitro studies of microbial pathogenesis in swine. Histochem. Cell Biol. 2006, 125, 293–305. [Google Scholar] [CrossRef]
- Brosnahan, A.J.; Brown, D.R. Porcine IPEC-J2 intestinal epithelial cells in microbiological investigations. Vet. Microbiol. 2012, 156, 229–237. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Georgana, I.; Hosmillo, M.; Jahun, A.S.; Emmott, E.; Sorgeloos, F.; Cho, K.-O.; Goodfellow, I.G. Porcine Sapovirus Protease Controls the Innate Immune Response and Targets TBK1. Viruses 2024, 16, 247. https://doi.org/10.3390/v16020247
Georgana I, Hosmillo M, Jahun AS, Emmott E, Sorgeloos F, Cho K-O, Goodfellow IG. Porcine Sapovirus Protease Controls the Innate Immune Response and Targets TBK1. Viruses. 2024; 16(2):247. https://doi.org/10.3390/v16020247
Chicago/Turabian StyleGeorgana, Iliana, Myra Hosmillo, Aminu S. Jahun, Edward Emmott, Frederic Sorgeloos, Kyoung-Oh Cho, and Ian G. Goodfellow. 2024. "Porcine Sapovirus Protease Controls the Innate Immune Response and Targets TBK1" Viruses 16, no. 2: 247. https://doi.org/10.3390/v16020247
APA StyleGeorgana, I., Hosmillo, M., Jahun, A. S., Emmott, E., Sorgeloos, F., Cho, K.-O., & Goodfellow, I. G. (2024). Porcine Sapovirus Protease Controls the Innate Immune Response and Targets TBK1. Viruses, 16(2), 247. https://doi.org/10.3390/v16020247




