Epidemiological and Molecular Investigation of Feline Panleukopenia Virus Infection in China
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Sample Collection and Virus Isolation
2.2. Viral and Hemagglutination Titers Determination
2.3. PCR and Sequence
2.4. Genetic Evolution Analysis
2.5. Data Collection and Geographical Distribution Analysis
3. Results
3.1. Isolation of FPV Strains
3.2. Determination of Viral and HA Titers
3.3. Genetic Distance Analysis
3.4. Key Amino Acid Sites Analysis
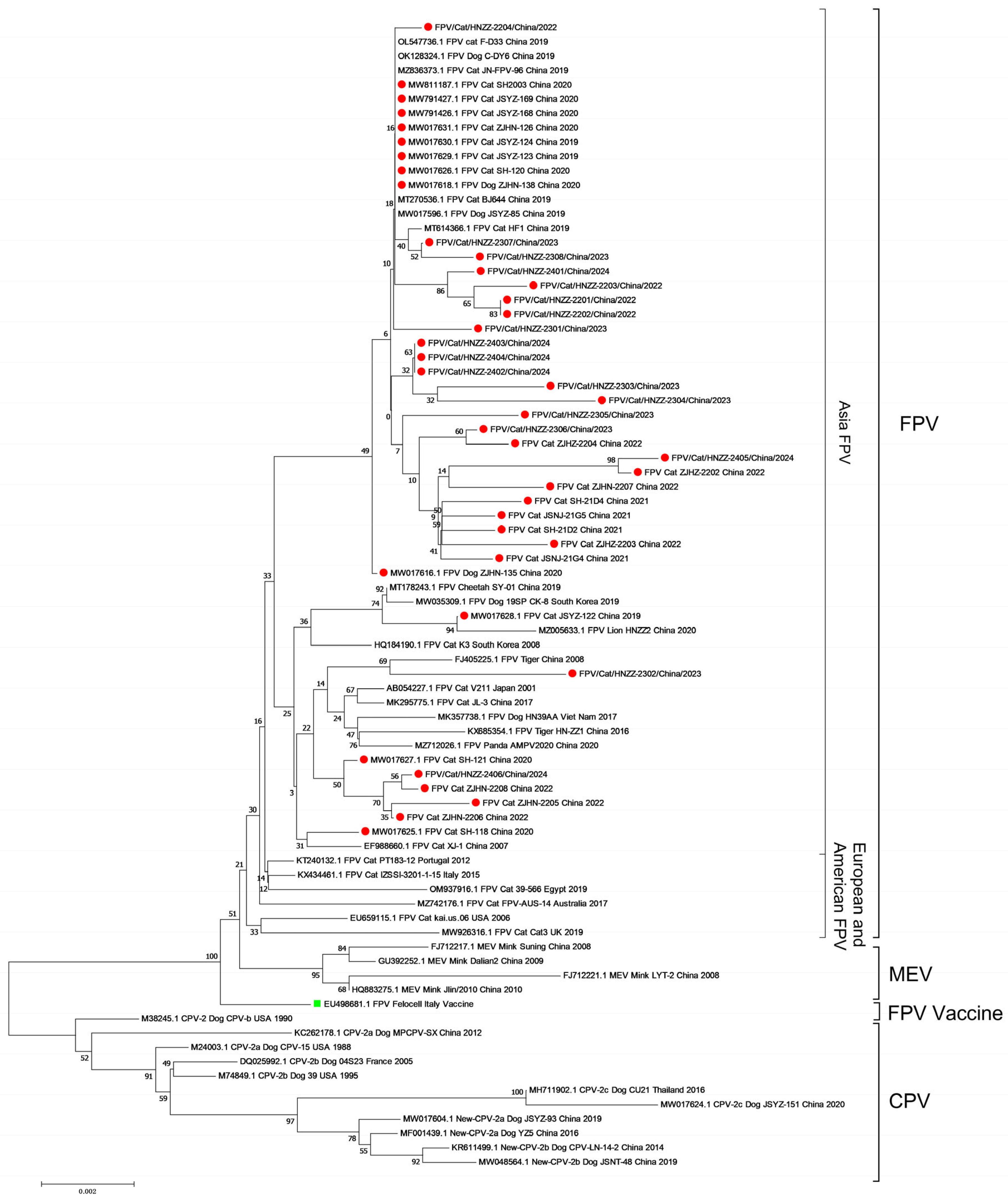
3.5. Phylogenetic Analysis
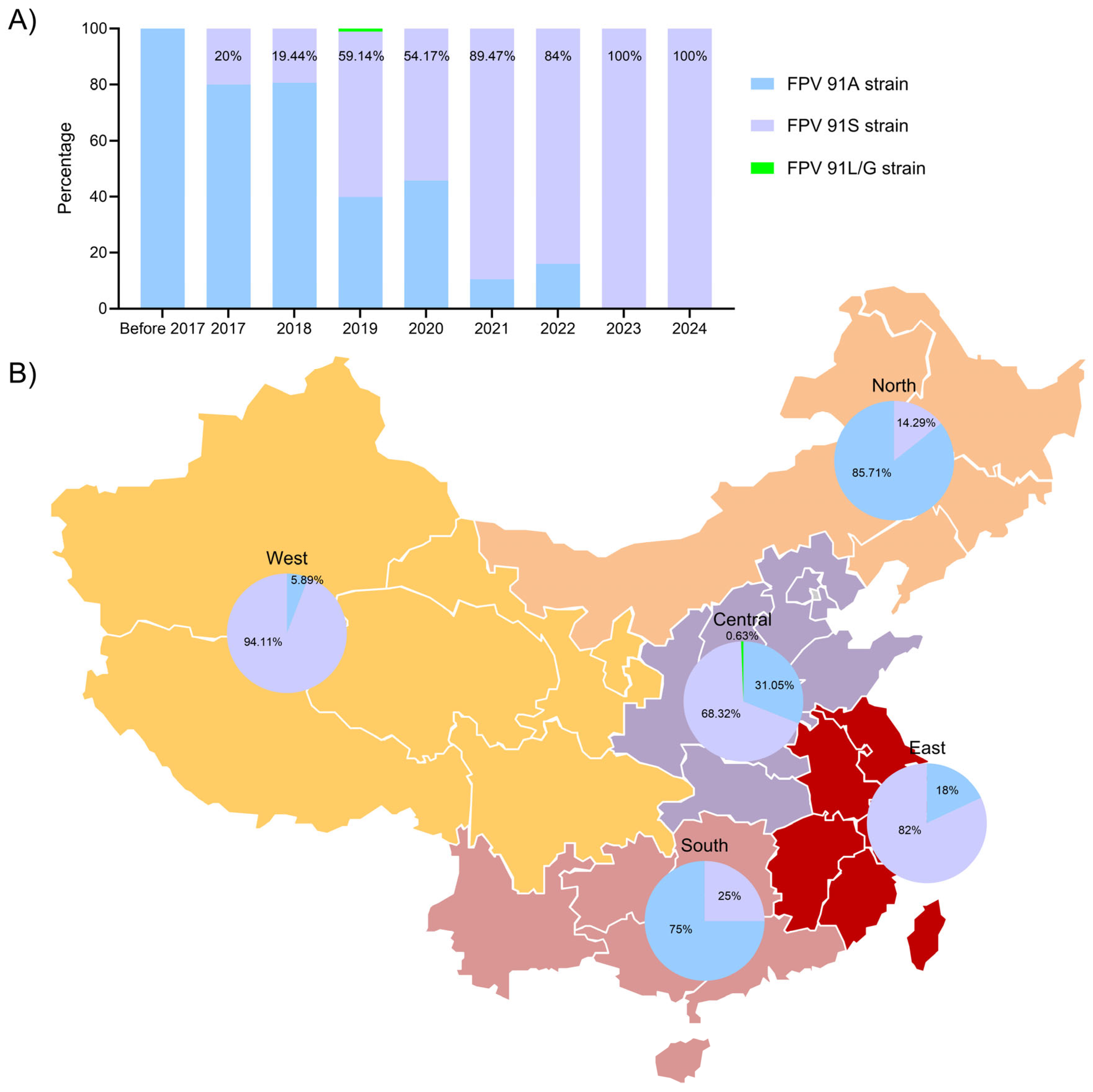
3.6. FPV Data and Geographical Distribution Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stuetzer, B.; Hartmann, K. Feline parvovirus infection and associated diseases. Vet. J. 2014, 201, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Barrs, V.R. Feline Panleukopenia: A Re-emergent Disease. Vet. Clin. N. Am. Small Anim. Pract. 2019, 49, 651–670. [Google Scholar] [CrossRef]
- Decaro, N.; Desario, C.; Miccolupo, A.; Campolo, M.; Parisi, A.; Martella, V.; Amorisco, F.; Lucente, M.S.; Lavazza, A.; Buonavoglia, C. Genetic analysis of feline panleukopenia viruses from cats with gastroenteritis. J. Gen. Virol. 2008, 89, 2290–2298. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Jiao, Y.; Li, L.; Yan, Y.; Fu, Z.; Liu, Z.; Hu, X.; Li, M.; Shi, Y.; He, J.; et al. A potential dual protection vaccine: Recombinant feline herpesvirus-1 expressing feline parvovirus VP2 antigen. Vet. Microbiol. 2024, 290, 109978. [Google Scholar] [CrossRef] [PubMed]
- Parrish, C.R. Host range relationships and the evolution of canine parvovirus. Vet. Microbiol. 1999, 69, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, H.K.; Nookala, M.; Thangamani, N.R.; Sivaprakasam, A.; Antony, P.X.; Thanislass, J.; Srinivas, M.V.; Pillai, R.M. Molecular characterisation of parvoviruses from domestic cats reveals emergence of newer variants in India. J. Feline Med. Surg. 2017, 19, 846–852. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Riaz, A.; Zubair, Z.; Saqib, M.; Ijaz, S.; Nawaz-Ul-Rehman, M.S.; Al-Qahtani, A.; Mubin, M. Molecular analysis of partial VP-2 gene amplified from rectal swab samples of diarrheic dogs in Pakistan confirms the circulation of canine parvovirus genetic variant CPV-2a and detects sequences of feline panleukopenia virus (FPV). Virol. J. 2018, 15, 45. [Google Scholar] [CrossRef] [PubMed]
- Diakoudi, G.; Desario, C.; Lanave, G.; Salucci, S.; Ndiana, L.A.; Zarea, A.A.K.; Fouad, E.A.; Lorusso, A.; Alfano, F.; Cavalli, A.; et al. Feline Panleukopenia Virus in Dogs from Italy and Egypt. Emerg. Infect. Dis. 2022, 28, 1933–1935. [Google Scholar] [CrossRef]
- Hoang, M.; Wu, C.N.; Lin, C.F.; Nguyen, H.T.T.; Le, V.P.; Chiou, M.T.; Lin, C.N. Genetic characterization of feline panleukopenia virus from dogs in Vietnam reveals a unique Thr101 mutation in VP2. PeerJ 2020, 8, e9752. [Google Scholar] [CrossRef]
- Chen, B.; Zhang, X.; Zhu, J.; Liao, L.; Bao, E. Molecular Epidemiological Survey of Canine Parvovirus Circulating in China from 2014 to 2019. Pathogens 2021, 10, 588. [Google Scholar] [CrossRef]
- Sassa, Y.; Yamamoto, H.; Mochizuki, M.; Umemura, T.; Horiuchi, M.; Ishiguro, N.; Miyazawa, T. Successive deaths of a captive snow leopard (Uncia uncia) and a serval (Leptailurus serval) by infection with feline panleukopenia virus at Sapporo Maruyama Zoo. J. Vet. Med. Sci. 2011, 73, 491–494. [Google Scholar] [CrossRef] [PubMed]
- Truyen, U.; Muller, T.; Heidrich, R.; Tackmann, K.; Carmichael, L.E. Survey on viral pathogens in wild red foxes (Vulpes vulpes) in Germany with emphasis on parvoviruses and analysis of a DNA sequence from a red fox parvovirus. Epidemiol. Infect. 1998, 121, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Kolangath, S.M.; Upadhye, S.V.; Dhoot, V.M.; Pawshe, M.D.; Bhadane, B.K.; Gawande, A.P.; Kolangath, R.M. Molecular investigation of Feline Panleukopenia in an endangered leopard (Panthera pardus)—A case report. BMC Vet. Res. 2023, 19, 56. [Google Scholar] [CrossRef] [PubMed]
- Studdert, M.J.; Kelly, C.M.; Harrigan, K.E. Isolation of panleucopaenia virus from lions. Vet. Rec. 1973, 93, 156–158. [Google Scholar] [CrossRef]
- Yang, S.; Wang, S.; Feng, H.; Zeng, L.; Xia, Z.; Zhang, R.; Zou, X.; Wang, C.; Liu, Q.; Xia, X. Isolation and characterization of feline panleukopenia virus from a diarrheic monkey. Vet. Microbiol. 2010, 143, 155–159. [Google Scholar] [CrossRef]
- Yi, S.; Liu, S.; Meng, X.; Huang, P.; Cao, Z.; Jin, H.; Wang, J.; Hu, G.; Lan, J.; Zhang, D.; et al. Feline Panleukopenia Virus with G299E Substitution in the VP2 Protein First Identified From a Captive Giant Panda in China. Front. Cell. Infect. Microbiol. 2021, 11, 820144. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Hu, H.; Lan, J.; Yang, Z.; Peng, Q.; Yan, L.; Luo, L.; Wu, L.; Lang, Y.; Yan, Q. Characterization of a fatal feline panleukopenia virus derived from giant panda with broad cell tropism and zoonotic potential. Front. Immunol. 2023, 14, 1237630. [Google Scholar] [CrossRef]
- Wang, K.; Du, S.; Wang, Y.; Wang, S.; Luo, X.; Zhang, Y.; Liu, C.; Wang, H.; Pei, Z.; Hu, G. Isolation and identification of tiger parvovirus in captive siberian tigers and phylogenetic analysis of VP2 gene. Infect. Genet. Evol. 2019, 75, 103957. [Google Scholar] [CrossRef]
- Wang, X.; Li, T.; Liu, H.; Du, J.; Zhou, F.; Dong, Y.; He, X.; Li, Y.; Wang, C. Recombinant feline parvovirus infection of immunized tigers in central China. Emerg. Microbes Infect. 2017, 6, e42. [Google Scholar] [CrossRef] [PubMed]
- Diao, F.; Zhao, Y.; Wang, J.; Wei, X.; Cui, K.; Liu, C.; Guo, S.; Jiang, S.; Xie, Z. Molecular characterization of feline panleukopenia virus isolated from mink and its pathogenesis in mink. Vet. Microbiol. 2017, 205, 92–98. [Google Scholar] [CrossRef]
- Allison, A.B.; Kohler, D.J.; Ortega, A.; Hoover, E.A.; Grove, D.M.; Holmes, E.C.; Parrish, C.R. Host-specific parvovirus evolution in nature is recapitulated by in vitro adaptation to different carnivore species. PLoS Pathog. 2014, 10, e1004475. [Google Scholar] [CrossRef]
- Chen, X.; Wang, J.; Zhou, Y.; Yue, H.; Zhou, N.; Tang, C. Circulation of heterogeneous Carnivore protoparvovirus 1 in diarrheal cats and prevalence of an A91S feline panleukopenia virus variant in China. Transbound. Emerg. Dis. 2022, 69, e2913–e2925. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, X.; Hao, Y.; Zhang, G.; Lyu, Y.; Wang, J.; Liu, W.; Qin, T. Characterization of the VP2 and NS1 genes from canine parvovirus type 2 (CPV-2) and feline panleukopenia virus (FPV) in Northern China. Front. Vet. Sci. 2022, 9, 934849. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, H.; Wang, L.; Spibey, N.; Liu, C.; Ding, H.; Liu, W.; Liu, Y.; Tian, K. Genetic characterization of parvoviruses in domestic cats in Henan province, China. Transbound. Emerg. Dis. 2018, 65, 1429–1435. [Google Scholar] [CrossRef]
- Pan, S.; Jiao, R.; Xu, X.; Ji, J.; Guo, G.; Yao, L.; Kan, Y.; Xie, Q.; Bi, Y. Molecular characterization and genetic diversity of parvoviruses prevalent in cats in Central and Eastern China from 2018 to 2022. Front. Vet. Sci. 2023, 10, 1218810. [Google Scholar] [CrossRef]
- Liu, C.; Liu, Y.; Qian, P.; Cao, Y.; Wang, J.; Sun, C.; Huang, B.; Cui, N.; Huo, N.; Wu, H.; et al. Molecular and serological investigation of cat viral infectious diseases in China from 2016 to 2019. Transbound. Emerg. Dis. 2020, 67, 2329–2335. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, J.; Bi, Z.; Tan, Y.; Lv, L.; Zhao, H.; Xia, X.; Zhu, Y.; Wang, Y.; Qian, J. Molecular epidemiology and genetic evolution of canine parvovirus in East China, during 2018–2020. Infect. Genet. Evol. 2021, 90, 104780. [Google Scholar] [CrossRef] [PubMed]
- Palermo, L.M.; Hueffer, K.; Parrish, C.R. Residues in the apical domain of the feline and canine transferrin receptors control host-specific binding and cell infection of canine and feline parvoviruses. J. Virol. 2003, 77, 8915–8923. [Google Scholar] [CrossRef] [PubMed]
- Inthong, N.; Sutacha, K.; Kaewmongkol, S.; Sinsiri, R.; Sribuarod, K.; Sirinarumitr, K.; Sirinarumitr, T. Feline panleukopenia virus as the cause of diarrhea in a banded linsang (Prionodon linsang) in Thailand. J. Vet. Med. Sci. 2019, 81, 1763–1768. [Google Scholar] [CrossRef]
- Parker, J.S.L.; Parrish, C.R. Canine parvovirus host range is determined by the specific conformation of an additional region of the capsid. J. Virol. 1997, 71, 9214–9222. [Google Scholar] [CrossRef]
- Domingues, C.F.; de Castro, T.X.; do Lago, B.V.; Garcia, R. Genetic characterization of the parvovirus full-length VP2 gene in domestic cats in Brazil. Res. Vet. Sci. 2024, 170, 105186. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Sun, Z.; Xue, X.; Pan, Y.; Zhen, S.; Liu, Y.; Zhan, J.; Jiang, L.; Zhang, J.; Zhu, H.; et al. China-origin G1 group isolate FPV072 exhibits higher infectivity and pathogenicity than G2 group isolate FPV027. Front. Vet. Sci. 2024, 11, 1328244. [Google Scholar] [CrossRef]
- Xue, H.; Hu, C.; Ma, H.; Song, Y.; Zhu, K.; Fu, J.; Mu, B.; Gao, X. Isolation of feline panleukopenia virus from Yanji of China and molecular epidemiology from 2021 to 2022. J. Vet. Sci. 2023, 24, e29. [Google Scholar] [CrossRef]
- Allison, A.B.; Organtini, L.J.; Zhang, S.; Hafenstein, S.L.; Holmes, E.C.; Parrish, C.R. Single Mutations in the VP2 300 Loop Region of the Three-Fold Spike of the Carnivore Parvovirus Capsid Can Determine Host Range. J. Virol. 2016, 90, 753–767. [Google Scholar] [CrossRef]
- Strassheim, M.L.; Gruenberg, A.; Veijalainen, P.; Sgro, J.Y.; Parrish, C.R. Two dominant neutralizing antigenic determinants of canine parvovirus are found on the threefold spike of the virus capsid. Virology 1994, 198, 175–184. [Google Scholar] [CrossRef]
- Agbandje-McKenna, M.; Llamas-Saiz, A.L.; Wang, F.; Tattersall, P.; Rossmann, M.G. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure 1998, 6, 1369–1381. [Google Scholar] [CrossRef] [PubMed]
- Bergeron, J.; Hebert, B.; Tijssen, P. Genome organization of the Kresse strain of porcine parvovirus: Identification of the allotropic determinant and comparison with those of NADL-2 and field isolates. J. Virol. 1996, 70, 2508–2515. [Google Scholar] [CrossRef]
- Chang, S.F.; Sgro, J.Y.; Parrish, C.R. Multiple amino acids in the capsid structure of canine parvovirus coordinately determine the canine host range and specific antigenic and hemagglutination properties. J. Virol. 1992, 66, 6858–6867. [Google Scholar] [CrossRef] [PubMed]
- Hueffer, K.; Parrish, C.R. Parvovirus host range, cell tropism and evolution. Curr. Opin. Microbiol. 2003, 6, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Callaway, H.M.; Feng, K.H.; Lee, D.W.; Allison, A.B.; Pinard, M.; McKenna, R.; Agbandje-McKenna, M.; Hafenstein, S.; Parrish, C.R. Parvovirus Capsid Structures Required for Infection: Mutations Controlling Receptor Recognition and Protease Cleavages. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Hafenstein, S.; Palermo, L.M.; Kostyuchenko, V.A.; Xiao, C.; Morais, M.C.; Nelson, C.D.; Bowman, V.D.; Battisti, A.J.; Chipman, P.R.; Parrish, C.R.; et al. Asymmetric binding of transferrin receptor to parvovirus capsids. Proc. Natl. Acad. Sci. USA 2007, 104, 6585–6589. [Google Scholar] [CrossRef]
- Agbandje, M.; McKenna, R.; Rossmann, M.G.; Strassheim, M.L.; Parrish, C.R. Structure determination of feline panleukopenia virus empty particles. Proteins 1993, 16, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Hueffer, K.; Govindasamy, L.; Agbandje-McKenna, M.; Parrish, C.R. Combinations of two capsid regions controlling canine host range determine canine transferrin receptor binding by canine and feline parvoviruses. J. Virol. 2003, 77, 10099–10105. [Google Scholar] [CrossRef] [PubMed]
- Govindasamy, L.; Hueffer, K.; Parrish, C.R.; Agbandje-McKenna, M. Structures of host range-controlling regions of the capsids of canine and feline parvoviruses and mutants. J. Virol. 2003, 77, 12211–12221. [Google Scholar] [CrossRef] [PubMed]
- Goodman, L.B.; Lyi, S.M.; Johnson, N.C.; Cifuente, J.O.; Hafenstein, S.L.; Parrish, C.R. Binding site on the transferrin receptor for the parvovirus capsid and effects of altered affinity on cell uptake and infection. J. Virol. 2010, 84, 4969–4978. [Google Scholar] [CrossRef]
- Shao, R.; Ye, C.; Zhang, Y.; Sun, X.; Cheng, J.; Zheng, F.; Cai, S.; Ji, J.; Ren, Z.; Zhong, L.; et al. Novel parvovirus in cats, China. Virus Res. 2021, 304, 198529. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.S.; Murphy, W.J.; Wang, D.; O’Brien, S.J.; Parrish, C.R. Canine and feline parvoviruses can use human or feline transferrin receptors to bind, enter, and infect cells. J. Virol. 2001, 75, 3896–3902. [Google Scholar] [CrossRef] [PubMed]
- Hueffer, K.; Parker, J.S.; Weichert, W.S.; Geisel, R.E.; Sgro, J.Y.; Parrish, C.R. The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J. Virol. 2003, 77, 1718–1726. [Google Scholar] [CrossRef]
- Ikeda, Y.; Mochizuki, M.; Naito, R.; Nakamura, K.; Miyazawa, T.; Mikami, T.; Takahashi, E. Predominance of canine parvovirus (CPV) in unvaccinated cat populations and emergence of new antigenic types of CPVs in cats. Virology 2000, 278, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Pollock, R.V.; Carmichael, L.E. Use of modified live feline panleukopenia virus vaccine to immunize dogs against canine parvovirus. Am. J. Vet. Res. 1983, 44, 169–175. [Google Scholar]
- Truyen, U.; Parrish, C.R. Canine and feline host ranges of canine parvovirus and feline panleukopenia virus: Distinct host cell tropisms of each virus in vitro and in vivo. J. Virol. 1992, 66, 5399–5408. [Google Scholar] [CrossRef]
- Steinel, A.; Munson, L.; van Vuuren, M.; Truyen, U. Genetic characterization of feline parvovirus sequences from various carnivores. J. Gen. Virol. 2000, 81, 345–350. [Google Scholar] [CrossRef] [PubMed]

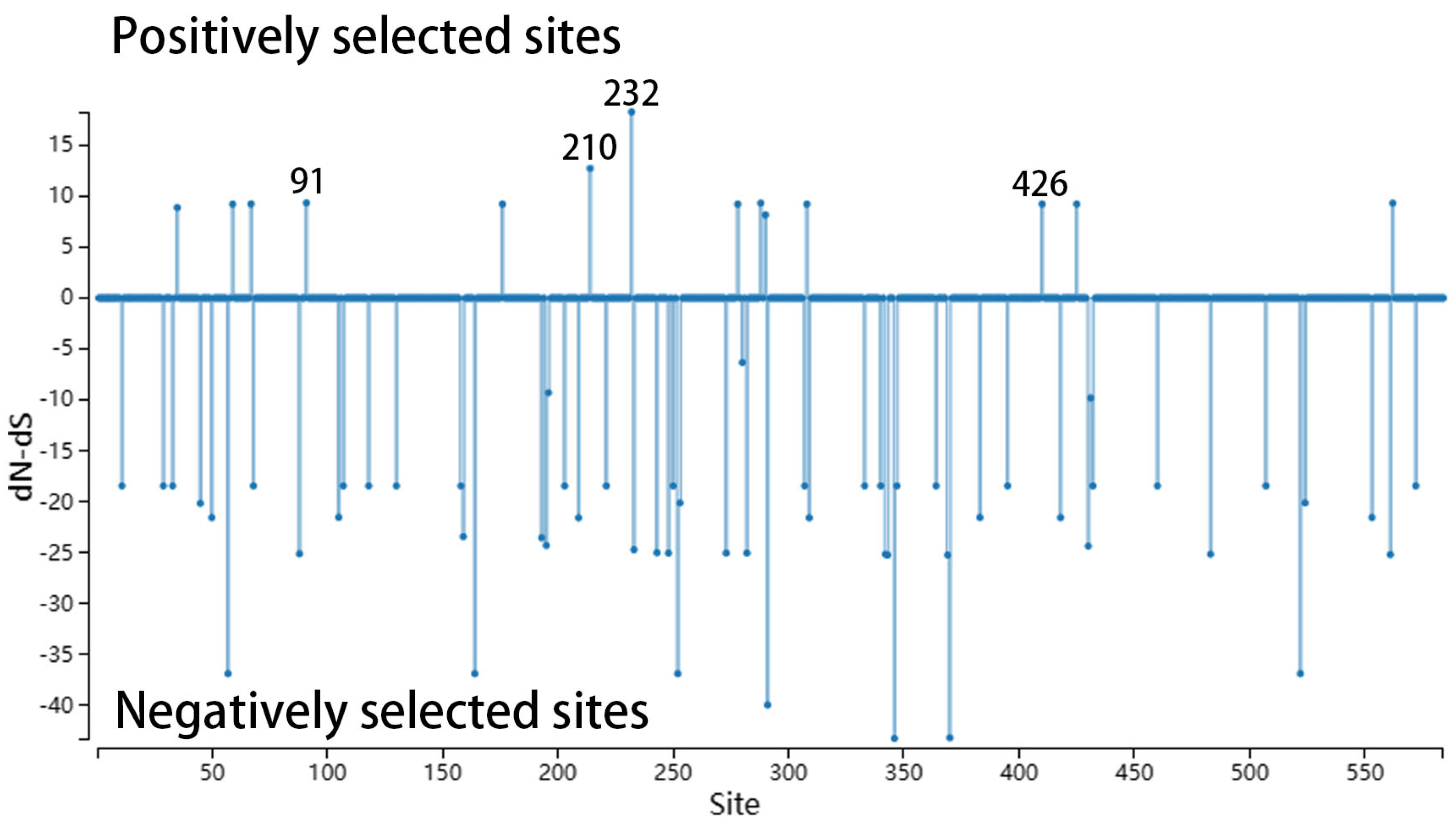
 ).
).
 ).
).
| Sample | Year | Origin | Host | Clinical Feature | Age | Vaccination (N/1 a/2 b/3 c) | GenBank Accession No. |
|---|---|---|---|---|---|---|---|
| JSYZ-85 | 2019 | China/Jiangsu | Dog | High fever, diarrhea, dehydration | 5 months | 2 d | MW017596.1 |
| JSYZ-122 | 2019 | China/Jiangsu | Cat | High fever, diarrhea | 4 months | 1 | MW017628.1 |
| JSYZ-123 | 2019 | China/Jiangsu | Cat | High fever, diarrhea, dehydration | 1 months | N | MW017629.1 |
| JSYZ-124 | 2019 | China/Jiangsu | Cat | High fever, vomit, diarrhea, dehydration | 2 months | N | MW017630.1 |
| ZJHN-135 | 2020 | China/Zhejiang | Dog | High fever, diarrhea, dehydration | 3 months | 1 d | MW017616.1 |
| ZJHN-138 | 2020 | China/Zhejiang | Dog | High fever, vomit | 1 months | N | MW017618.1 |
| SH-118 | 2020 | China/Shanghai | Cat | High fever | 4 months | 2 | MW017625.1 |
| SH-120 | 2020 | China/Shanghai | Cat | High fever, vomit, diarrhea | 3 months | N | MW017626.1 |
| SH-121 | 2020 | China/Shanghai | Cat | High fever, diarrhea | 3 months | N | MW017627.1 |
| ZJHN-126 | 2020 | China/Zhejiang | Cat | High fever, vomit, diarrhea, dehydration | 3 months | 1 | MW017631.1 |
| JSYZ-168 | 2020 | China/Jiangsu | Cat | High fever, vomit, diarrhea | 4 months | 1 | MW791426.1 |
| JSYZ-169 | 2020 | China/Jiangsu | Cat | High fever, diarrhea, dehydration | 3 months | 1 | MW791427.1 |
| SH-21D2 | 2021 | China/Shanghai | Cat | High fever, vomit | 4 months | 2 | OP796706 |
| SH-21D4 | 2021 | China/Shanghai | Cat | High fever | 4 months | 1 | OP796707 |
| JSNJ-21G4 | 2021 | China/Jiangsu | Cat | High fever, vomit, diarrhea | 3 months | N | OP796708 |
| JSNJ-21G5 | 2021 | China/Jiangsu | Cat | High fever, vomit, diarrhea | 6 months | 3 | OP796709 |
| HNZZ-2201 | 2022 | China/Henan | Cat | High fever, diarrhea, dehydration | 3 months | N | PQ560975 |
| HNZZ-2202 | 2022 | China/Henan | Cat | High fever, diarrhea | 6 months | 2 | PQ560976 |
| HNZZ-2203 | 2022 | China/Henan | Cat | High fever, vomit, diarrhea | 3 months | 1 | PQ560977 |
| HNZZ-2204 | 2022 | China/Henan | Cat | High fever, diarrhea | 5 months | 2 | PQ560978 |
| ZJHZ-2202 | 2022 | China/Zhejiang | Cat | High fever, diarrhea | 5 months | N | OP796710 |
| ZJHZ-2203 | 2022 | China/Zhejiang | Cat | High fever, vomit, diarrhea | 5 months | 2 | OP796711 |
| ZJHZ-2204 | 2022 | China/Zhejiang | Cat | High fever, diarrhea, dehydration | 4 months | N | OP796712 |
| ZJHZ-2205 | 2022 | China/Zhejiang | Cat | High fever, diarrhea, dehydration | 8 months | 3 | OP796713 |
| ZJHZ-2206 | 2022 | China/Zhejiang | Cat | High fever, vomit, diarrhea, dehydration | 6 months | 2 | OP796714 |
| ZJHZ-2207 | 2022 | China/Zhejiang | Cat | High fever, diarrhea, dehydration | 6 months | N | OP796715 |
| ZJHZ-2208 | 2022 | China/Zhejiang | Cat | High fever, diarrhea | 11 months | 3 | OP796716 |
| HNZZ-2301 | 2023 | China/Henan | Cat | High fever | 4 months | 2 | PQ560979 |
| HNZZ-2302 | 2023 | China/Henan | Cat | High fever, vomit, diarrhea | 3 months | N | PQ560980 |
| HNZZ-2303 | 2023 | China/Henan | Cat | High fever, diarrhea | 3 months | N | PQ560981 |
| HNZZ-2304 | 2023 | China/Henan | Cat | High fever, vomit, diarrhea, dehydration | 3 months | 1 | PQ560982 |
| HNZZ-2305 | 2023 | China/Henan | Cat | High fever, vomit, diarrhea | 4 months | 1 | PQ560983 |
| HNZZ-2306 | 2023 | China/Henan | Cat | High fever | 3 months | N | PQ560984 |
| HNZZ-2307 | 2023 | China/Henan | Cat | High fever | 3 months | N | PQ560985 |
| HNZZ-2308 | 2023 | China/Henan | Cat | High fever, vomit, diarrhea, dehydration | 6 months | 2 | PQ560986 |
| HNZZ-2401 | 2024 | China/Henan | Cat | High fever, diarrhea | 4 months | 1 | PQ560987 |
| HNZZ-2402 | 2024 | China/Henan | Cat | High fever, diarrhea, dehydration | 1 months | N | PQ560988 |
| HNZZ-2403 | 2024 | China/Henan | Cat | High fever, vomit, diarrhea, dehydration | 2 months | N | PQ560989 |
| HNZZ-2404 | 2024 | China/Henan | Cat | High fever, diarrhea, dehydration | 2 months | N | PQ560990 |
| HNZZ-2405 | 2024 | China/Henan | Cat | High fever | 11 months | 3 | PQ560991 |
| HNZZ-2406 | 2024 | China/Henan | Cat | High fever, vomit, diarrhea | 4 months | 2 | PQ560992 |
| Strain | Virus or Genotype | 35 | 59 | 67 | 80 | 91 | 93 | 101 | 103 | 176 | 196 | 214 | 232 | 278 | 280 | 288 | 305 | 323 | 410 | 425 | 426 | 564 | 568 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SH-21D2 | FPV | S | V | R | K | S | K | T | V | A | D | W | V | T | Q | P | D | D | P | T | N | N | A |
| SH-21D4 | FPV | S | A | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | S | T | N | N | A |
| JSNJ-21G4 | FPV | S | V | R | K | S | K | T | V | A | G | W | G | T | Q | P | D | D | P | T | N | N | A |
| JSNJ-21G5 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | A | Q | P | D | D | P | T | N | N | A |
| HNZZ-2201 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | Y | D | P | T | N | N | A |
| HNZZ-2202 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | Y | D | P | T | N | N | A |
| HNZZ-2203 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | Y | D | P | T | N | N | A |
| HNZZ-2204 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| ZJHZ-2202 | FPV | S | V | G | K | S | K | T | V | A | G | W | V | T | Q | L | D | D | P | S | N | N | A |
| ZJHZ-2203 | FPV | S | V | R | K | S | K | T | V | T | G | G | V | T | Q | P | D | D | P | T | N | N | A |
| ZJHZ-2204 | FPV | W | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| ZJHZ-2205 | FPV | W | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| ZJHZ-2206 | FPV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| ZJHZ-2207 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | R | P | D | D | P | T | N | N | A |
| ZJHZ-2208 | FPV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| HNZZ-2301 | FPV | S | V | R | K | S | K | T | V | A | G | W | G | T | Q | P | D | D | P | T | N | N | A |
| HNZZ-2302 | FPV | S | V | R | K | S | K | I | V | A | G | W | G | T | Q | P | D | D | P | T | N | N | A |
| HNZZ-2303 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| HNZZ-2304 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| HNZZ-2305 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| HNZZ-2306 | FPV | W | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| HNZZ-2307 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| HNZZ-2308 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| HNZZ-2401 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| HNZZ-2402 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| HNZZ-2403 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| HNZZ-2404 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| HNZZ-2405 | FPV | S | V | G | K | S | K | T | V | A | G | W | V | T | Q | L | D | D | P | S | N | N | A |
| HNZZ-2406 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| JSYZ-85 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| ZJHN-135 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| ZJHN-138 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| SH-118 | FPV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| SH-120 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| SH-121 | FPV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| JSYZ-122 | FPV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| JSYZ-123 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| JSYZ-124 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| ZJHN-126 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| JSYZ-168 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| JSYZ-169 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| Felocell (Vaccine) | FPV | S | V | R | K | A | K | T | V | A | G | W | I | T | Q | P | D | D | P | T | N | N | A |
| XJ-1 | FPV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| JL-3 | FPV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| BJ644 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| HF1 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| F-D33 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| JN-FPV-96 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| SH2003 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| V211 | FPV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| K3 | FPV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| C-DY6 | FPV | S | V | R | K | S | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| HN39AA | FPV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| 19SP_CK-8 | FPV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| Tiger | FPV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| HN-ZZ1 | FPV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| AMPV2020 | FPV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| SY-01/2019 | FPV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| HNZZ2 | FPV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| Cat 3 | FPV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| kai.us.06 | FPV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| AUS-14 | FPV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| 39–566 | FPV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| PT183/12 | FPV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| IZSSI_3201_1_15 | FPV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| Suning | MEV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| LYT-2 | MEV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | K | N | A |
| Dalian2 | MEV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| Jlin/2010 | MEV | S | V | R | K | A | K | T | V | A | G | W | V | T | Q | P | D | D | P | T | N | N | A |
| CPV-b | CPV-2 | S | V | R | R | A | N | I | A | A | G | W | I | T | Q | P | D | N | P | T | N | S | G |
| CPV-15 | CPV-2a | S | V | R | R | A | N | T | A | A | G | W | I | T | Q | P | Y | N | P | T | N | S | G |
| MPCPV-SX | CPV-2a | S | V | R | R | V | N | T | A | A | G | W | I | T | Q | P | D | N | P | T | N | S | G |
| 39 | CPV-2b | S | V | R | R | A | N | T | A | A | G | W | I | T | Q | P | Y | N | P | T | D | S | G |
| 04S23 | CPV-2b | S | V | R | R | A | N | T | A | A | G | W | I | T | Q | P | Y | N | P | T | D | S | G |
| YZ5 | New CPV-2a | S | V | R | R | A | N | T | A | A | G | W | I | T | Q | P | Y | N | P | T | N | S | G |
| JSYZ-93 | New CPV-2a | S | V | R | R | A | N | T | A | A | G | W | I | T | Q | P | Y | N | P | T | N | S | G |
| JSNT-48 | New CPV-2b | S | V | R | R | A | N | T | A | A | G | W | I | T | Q | P | Y | N | P | T | D | S | G |
| LN-14-2 | New CPV-2b | S | V | R | R | A | N | T | A | A | G | W | I | T | Q | P | Y | N | P | T | D | S | G |
| CU21 | CPV-2c | S | V | R | R | A | N | T | A | A | G | W | I | T | Q | P | Y | N | P | T | E | S | G |
| JSYZ-151 | CPV-2c | S | V | R | R | A | N | T | A | A | G | W | I | T | Q | P | Y | N | P | T | E | S | G |
 well-recognized mutations compared with vaccine FPV strain.
well-recognized mutations compared with vaccine FPV strain.  novel mutations compared with vaccine FPV strain.
novel mutations compared with vaccine FPV strain.  well-recognized mutations compared with reference CPV strain.
well-recognized mutations compared with reference CPV strain.Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, Y.; Tang, Z.; Wang, K.; Geng, Z.; Yang, S.; Guo, J.; Chen, Y.; Wang, J.; Fan, Z.; Chen, P.; et al. Epidemiological and Molecular Investigation of Feline Panleukopenia Virus Infection in China. Viruses 2024, 16, 1967. https://doi.org/10.3390/v16121967
Wen Y, Tang Z, Wang K, Geng Z, Yang S, Guo J, Chen Y, Wang J, Fan Z, Chen P, et al. Epidemiological and Molecular Investigation of Feline Panleukopenia Virus Infection in China. Viruses. 2024; 16(12):1967. https://doi.org/10.3390/v16121967
Chicago/Turabian StyleWen, Yinghui, Zhengxu Tang, Kunli Wang, Zhengyang Geng, Simin Yang, Junqing Guo, Yongzhen Chen, Jiankun Wang, Zhiyu Fan, Pengju Chen, and et al. 2024. "Epidemiological and Molecular Investigation of Feline Panleukopenia Virus Infection in China" Viruses 16, no. 12: 1967. https://doi.org/10.3390/v16121967
APA StyleWen, Y., Tang, Z., Wang, K., Geng, Z., Yang, S., Guo, J., Chen, Y., Wang, J., Fan, Z., Chen, P., & Qian, J. (2024). Epidemiological and Molecular Investigation of Feline Panleukopenia Virus Infection in China. Viruses, 16(12), 1967. https://doi.org/10.3390/v16121967






