First Description of Loreto Virus in Three Culicidae Species from the Atlantic Forest, Bahia, Brazil
Abstract
1. Introduction
2. Materials and Methods
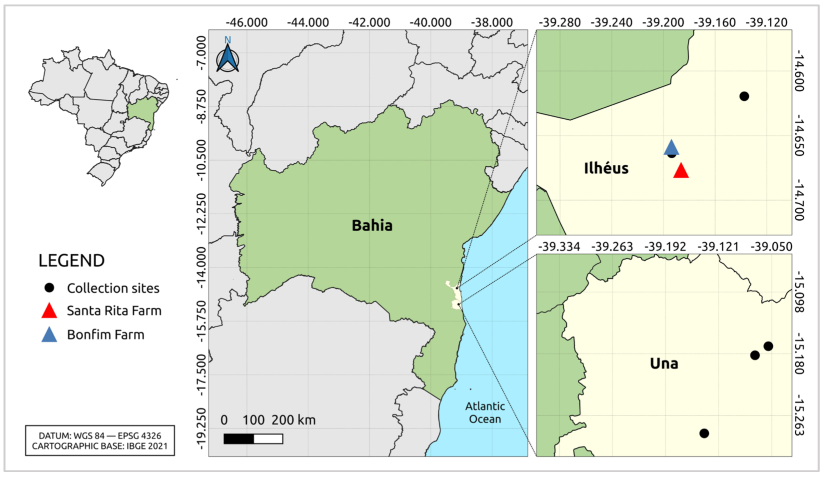
2.1. Sample Collection
2.2. Virus Isolation in Cell Culture
2.3. RNA Extraction and Sequencing
2.4. Sequencing Analysis
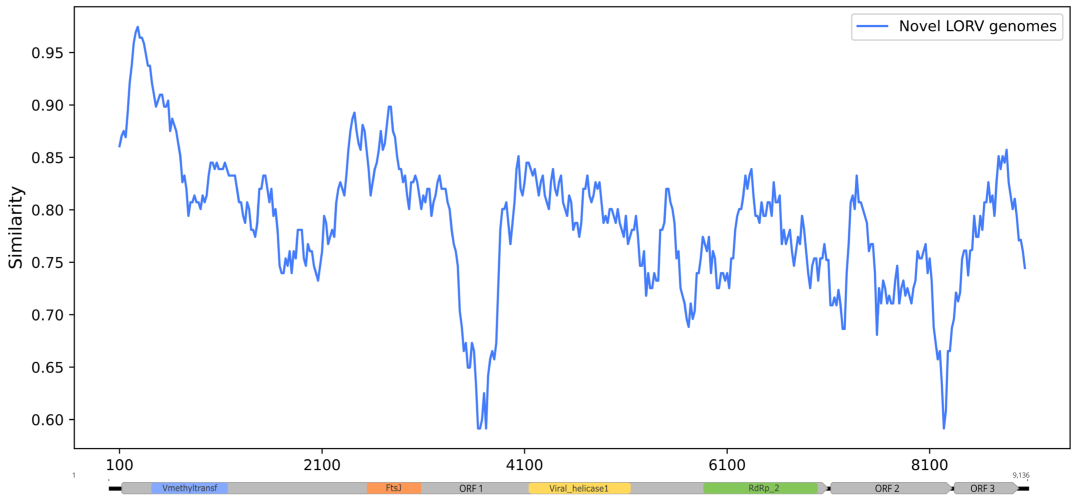
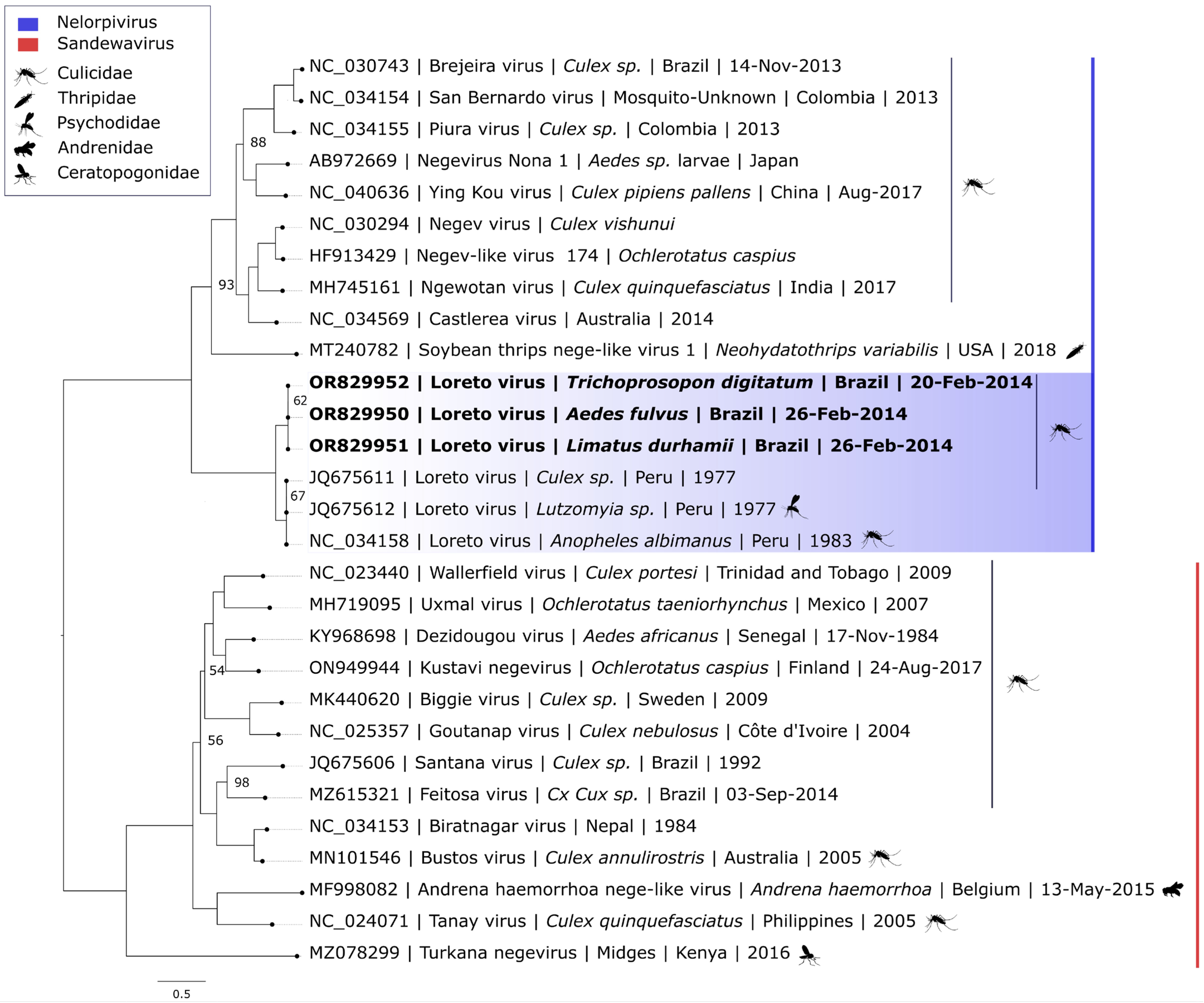
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vasilakis, N.; Forrester, N.L.; Palacios, G.; Nasar, F.; Savji, N.; Rossi, S.L.; Guzman, H.; Wood, T.G.; Popov, V.; Gorchakov, R.; et al. Negevirus: A Proposed New Taxon of Insect-Specific Viruses with Wide Geographic Distribution. J. Virol. 2013, 87, 2475–2488. [Google Scholar] [CrossRef] [PubMed]
- Suvanto, M.T.; Truong Nguyen, P.; Uusitalo, R.; Korhonen, E.M.; Faolotto, G.; Vapalahti, O.; Huhtamo, E.; Smura, T. A Novel Negevirus Isolated from Aedes Vexans Mosquitoes in Finland. Arch. Virol. 2020, 165, 2989–2992. [Google Scholar] [CrossRef] [PubMed]
- da Silva Ribeiro, A.C.; Martins, L.C.; da Silva, S.P.; de Almeida Medeiros, D.B.; Miranda, K.K.P.; Nunes Neto, J.P.; de Oliveira Monteiro, H.A.; do Nascimento, B.L.S.; Junior, J.W.R.; Cruz, A.C.R.; et al. Negeviruses Isolated from Mosquitoes in the Brazilian Amazon. Virol. J. 2022, 19, 17. [Google Scholar] [CrossRef] [PubMed]
- Alger, K.; Caldas, M. The Declining Cocoa Economy and the Atlantic Forest of Southern Bahia, Brazil: Conservation Attitudes of Cocoa Planters. Environmentalist 1994, 14, 107–119. [Google Scholar] [CrossRef]
- Sollberg, I.; Schiavetti, A.; Moraes, M.E.B. Agricultural Management in the Una’s Wildlife Refuge: A Perspective of Conservation by Agroforestry. Rev. Árvore 2014, 38, 241–250. [Google Scholar] [CrossRef]
- Consoli, R.A.G.B.; de Oliveira, R.L. Principais Mosquitos de Importância Sanitária no Brasil; Editora FIOCRUZ: Rio de Janeiro, Brazil, 1994; ISBN 978-85-85676-03-2. [Google Scholar]
- Babraham Bioinformatics—Trim Galore! Available online:. Available online: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 19 January 2023).
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for Clustering the next-Generation Sequencing Data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and Accurate Filtering of Ribosomal RNAs in Metatranscriptomic Data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Huson, D.H.; Auch, A.F.; Qi, J.; Schuster, S.C. MEGAN Analysis of Metagenomic Data. Genome Res. 2007, 17, 377–386. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Guo, X.; Peng, H.; Lu, Y.; Zeng, X.; Dai, K.; Zuo, S.; Zhou, H.; Zhang, J.; Tong, Y. Complete Genome Sequence of a Novel Negevirus Isolated from Culex Tritaeniorhynchus in China. Arch. Virol. 2019, 164, 907–911. [Google Scholar] [CrossRef] [PubMed]
- da Silva Ferreira, R.; de Toni Aquino da Cruz, L.C.; de Souza, V.J.; da Silva Neves, N.A.; de Souza, V.C.; Filho, L.C.F.; da Silva Lemos, P.; de Lima, C.P.S.; Naveca, F.G.; Atanaka, M.; et al. Insect-Specific Viruses and Arboviruses in Adult Male Culicids from Midwestern Brazil. Infect. Genet. Evol. 2020, 85, 104561. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Lin, X.-D.; Tian, J.-H.; Chen, L.-J.; Chen, X.; Li, C.-X.; Qin, X.-C.; Li, J.; Cao, J.-P.; Eden, J.-S.; et al. Redefining the Invertebrate RNA Virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Kondo, H.; Chiba, S.; Maruyama, K.; Andika, I.B.; Suzuki, N. A Novel Insect-Infecting Virga/Nege-like Virus Group and Its Pervasive Endogenization into Insect Genomes. Virus Res. 2019, 262, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Bojko, J.; Jennings, L.A.; Behringer, D.C. A Novel Positive Single-Stranded RNA Virus from the Crustacean Parasite, Probopyrinella Latreuticola (Peracarida: Isopoda: Bopyridae). J. Invertebr. Pathol. 2020, 177, 107494. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, K.; Song, C.; Wang, H.; Sakaguchi, M.; Chalkiadaki, C.; Miyazaki, N.; Nabeshima, T.; Morita, K.; Inoue, S.; Murata, K. Structure and Its Transformation of Elliptical Nege-like Virus Tanay Virus. J. Gen. Virol. 2023, 104, 001863. [Google Scholar] [CrossRef]
- Jancarova, M.; Polanska, N.; Volf, P.; Dvorak, V. The Role of Sand Flies as Vectors of Viruses Other than Phleboviruses. J. Gen. Virol. 2023, 104, 001837. [Google Scholar] [CrossRef]
- Hernández-Rodríguez, J.L.; Granados-Echegoyen, C.A.; Ortega-Morales, B.O.; Ibáñez-Bernal, S.; Pérez-Pacheco, R.; Chan-Bacab, M.; Alonso-Hernández, N.; Pérez-Rentería, C.; Huerta-Jiménez, H. First Record of Limatus Durhamii Theobald (Diptera: Culicidae) in Campeche, Mexico. Fla. Entomol. 2018, 101, 712–715. [Google Scholar] [CrossRef]
- Mangudo, C.; Campos, R.E.; Rossi, G.C.; Gleiser, R.M. Snail Shells as Larval Habitat of Limatus Durhamii (Diptera: Culicidae) in the Yungas of Argentina. Acta Trop. 2017, 167, 204–207. [Google Scholar] [CrossRef]
- Guimarães, A.E.; Gentile, C.; Lopes, C.M.; Mello, R.P. Ecology of Mosquitoes (Diptera: Culicidae) in Areas of Serra Do Mar State Park, State of São Paulo, Brazil. II—Habitat Distribution. Mem. Inst. Oswaldo Cruz 2000, 95, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Palermo, P.M.; Aguilar, P.V.; Sanchez, J.F.; Zorrilla, V.; Flores-Mendoza, C.; Huayanay, A.; Guevara, C.; Lescano, A.G.; Halsey, E.S. Identification of Blood Meals from Potential Arbovirus Mosquito Vectors in the Peruvian Amazon Basin. Am. J. Trop. Med. Hyg. 2016, 95, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, R.S.G.; Hutchings, R.W.; Menezes, I.S.; Sallum, M.A.M. Mosquitoes (Diptera: Culicidae) from the Southwestern Brazilian Amazon: Liberdade and Gregório Rivers. J. Med. Entomol. 2020, 57, 1793–1811. [Google Scholar] [CrossRef] [PubMed]
- Zavortink, T.J.; Roberts, D.R.; Hoch, A.L. Trichoprosopon Digitatum—Morphology, Biology, and Potential Medical Importance. Mosq. Syst. 1983, 15, 141–149. [Google Scholar]
- Carapeta, S.; do Bem, B.; McGuinness, J.; Esteves, A.; Abecasis, A.; Lopes, Â.; de Matos, A.P.; Piedade, J.; de Almeida, A.P.G.; Parreira, R. Negeviruses Found in Multiple Species of Mosquitoes from Southern Portugal: Isolation, Genetic Diversity, and Replication in Insect Cell Culture. Virology 2015, 483, 318–328. [Google Scholar] [CrossRef]
- Carvalho, V.L.; Long, M.T. Insect-Specific Viruses: An Overview and Their Relationship to Arboviruses of Concern to Humans and Animals. Virology 2021, 557, 34–43. [Google Scholar] [CrossRef]
- Patterson, E.I.; Kautz, T.F.; Contreras-Gutierrez, M.A.; Guzman, H.; Tesh, R.B.; Hughes, G.L.; Forrester, N.L. Negeviruses Reduce Replication of Alphaviruses during Coinfection. J. Virol. 2021, 95, e0043321. [Google Scholar] [CrossRef]
- Agboli, E.; Schulze, J.; Jansen, S.; Cadar, D.; Sreenu, V.B.; Leggewie, M.; Altinli, M.; Badusche, M.; Jöst, H.; Börstler, J.; et al. Interaction of Mesonivirus and Negevirus with Arboviruses and the RNAi Response in Culex Tarsalis-Derived Cells. Parasites Vectors 2023, 16, 361. [Google Scholar] [CrossRef]
| Identities (%) | |||||||
|---|---|---|---|---|---|---|---|
| ORF1 | ORF2 | ORF3 * | |||||
| Sample | Host | nt | aa | nt | Aa | nt | aa |
| OR829950 | Ae. fulvus | 82.549 | 93.753 | 77.363 | 87.531 | 82.439 | 92.683 |
| OR829951 | L. durrhamii | 82.563 | 93.753 | 77.363 | 87.531 | 82.771 | 91.979 |
| OR829952 | Tr. digitatum | 82.563 | 93.753 | 77.363 | 87.531 | 82.343 | 92.574 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Paz, T.Y.B.; Hernández, L.H.A.; Silva, F.S.d.; Cruz, A.C.R.; da Silva, S.P.; Fuzii, H.T.; Massafra, J.M.V.; Vianez Júnior, J.L.S.G.; Deem, S.L.; Oliveira, L.d.C.; et al. First Description of Loreto Virus in Three Culicidae Species from the Atlantic Forest, Bahia, Brazil. Viruses 2024, 16, 1674. https://doi.org/10.3390/v16111674
da Paz TYB, Hernández LHA, Silva FSd, Cruz ACR, da Silva SP, Fuzii HT, Massafra JMV, Vianez Júnior JLSG, Deem SL, Oliveira LdC, et al. First Description of Loreto Virus in Three Culicidae Species from the Atlantic Forest, Bahia, Brazil. Viruses. 2024; 16(11):1674. https://doi.org/10.3390/v16111674
Chicago/Turabian Styleda Paz, Thito Y. Bezerra, Leonardo H. Almeida Hernández, Fábio Silva da Silva, Ana C. Ribeiro Cruz, Sandro Patroca da Silva, Hellen Thais Fuzii, Janaina M. Vasconcelos Massafra, João L. S. G. Vianez Júnior, Sharon L. Deem, Leonardo de Carvalho Oliveira, and et al. 2024. "First Description of Loreto Virus in Three Culicidae Species from the Atlantic Forest, Bahia, Brazil" Viruses 16, no. 11: 1674. https://doi.org/10.3390/v16111674
APA Styleda Paz, T. Y. B., Hernández, L. H. A., Silva, F. S. d., Cruz, A. C. R., da Silva, S. P., Fuzii, H. T., Massafra, J. M. V., Vianez Júnior, J. L. S. G., Deem, S. L., Oliveira, L. d. C., De Vleeschouwer, K. M., & Catenacci, L. S. (2024). First Description of Loreto Virus in Three Culicidae Species from the Atlantic Forest, Bahia, Brazil. Viruses, 16(11), 1674. https://doi.org/10.3390/v16111674









