Antiviral Peptide-Generative Pre-Trained Transformer (AVP-GPT): A Deep Learning-Powered Model for Antiviral Peptide Design with High-Throughput Discovery and Exceptional Potency
Abstract
1. Introduction
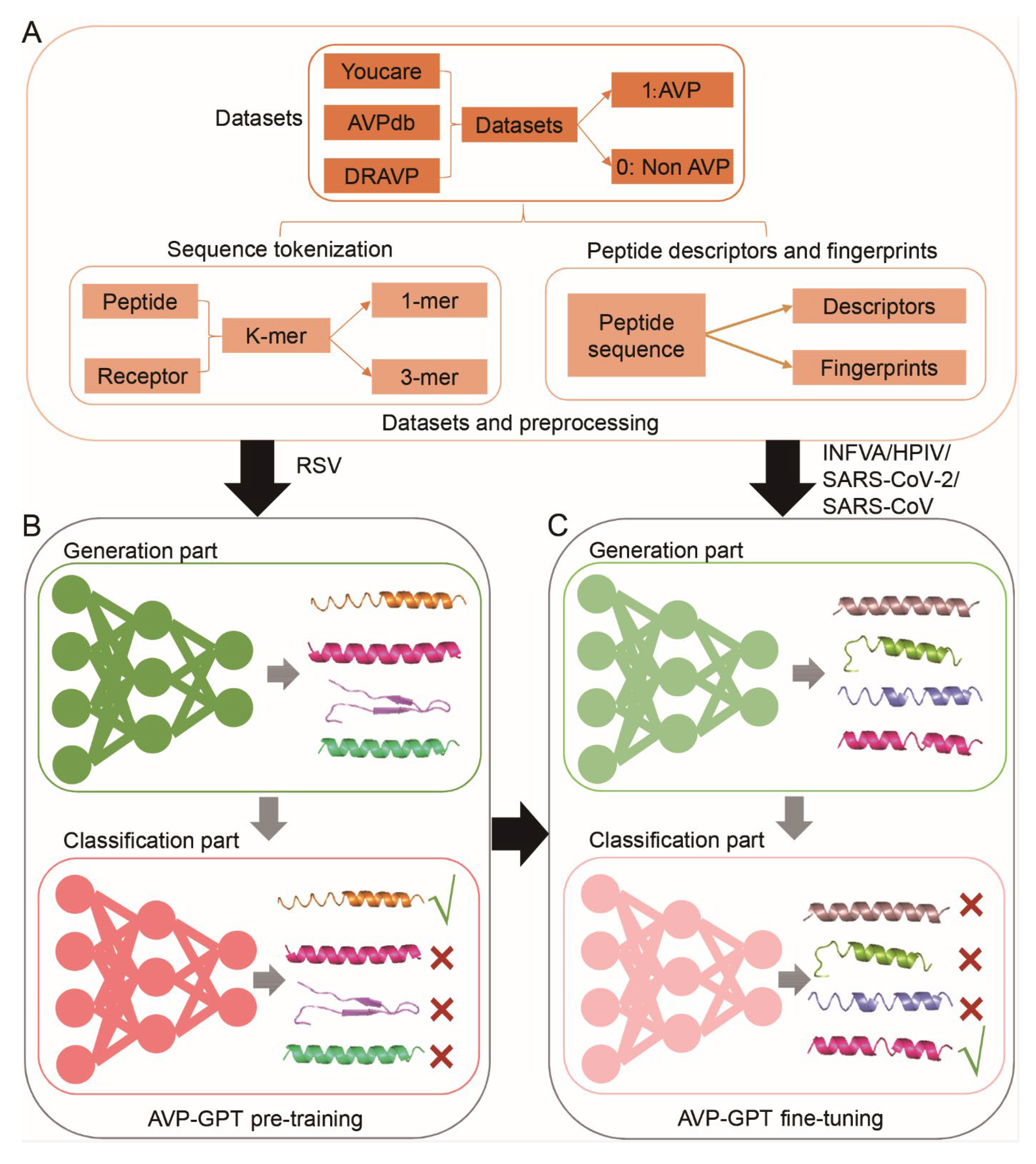
2. Materials and Methods
2.1. Datasets
2.2. Sequence Tokenization
2.3. Peptide Descriptors
- Amino acid composition (AAC): the overall frequency of each amino acid type present in the peptide sequence [6].
- Dipeptide composition (DPC): the frequency of each unique dipeptide (two consecutive amino acids) combination within the sequence [6].
- CKSAAGP: a descriptor set capturing properties like aliphatic, aromatic, and charge distribution [6].
- Pseudo-amino acid composition (PAAC): a broader representation of the amino acid composition that considers additional features beyond just the amino acid type [14].
- Physicochemical features (PHYC): a set of descriptors that encode various physicochemical properties of the peptide, such as alpha-helical propensity, isoelectric point, and hydrophobicity [6].
2.4. Peptide Fingerprints
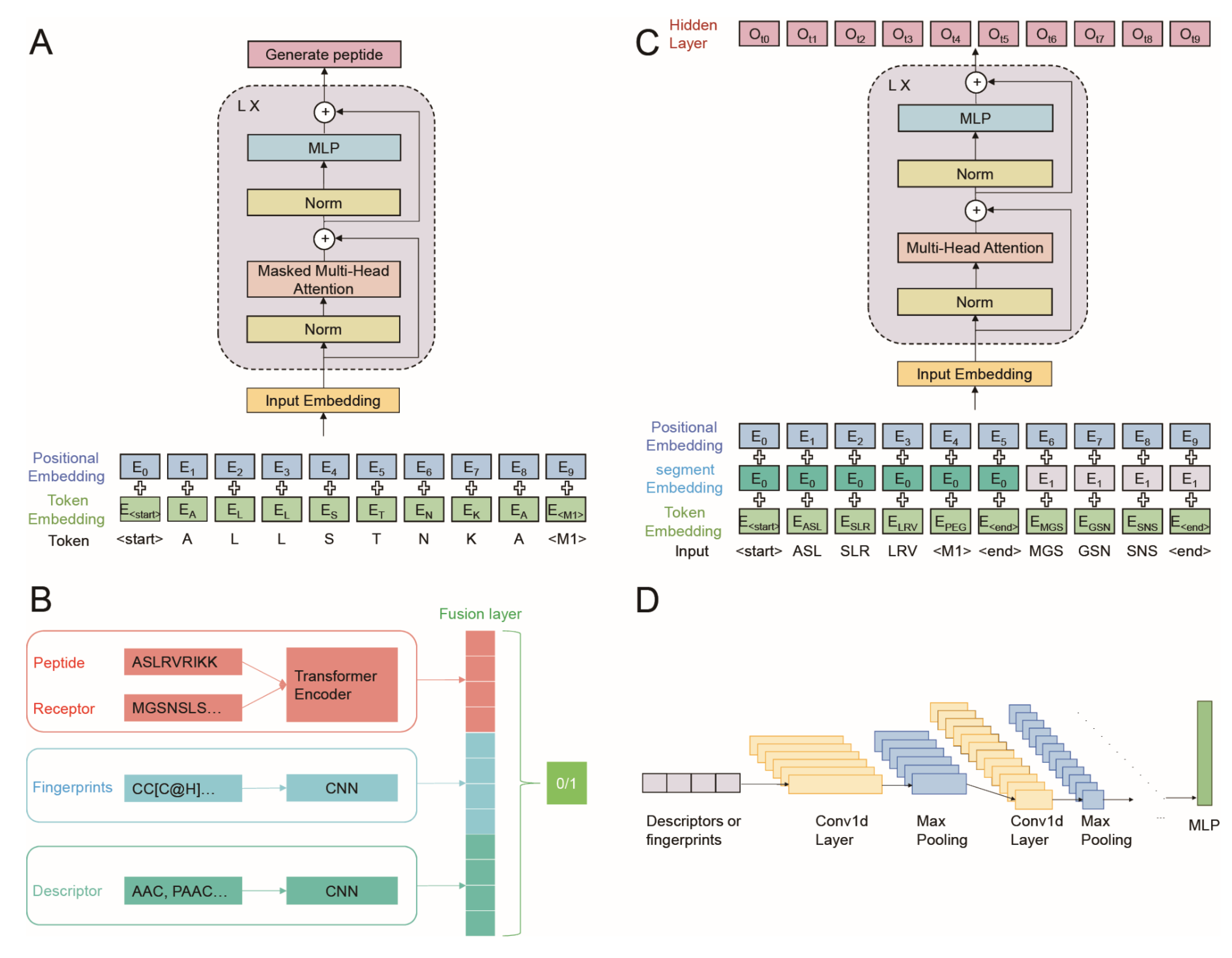
2.5. Generation Part of AVP-GPT Architectures
- Input embedding: set as learnable parameters, positional encodings were added to the input embedding of sequence tokens and modification tokens.
- Masked multi-head attention: this layer allows each output to only pay attention to the past tokens.
- Layer normalization and residual connections: these techniques are applied after each block to improve training stability and gradient flow.
- Peptide generation process, which included the following:
- A “<start>” token is fed as input.
- The model iteratively predicts the next amino acid and modification (if applicable) based on the current sequence.
- The temperature parameter controls the randomness of the generated sequence.
- Top-k sampling selects the next token based on the top-k most probable options.
- Max_new_tokens defines the maximum length of the generated peptide.
- Loss function: cross-entropy with ignore_index = 0 is used to calculate the loss during training, ignoring padded “<pad>” tokens.
2.6. Classification Part of AVP-GPT Architectures
- Transformer encoder: handles peptide sequences and receptor sequences with segment embeddings and positional encodings added to the input embedding (Figure 2C).
- CNN models: two separate CNN models process peptide descriptors and peptide fingerprints.
- Fusion layers: the hidden outputs from the transformer encoder and both CNN models are concatenated in the fusion layers (Figure 2D).
- Binary classification: the final output predicts whether the peptide is an AVP (antiviral) or not.
2.7. Improving Robustness
2.8. AVP-GPT Pre-Training
2.8.1. Generation Part
- Transformer decoders: 3.
- Self-attention heads: 3.
- Dropout rate: 0.5.
- Learning rate: 0.0003.
- Output dimension: 26 (number of tokens).
- Maximum generated tokens: 36 (peptide length limit).
- Temperature: 1 (controls randomness).
- Top-k sampling: 24 (considers top 24 probable next tokens). These are used during inference only.
2.8.2. Classification Part
- Transformer encoders: 2.
- Self-attention heads: 3.
- Dropout rate: 0.5.
- Output dimension: 64 (for transformer encoders).
- CNN layers (peptide descriptors): 3 layers with kernel size 3, dropout 0.5, and output dim 64.
- CNN layers (peptide fingerprints): 3 layers with kernel size 3, dropout 0.5, and output dim 16.
2.9. Training Setup
2.10. Evaluation Methods
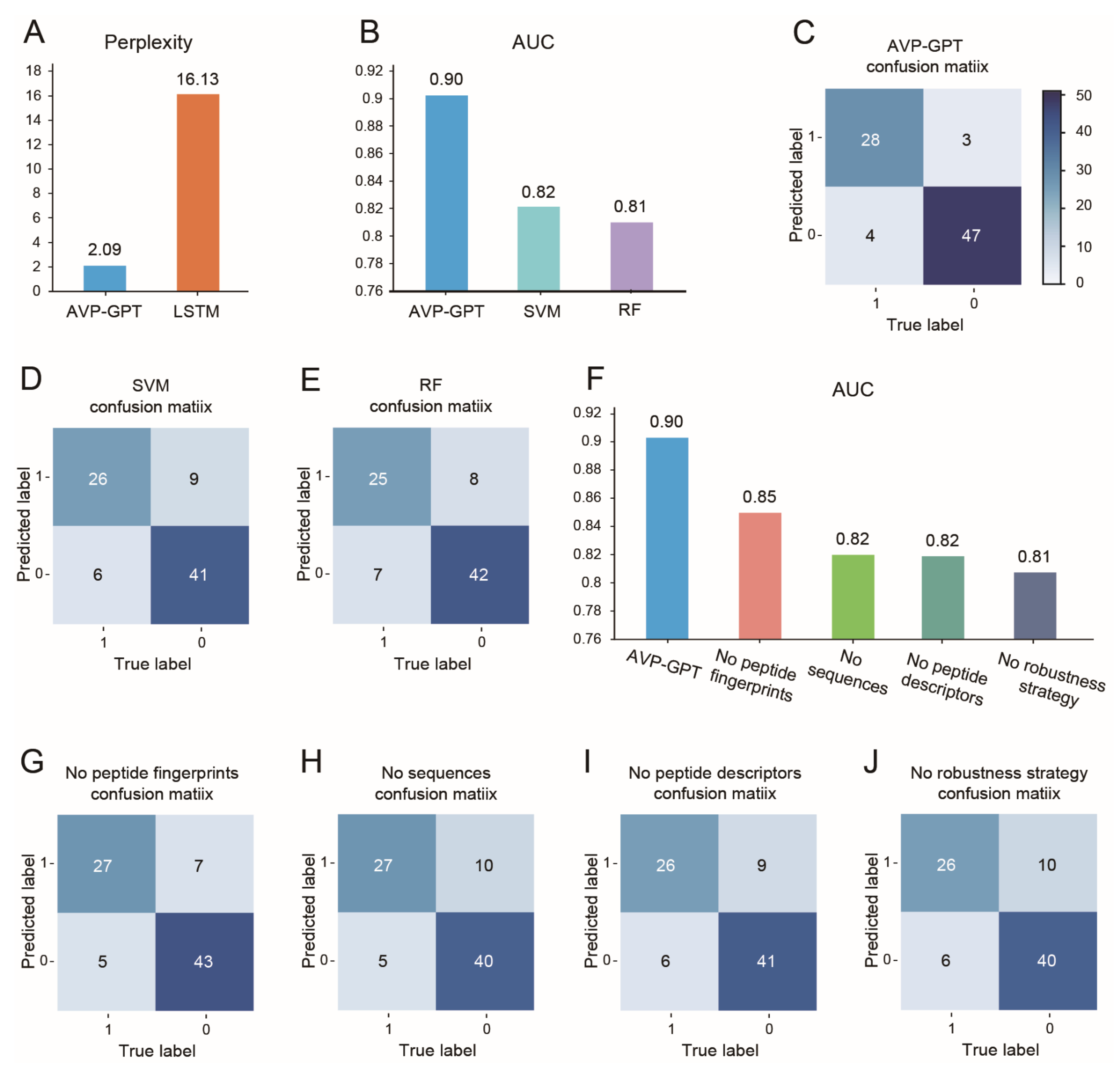
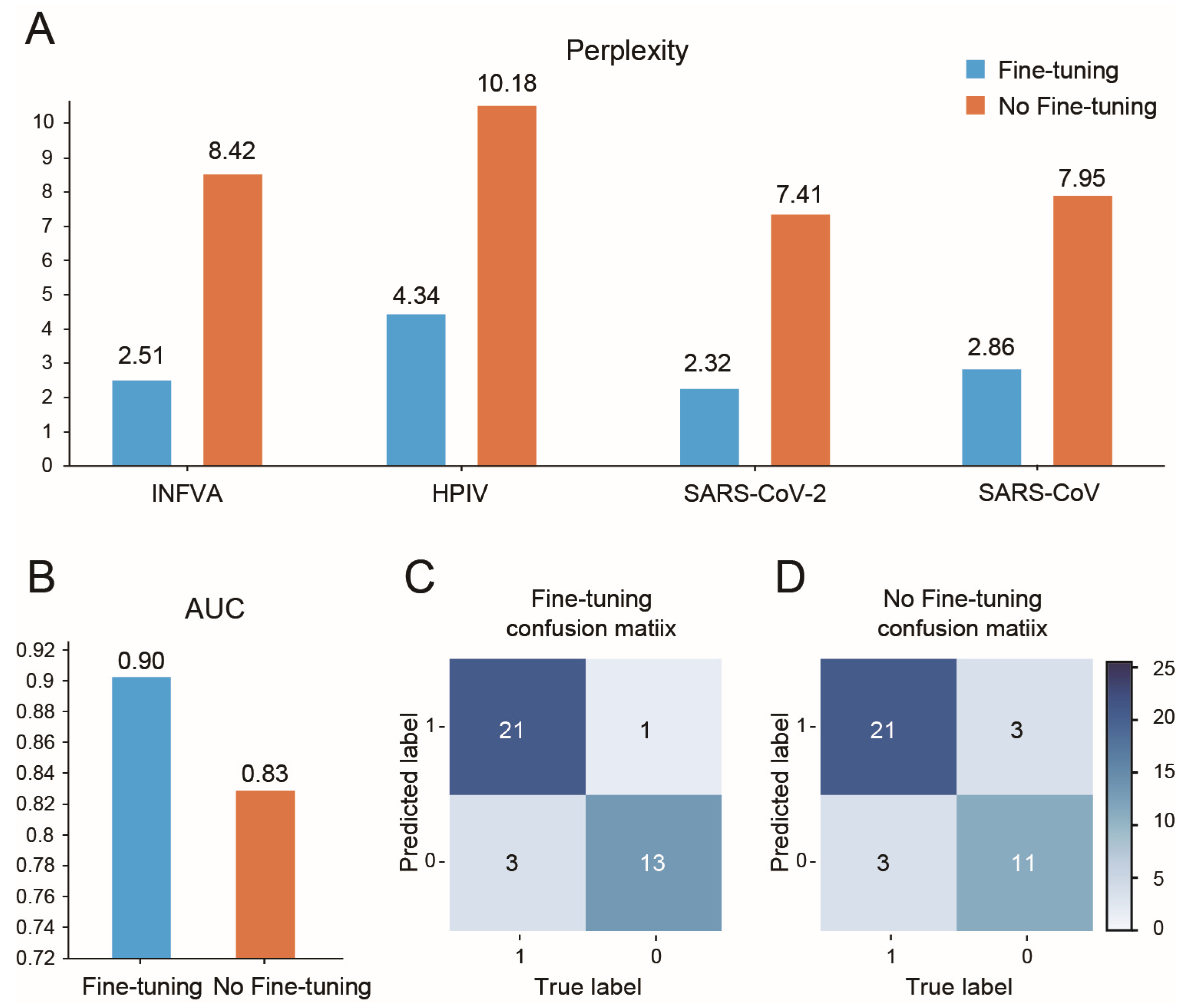
- Perplexity: Used to compare AVP-GPT’s generation performance to an LSTM model, a common approach for peptide generation. Lower perplexity indicates better performance. Perplexity is calculated as the exponent of the loss obtained from the model.
- Area under the curve (AUC): Used to evaluate the classification performance of AVP-GPT compared to an RF and an SVM model, widely used methods for antiviral activity prediction. Higher AUC signifies better classification ability.
- Confusion matrix: We used the confusion matrix to evaluate the classification performance of all models. The confusion matrix provides a detailed breakdown of true positive, true negative, false positive, and false negative predictions, allowing us to assess the model’s accuracy, sensitivity, specificity, and other relevant metrics.
- Ablation study: Conducted to assess the importance of different components and strategies within the AVP-GPT classification part.
2.11. AVP-GPT Fine-Tuning
2.12. Software Implementation of AVP-GPT
2.13. Comparison Study
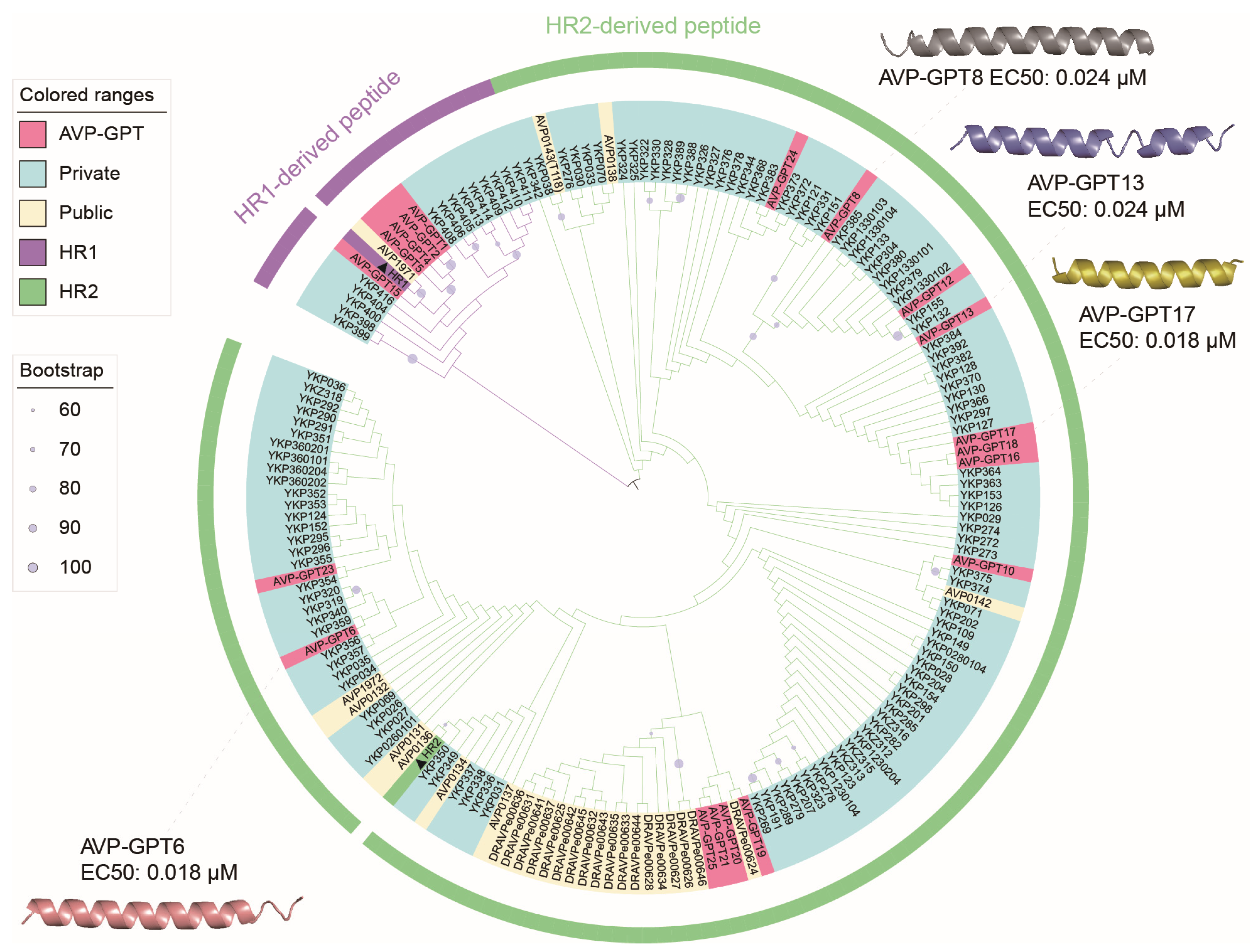
2.14. Phylogenetic Analysis of RSV Peptides
- Sequence alignment: The MUSCLE program (v5.1) [18] was employed to align all 179 peptide sequences.
- Phylogenetic tree construction: Phylogenetic trees were built using RaxML software (v8.2.12) with the PROTGAMMAGTR model [19]. Additionally, bootstrap analysis with 1000 replicates was performed to assess the robustness of the inferred evolutionary relationships.
- Visualization and annotation: The resulting phylogenetic trees were visualized and annotated using the Interactive Tree of Life (iTOL) platform [20].
- Structure prediction: Finally, the predicted structures of the peptides were obtained using the HelixFold-Single tool [21]. This helps to understand the potential functional roles of the AVPs based on their structural features.
2.15. In Vitro Study
- Peptide synthesis: The identified AVP candidates were synthesized using solid-phase peptide synthesis (SPPS) [22] by a commercial vendor, Chinese Peptide Company (CPC, Hangzhou, China). SPPS was chosen for its versatility and ability to produce peptides with various modifications and lengths. Crude peptides were analyzed by UPLC/MS to verify their purity and molecular weight. HPLC preparative was used to purify the peptides to high purity levels. Peptide sequencing was conducted by Shanghai Weipu Company (Shanghai, China) to confirm the correct amino acid sequence.
- Antiviral activity assay: The antiviral activity of the synthesized peptides against RSV and INFVA was evaluated by WuXi AppTec using a plaque reduction assay with the RSV strain A Long and INFV strain A/California/07/2009. HEp-2 cells were seeded in a 96-well plate overnight and infected with RSV, while MDCK cells were seeded and infected with INFVA. Peptides were serially diluted and added to the infected cells, followed by a 2 h incubation period. The medium was then replaced with fresh medium containing the same peptide concentration. Plates were incubated for 24 h at 37 °C. To quantify viral replication, the cells were fixed, permeabilized, and stained with RSV-specific or INFVA-specific antibodies and a secondary antibody. TrueBlue solution was added, and the number of plaques was counted using a microplate imaging counter.
- Cytotoxic assay: Cytotoxicity experiments were conducted in parallel with antiviral experiments to assess the potential toxicity of the peptides. HEp-2 or MDCK cells were seeded into microplates at a density of 30,000 cells per well and cultured overnight in a 5% CO2, 37 °C incubator. The diluted test samples were added to the cells, and a cell control (cells, no compound treatment) and a culture medium control (culture medium only, no cell or compound treatment) were set up. The final concentration of DMSO in the culture medium was 0.5%, which is a commonly used concentration that does not significantly affect cell viability. Cells were cultured for 1 day in a 5% CO2, 37 °C incubator. Cell viability was detected using the CCK8 assay.
3. Results and Discussion
3.1. Workflow of the AVP-GPT
3.2. AVP Generation in Pre-Training
3.3. AVP Identification in Pre-Training
- 0.6 ≤ AUC < 0.7: poor;
- 0.7 ≤ AUC < 0.8: fair;
- 0.8 ≤ AUC < 0.9: considerable;
- 0.9 ≤ AUC: excellent.
3.4. Ablation Study in Pre-Training
3.5. AVP-GPT Fine-Tuning Study
3.6. Comparison Study and Phylogenetic Analysis
3.7. Result of In Vitro Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tripathi, N.M.; Bandyopadhyay, A. High throughput virtual screening (HTVS) of peptide library: Technological advancement in ligand discovery. Eur. J. Med. Chem. 2022, 243, 114766. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Pan, Y.; Jiang, S.; Lu, L. Respiratory syncytial virus entry inhibitors targeting the F protein. Viruses 2013, 5, 211–225. [Google Scholar] [CrossRef]
- López-Martínez, R.; Ramírez-Salinas, G.L.; Correa-Basurto, J.; Barrón, B.L. Inhibition of influenza A virus infection in vitro by peptides designed in silico. PLoS ONE 2013, 8, e76876. [Google Scholar] [CrossRef]
- Muller, A.T.; Hiss, J.A.; Schneider, G. Recurrent neural network model for constructive peptide design. J. Chem. Inf. Model. 2018, 58, 472–479. [Google Scholar] [CrossRef]
- Ali, F.; Kumar, H.; Alghamdi, W.; Kateb, F.A.; Alarfaj, F.K. Recent advances in machine learning-based models for prediction of antiviral peptides. Arch. Comput. Methods Eng. 2023, 30, 4033–4044. [Google Scholar] [CrossRef]
- Pang, Y.; Yao, L.; Jhong, J.H.; Wang, Z.; Lee, T.Y. AVPIden: A new scheme for identification and functional prediction of antiviral peptides based on machine learning approaches. Brief. Bioinform. 2021, 22, bbab263. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention is all you need. In Proceedings of the 31st Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017; Volume 30. Available online: https://user.phil.hhu.de/~cwurm/wp-content/uploads/2020/01/7181-attention-is-all-you-need.pdf (accessed on 20 October 2024).
- Devlin, J.; Chang, M.W.; Lee, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Radford, A.; Narasimhan, K.; Salimans, T. Improving Language Understanding by Generative Pre-Training. 2018. Available online: https://www.mikecaptain.com/resources/pdf/GPT-1.pdf (accessed on 20 October 2024).
- Qureshi, A.; Thakur, N.; Tandon, H.; Kumar, M. AVPdb: A database of experimentally validated antiviral peptides targeting medically important viruses. Nucleic Acids Res. 2014, 42, D1147–D1153. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, Y.; Sun, X.; Ma, T.; Lao, X.; Zheng, H. DRAVP: A comprehensive database of antiviral peptides and proteins. Viruses 2023, 15, 820. [Google Scholar] [CrossRef]
- Ji, Y.; Zhou, Z.; Liu, H.; Davuluri, R.V. DNABERT: Pre-trained Bidirectional Encoder Representations from Transformers model for DNA-language in genome. Bioinformatics 2021, 37, 2112–2120. [Google Scholar] [CrossRef]
- Singh, M.; Berger, B.; Kim, P.S. LearnCoil-VMF: Computational evidence for coiled-coil-like motifs in many viral membrane-fusion proteins. J. Mol. Biol. 1999, 290, 1031–1041. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C. Prediction of protein cellular attributes using pseudo-amino acid composition. Proteins Struct. Funct. Bioinform. 2001, 43, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Dozat, T. Incorporating Nesterov Momentum into Adam. 2016. Available online: https://openreview.net/forum?id=OM0jvwB8jIp57ZJjtNEZ (accessed on 20 October 2024).
- Paszke, A.; Gross, S.; Chintala, S.; Chanan, G.; Yang, E.; DeVito, Z.; Lin, Z.; Desmaison, A.; Antiga, L.; Lerer, A. Automatic differentiation in pytorch. Openreview 2017. Available online: https://openreview.net/pdf?id=BJJsrmfCZ (accessed on 20 October 2024).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Edgar, R.C. Muscle5: High-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat. Commun. 2022, 13, 6968. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Fang, X.; Wang, F.; Liu, L.; He, J.; Lin, D.; Xiang, Y.; Zhu, K.; Zhang, X.; Wu, H.; Li, H.; et al. A method for multiple-sequence-alignment-free protein structure prediction using a protein language model. Nat. Mach. Intell. 2023, 5, 1087–1096. [Google Scholar] [CrossRef]
- Gaillard, V.; Galloux, M.; Garcin, D.; Eléouët, J.-F.; Le Goffic, R.; Larcher, T.; Rameix-Welti, M.-A.; Boukadiri, A.; Héritier, J.; Segura, J.-M.; et al. A short double-stapled peptide inhibits respiratory syncytial virus entry and spreading. Antimicrob. Agents Chemother. 2017, 61, 02241-16. [Google Scholar] [CrossRef]
- Radford, A.; Wu, J.; Child, R.; Luan, D.; Amodei, D.; Sutskever, I. Language models are unsupervised multitask learners. OpenAI Blog 2019, 1, 9. [Google Scholar]
- Hao, Y.; Mendelsohn, S.; Sterneck, R.; Martinez, R.; Frank, R. Probabilistic predictions of people perusing: Evaluating metrics of language model performance for psycholinguistic modeling. arXiv 2020, arXiv:2009.03954. [Google Scholar]
- Moret, M.; Grisoni, F.; Katzberger, P.; Schneider, G. Perplexity-based molecule ranking and bias estimation of chemical language models. J. Chem. Inf. Model. 2022, 62, 1199–1206. [Google Scholar] [CrossRef] [PubMed]
- Sulam, J.; Ben-Ari, R.; Kisilev, P. Maximizing AUC with Deep Learning for Classification of Imbalanced Mammogram Datasets. In Proceedings of the Eurographics Workshop on Visual Computing for Biology and Medicine (VCBM), Bremen, Germany, 7–8 September 2017; pp. 131–135. [Google Scholar]
- Liu, M.; Yuan, Z.; Ying, Y.; Yang, T. Stochastic auc maximization with deep neural networks. arXiv 2019, arXiv:1908.10831. [Google Scholar]
- Carrington, A.M.; Manuel, D.G.; Fieguth, P.W.; Ramsay, T.; Osmani, V.; Wernly, B.; Bennett, C.; Hawken, S.; McInnes, M.; Magwood, O.; et al. Deep ROC analysis and AUC as balanced average accuracy to improve model selection, understanding and interpretation. arXiv 2021, arXiv:2103.11357. [Google Scholar]
- Çorbacıoğlu, Ş.K.; Aksel, G. Receiver operating characteristic curve analysis in diagnostic accuracy studies: A guide to interpreting the area under the curve value. Turk. J. Emerg. Med. 2023, 23, 195. [Google Scholar] [CrossRef]
- Stahlschmidt, S.R.; Ulfenborg, B.; Synnergren, J. Multimodal deep learning for biomedical data fusion: A review. Brief. Bioinform. 2022, 23, bbab569. [Google Scholar] [CrossRef]
- Han, X.; Zhang, Z.; Ding, N.; Gu, Y.; Liu, X.; Huo, Y.; Qiu, J.; Yao, Y.; Zhang, A.; Zhang, L.; et al. Pre-trained models: Past, present and future. AI Open 2021, 2, 225–250. [Google Scholar] [CrossRef]
- Lambert, D.M.; Barney, S.; Lambert, A.L.; Guthrie, K.; Medinas, R.; E Davis, D.; Bucy, T.; Erickson, J.; Merutka, G.; Petteway, S.R. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc. Natl. Acad. Sci. USA 1996, 93, 2186–2191. [Google Scholar] [CrossRef]
| Data Category | Generation Part (AVP Sequence Creation) | Classification Part (AVP Identification) |
|---|---|---|
| Private AVPs | 132 | 132 |
| Private non-AVPs | Not Available (NA) | 238 |
| Public AVPs | 28 | 28 |
| Public non-AVPs | Not Available (NA) | 14 |
| Total | 160 | 412 |
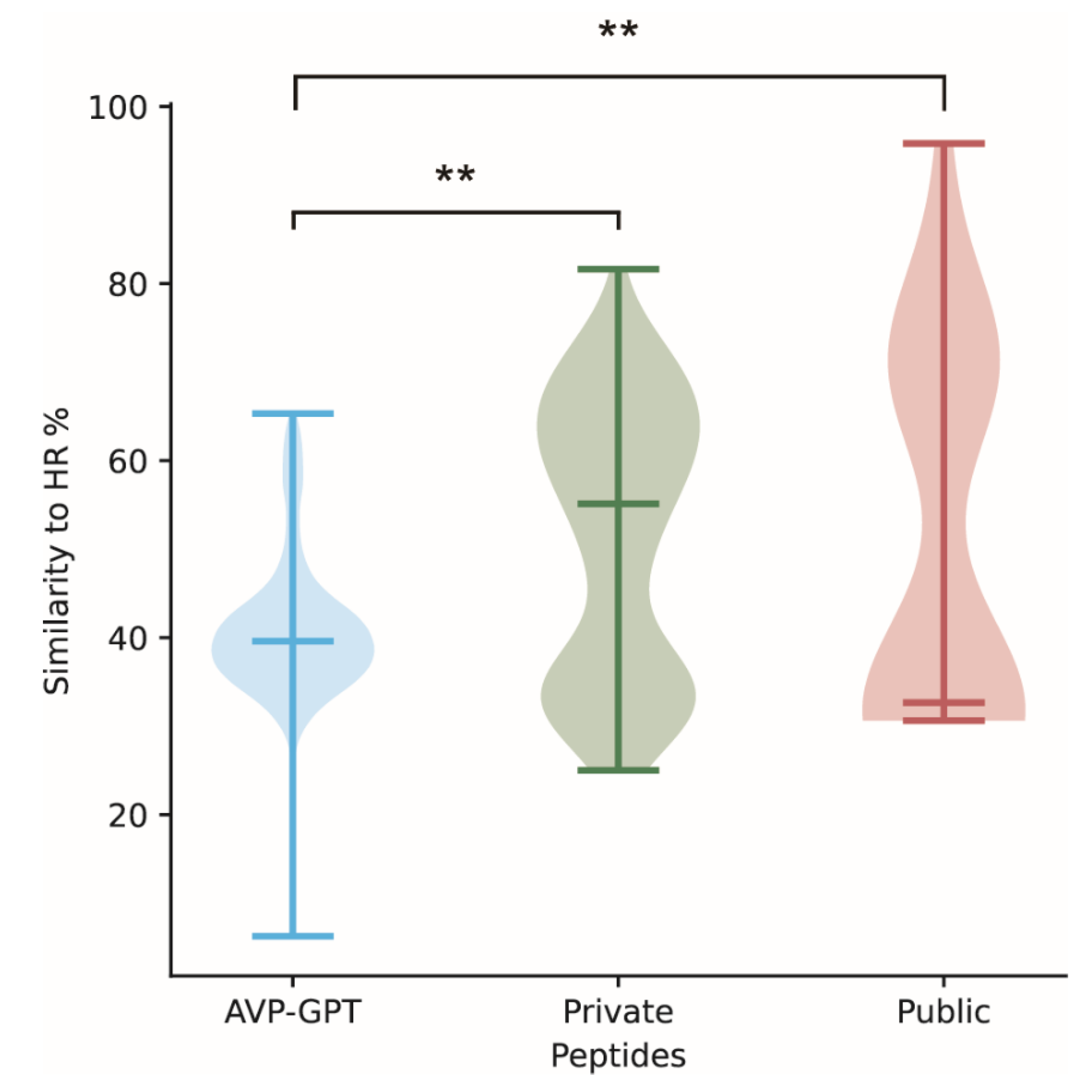
| Category | Similarity to HR (%) | Alpha-Helical Content (%) | Beta-Sheet Content (%) |
|---|---|---|---|
| AVP-GPT peptides | 6.25–65.31 | 86.44 | 0.28 |
| Private peptides | 25.00–81.63 | 82.16 | 0.74 |
| Public peptides | 30.61–95.83 | 45.24 | 0.00 |
| ID | EC50 (μM) | CC50 (μM) |
|---|---|---|
| AVP-GPT1 | 1.281 | >8 |
| AVP-GPT2 | 0.418 | >8 |
| AVP-GPT3 | >8 | >8 |
| AVP-GPT4 | 1.246 | >8 |
| AVP-GPT5 | 0.698 | >8 |
| AVP-GPT6 | 0.018 | 4.907 |
| AVP-GPT7 | >8 | >8 |
| AVP-GPT8 | 0.024 | 1.074 |
| AVP-GPT9 | >8 | >8 |
| AVP-GPT10 | 0.4522 | >8 |
| AVP-GPT11 | >8 | >8 |
| AVP-GPT12 | 6.649 | 5.813 |
| AVP-GPT13 | 0.024 | 4.257 |
| AVP-GPT14 | >2 | 4.167 |
| AVP-GPT15 | 5.472 | 3.372 |
| AVP-GPT16 | 0.096 | >8 |
| AVP-GPT17 | 0.018 | 1.428 |
| AVP-GPT18 | 0.099 | >8 |
| AVP-GPT19 | 2.219 | >8 |
| AVP-GPT20 | 0.296 | >8 |
| AVP-GPT21 | 5.037 | >8 |
| AVP-GPT22 | >2 | 1.519 |
| AVP-GPT23 | 5.529 | 5.138 |
| AVP-GPT24 | 6.823 | >8 |
| AVP-GPT25 | 5.226 | 4.022 |
| Category | Low Antiviral Rate (EC50/IC50 < 10 uM, %) | High Antiviral Rate (EC50/IC50 < 0.03 uM, %) |
|---|---|---|
| AVP-GPT peptides | 76.00 | 16.00 |
| Private peptides | 35.68 | 5.41 |
| Public peptides | 66.67 | 0.00 |
| ID | EC50 (μM) | CC50 (μM) |
|---|---|---|
| AVP-GPT26 | 13.30 | 9.29 |
| AVP-GPT27 | 2.36 | >40 |
| AVP-GPT28 | 1.27 | >40 |
| AVP-GPT29 | 11.19 | >40 |
| AVP-GPT30 | 3.98 | >40 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, H.; Song, G. Antiviral Peptide-Generative Pre-Trained Transformer (AVP-GPT): A Deep Learning-Powered Model for Antiviral Peptide Design with High-Throughput Discovery and Exceptional Potency. Viruses 2024, 16, 1673. https://doi.org/10.3390/v16111673
Zhao H, Song G. Antiviral Peptide-Generative Pre-Trained Transformer (AVP-GPT): A Deep Learning-Powered Model for Antiviral Peptide Design with High-Throughput Discovery and Exceptional Potency. Viruses. 2024; 16(11):1673. https://doi.org/10.3390/v16111673
Chicago/Turabian StyleZhao, Huajian, and Gengshen Song. 2024. "Antiviral Peptide-Generative Pre-Trained Transformer (AVP-GPT): A Deep Learning-Powered Model for Antiviral Peptide Design with High-Throughput Discovery and Exceptional Potency" Viruses 16, no. 11: 1673. https://doi.org/10.3390/v16111673
APA StyleZhao, H., & Song, G. (2024). Antiviral Peptide-Generative Pre-Trained Transformer (AVP-GPT): A Deep Learning-Powered Model for Antiviral Peptide Design with High-Throughput Discovery and Exceptional Potency. Viruses, 16(11), 1673. https://doi.org/10.3390/v16111673






