Advancements in Bacteriophages for the Fire Blight Pathogen Erwinia amylovora
Abstract
1. Introduction
2. Characterization of E. amylovora Phages
2.1. Isolation of Phages
2.2. Morphological Features
2.3. Host Ranges
2.3.1. The Impact of Bacterial EPSs on Host Preference
2.3.2. The Impact of Phage Source Influences Host Preference
3. Genome Analysis of E. amylovora Phages
4. Infection Mechanism of E. amylovora Phages
4.1. Lytic Activity
4.2. Variability in Infection Manner of E. amylovora Phages
4.3. Synergistic Interaction
5. Tolerance to Environmental Stresses
6. Application
6.1. Phage Cocktails
6.1.1. Development of Cocktail Formulation Methods
6.1.2. Formulation and Commercial Preparation
6.2. Phage–Carrier System
6.3. Prospects of Filamentous Phage Applications
6.4. Factors Affecting the Efficacy of Phage Applications
7. Challenges and Perspectives
7.1. Risks of Genetic Engineering
7.2. Risks for Plant, Soil, and Microbiome
7.3. Phage–Plant Interactions
7.4. Challenges in Field Application
7.5. Perspectives on Future Directions
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bonn, W.G.; Zwet, T.V.D. Distribution and economic importance of fire blight. In Fire Blight: The Disease and Its Causative Agent, Erwinia amylovora; CABI Publishing: Wallingford, UK, 2000; pp. 37–53. [Google Scholar]
- Vanneste, J. Fire Blight: The Disease and Its Causative Agent, Erwinia amylovora; CABI Publishing: Wallingford, UK, 2000. [Google Scholar]
- Rhim, S.; Völksch, B.; Gardan, L.; Paulin, J.; Langlotz, C.; Kim, W.; Geider, K. Erwinia pyrifoliae, an Erwinia species different from Erwinia amylovora, causes a necrotic disease of Asian pear trees. Plant Pathol. 1999, 48, 514–520. [Google Scholar] [CrossRef]
- Jock, S.; Rodoni, B.; Gillings, M.; Kim, W.S.; Copes, C.; Merriman, P.; Geider, K. Screening of ornamental plants from the Botanic Gardens of Melbourne and Adelaide for the occurrence of Erwinia amylovora. Austral. Plant Pathol. 2000, 29, 120–128. [Google Scholar] [CrossRef]
- Gill, J.; Svircev, A.; Smith, R.; Castle, A. Bacteriophages of Erwinia amylovora. Appl. Environ. Microbiol. 2003, 69, 2133–2138. [Google Scholar] [CrossRef] [PubMed]
- Bahadou, S.A.; Ouijja, A.; Tahiri, A.; Lahlali, R. Fire blight (Erwinia amylovora) disease in Morocco: Current status and action for its management. Rev. Mar. Sci. Agron. Vét. 2020, 1, 203379773. [Google Scholar]
- Besarab, N.V.; Akhremchuk, A.E.; Zlatohurska, M.A.; Romaniuk, L.V.; Valentovich, L.N.; Tovkach, F.I.; Lagonenko, A.L.; Evtushenkov, A.N. Isolation and characterization of Hena1-a novel Erwinia amylovora bacteriophage. FEMS Microbiol. Lett. 2020, 367, fnaa070. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kim, B.; Song, S.; Lee, Y.W.; Roh, E. Isolation of nine bacteriophages shown effective against Erwinia amylovora in Korea. Plant Pathol. J. 2022, 38, 248–253. [Google Scholar] [CrossRef]
- Xi, H.; Fu, B.; Sheng, Q.; Luo, M.; Sun, L. Isolation and characterization of a lytic bacteriophage RH-42-1 of Erwinia amylovora from orchard soil in China. Viruses 2024, 16, 509. [Google Scholar] [CrossRef]
- Akremi, I.; Holtappels, D.; Brabra, W.; Jlidi, M.; Hadj Ibrahim, A.; Ben Ali, M.; Fortuna, K.; Ahmed, M.; Van Meerbeek, B.; Rhouma, A.; et al. First report of filamentous phages isolated from Tunisian orchards to control Erwinia amylovora. Microorganisms 2020, 8, 1762. [Google Scholar] [CrossRef]
- Emmett, B.J.; Baker, L.A.E. Insect transmission of fireblight. Plant Pathol. 1971, 20, 41–45. [Google Scholar] [CrossRef]
- Johnson, K.B.; Stockwell, V.O. Management of fire blight: A case study in microbial ecology. Ann. Rev. Phytopathol. 1998, 36, 227–248. [Google Scholar] [CrossRef]
- Kamber, T.; Pothier, J.F.; Pelludat, C.; Rezzonico, F.; Duffy, B.; Smits, T.H.M. Role of the Type VI secretion systems during disease interactions of Erwinia amylovora with its plant host. BMC Genom. 2017, 18, 628. [Google Scholar] [CrossRef] [PubMed]
- Bogdanove, A.J.; Bauer, D.W.; Beer, S.V. Erwinia amylovora secretes DspE, a pathogenicity factor and functional AvrE homolog, through the Hrp (Type III Secretion) pathway. J. Bacteriol. 1998, 180, 2244–2247. [Google Scholar] [CrossRef] [PubMed]
- Biosca, E.G.; Delgado Santander, R.; Morán, F.; Figàs-Segura, À.; Vázquez, R.; Català-Senent, J.F.; Álvarez, B. First European Erwinia amylovora lytic bacteriophage cocktails effective in the host: Characterization and prospects for fire blight biocontrol. Biology 2024, 13, 176. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Tian, Y.; Wang, L.; Geng, G.; Zhao, W.; Hu, B.; Zhao, Y. Fire blight disease, a fast-approaching threat to apple and pear production in China. J. Integr. Agric. 2019, 18, 815–820. [Google Scholar] [CrossRef]
- Slack, S.M.; Walters, K.J.; Outwater, C.A.; Sundin, G.W. Effect of kasugamycin, oxytetracycline, and streptomycin on in-orchard population dynamics of Erwinia amylovora on apple flower stigmas. Plant Dis. 2021, 105, 1843–1850. [Google Scholar] [CrossRef]
- Sundin, G.W.; Wang, N. Antibiotic resistance in plant-pathogenic bacteria. Ann. Rev. Phytopathol. 2018, 56, 161–180. [Google Scholar] [CrossRef]
- Wierup, M. The Swedish experience of the 1986 Year ban of antimicrobial growth promoters, with special reference to animal health, disease prevention, productivity, and usage of antimicrobials. Microb. Drug Resist. 2001, 7, 183–190. [Google Scholar] [CrossRef]
- Li, D.; Yu, S.; Zhang, Y.; Lin, C.; Li, L.; Wang, S. Antibacterial activity of four alternative bactericides and their field control efficacies against pear fire blight pathogen. J. Plant Protect. 2023, 50, 1368–1376. [Google Scholar]
- Boule, J.; Sholberg, P.L.; Lehman, S.M.; O’Gorman, D.T.; Svircev, A.M. Isolation and characterization of eight bacteriophages infecting Erwinia amylovora and their potential as biological control agents in British Columbia, Canada. Can. J. Plant Pathol. 2011, 33, 308–317. [Google Scholar] [CrossRef]
- Roach, D.R.; Sjaarda, D.R.; Sjaarda, C.P.; Ayala, C.J.; Howcroft, B.; Castle, A.J.; Svircev, A.M. Absence of lysogeny in wild populations of Erwinia amylovora and Pantoea agglomerans. Microb. Biotechnol. 2015, 8, 510–518. [Google Scholar] [CrossRef]
- Schwarczinger, I.; Nagy, J.K.; Kunstler, A.; Szabo, L.; Geider, K.; Kiraly, L.; Pogany, M. Characterization of Myovirus and Podovirus family bacteriophages of Erwinia amylovora from Hungary—Potential of application in biological control of fire blight. Eur. J. Plant Pathol. 2017, 149, 639–652. [Google Scholar] [CrossRef]
- Sharma, R.; Pielstick, B.A.; Bell, K.A.; Nieman, T.B.; Stubbs, O.A.; Yeates, E.L.; Baltrus, D.A.; Grose, J.H. A novel, highly related jumbo family of bacteriophages that were isolated against Erwinia. Front. Microbiol. 2019, 10, 1533. [Google Scholar] [CrossRef] [PubMed]
- Doemoetoer, D.; Becsagh, P.; Rakhely, G.; Schneider, G.; Kovacs, T. Complete genomic sequence of Erwinia amylovora phage PhiEaH2. J. Virol. 2012, 86, 10899. [Google Scholar] [CrossRef] [PubMed]
- Meczker, K.; Dömötör, D.; Vass, J.; Rákhely, G.; Schneider, G.; Kovács, T. The genome of the Erwinia amylovora phage PhiEaH1 reveals greater diversity and broadens the applicability of phages for the treatment of fire blight. FEMS Microbiol. Lett. 2014, 350, 25–27. [Google Scholar] [CrossRef] [PubMed]
- Lehman, S.M. Development of a Bacteriophage-Based Biopesticide for Fire Blight. Ph.D. Thesis, Brock University, St. Catharines, ON, Canada, 2007. [Google Scholar]
- Balogh, B.; Jones, J.B.; Iriarte, F.B.; Momol, M.T. Phage therapy for plant disease control. Curr. Pharm. Biotechnol. 2010, 11, 48–57. [Google Scholar] [CrossRef]
- Kering, K.K.; Kibii, B.J.; Wei, H. Biocontrol of phytobacteria with bacteriophage cocktails. Pest Manag. Sci. 2019, 75, 1775–1781. [Google Scholar] [CrossRef]
- Erskine, J. Characteristics of Erwinia amylovora bacteriophage and its possible role in epidemiology of fire blight. Can. J. Microbiol. 1973, 19, 837. [Google Scholar] [CrossRef]
- Kim, S.G.; Roh, E.; Park, J.; Giri, S.S.; Kwon, J.; Kim, S.W.; Kang, J.W.; Lee, S.B.; Jung, W.J.; Lee, Y.M.; et al. The bacteriophage pEp_SNUABM_08 is a novel singleton Siphovirus with high host specificity for Erwinia pyrifoliae. Viruses 2021, 13, 1231. [Google Scholar] [CrossRef]
- Zlatohurska, M.; Gorb, T.; Romaniuk, L.; Shenderovska, N.; Faidiuk, Y.; Zhuminska, G.; Hubar, Y.; Hubar, O.; Kropinski, A.M.; Kushkina, A.; et al. Broad-host-range lytic Erwinia phage key with exopolysaccharide degrading activity. Virus Res. 2023, 329, 199088. [Google Scholar] [CrossRef]
- Clokie, M.R.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef]
- Jurczak-Kurek, A.; Gąsior, T.; Nejman-Faleńczyk, B.; Bloch, S.; Dydecka, A.; Topka, G.; Necel, A.; Jakubowska-Deredas, M.; Narajczyk, M.; Richert, M.; et al. Biodiversity of bacteriophages: Morphological and biological properties of a large group of phages isolated from urban sewage. Sci. Rep. 2016, 6, 34338. [Google Scholar] [CrossRef]
- Jones, J.B.; Jackson, L.E.; Balogh, B.; Obradovic, A.; Iriarte, F.B.; Momol, M.T. Bacteriophages for plant disease control. Ann. Rev. Phytopathol. 2007, 45, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Mueller, I.; Gernold, M.; Schneider, B.; Geider, K. Expression of lysozymes from Erwinia amylovora phages and Erwinia genomes and inhibition by a bacterial protein. J. Mol. Microb. Biotechnol. 2012, 22, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Berg, J.A.; Beatty, N.J.; Choi, M.C.; Cowger, A.E.; Cozzens, B.J.R.; Duncan, S.G.; Fajardo, C.P.; Ferguson, H.P.; Galbraith, T.; et al. Genome sequences of nine Erwinia amylovora bacteriophages. Microbiol. Resour. Announc. 2018, 7, e00944-18. [Google Scholar] [CrossRef]
- Esplin, I.N.D.; Berg, J.A.; Sharma, R.; Allen, R.C.; Arens, D.K.; Ashcroft, C.R.; Bairett, S.R.; Beatty, N.J.; Bickmore, M.; Bloomfield, T.J.; et al. Genome sequences of 19 novel Erwinia amylovora bacteriophages. Genome Announc. 2017, 5, e00931-17. [Google Scholar] [CrossRef] [PubMed]
- Mueller, I.; Kube, M.; Reinhardt, R.; Jelkmann, W.; Geider, K. Complete genome sequences of three Erwinia amylovora phages isolated in north America and a bacteriophage induced from an Erwinia tasmaniensis strain. J. Bacteriol. 2011, 193, 795–796. [Google Scholar] [CrossRef] [PubMed]
- Besarab, N.V.; Letarov, A.V.; Kulikov, E.E.; Babenko, V.V.; Belalov, I.S.; Lagonenko, A.L.; Golomidova, A.K.; Evtushenkov, A.N. Two novel Erwinia amylovora bacteriophages, Loshitsa2 and Micant, isolated in Belarus. Arch. Virol. 2022, 167, 2633–2642. [Google Scholar] [CrossRef]
- Besarab, N.V.; Letarova, M.A.; Babenko, V.V.; Belalov, I.S.; Golomidova, A.K.; Kulikov, E.E.; Lagonenko, A.L.; Evtushenkov, A.N.; Letarov, A.V. The metastable associations of bacteriophages and Erwinia amylovora. Arch. Microbiol. 2023, 205, 214. [Google Scholar] [CrossRef]
- Lagonenko, A.L.; Sadovskaya, O.; Valentovich, L.N.; Evtushenkov, A.N. Characterization of a new ViI-like Erwinia amylovora bacteriophage phiEa2809. FEMS Microbiol. Lett. 2015, 362, fnv031. [Google Scholar] [CrossRef]
- Chen, N.; Yu, C.; Cui, B.; Ren, C.; Yang, L.; Dong, Y.; Liu, L.; Zheng, Y. Isolation, genome determination and lysis function analysis of phage Kuerle of Erwinia amylovora. Sci. Agric. Sin. 2024, 57, 295–305. [Google Scholar]
- Sabri, M.; El Handi, K.; Valentini, F.; De Stradis, A.; Achbani, E.H.; Benkirane, R.; Resch, G.; Elbeaino, T. Identification and characterization of Erwinia phage IT22: A new bacteriophage-based biocontrol against Erwinia amylovora. Viruses 2022, 14, 2455. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Lee, S.B.; Giri, S.S.; Kim, H.J.; Kim, S.W.; Kwon, J.; Park, J.; Roh, E.; Park, S.C. Characterization of novel Erwinia amylovora jumbo bacteriophages from Eneladusvirus genus. Viruses 2020, 12, 1373. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Lee, S.B.; Jo, S.J.; Cho, K.; Park, J.K.; Kwon, J.; Giri, S.S.; Kim, S.W.; Kang, J.W.; Jung, W.J.; et al. Phage cocktail in combination with kasugamycin as a potential treatment for fire blight caused by Erwinia amylovora. Antibiotics 2022, 11, 1566. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.J.; Giri, S.S.; Lee, Y.M.; Park, J.H.; Hwang, M.H.; Lee, S.B.; Jung, W.J.; Kim, S.G.; Roh, E.; Park, S.C. Genomic insights into novel Erwinia bacteriophages: Unveiling their Henunavirus membership and host infection strategies. Curr. Microbiol. 2024, 81, 204. [Google Scholar] [CrossRef] [PubMed]
- Choe, J.; Kim, B.; Park, M.K.; Roh, E. Biological and genetic characterizations of a novel lytic ΦFifi106 against indigenous Erwinia amylovora and evaluation of the control of fire blight in apple plants. Biology 2023, 12, 1060. [Google Scholar] [CrossRef]
- Park, J.; Lee, G.M.; Kim, D.; Park, D.H.; Oh, C.S. Characterization of the lytic bacteriophage phiEaP-8 effective against both Erwinia amylovora and Erwinia pyrifoliae causing severe diseases in apple and pear. Plant Pathol. J. 2018, 34, 445–450. [Google Scholar] [CrossRef]
- Jo, S.J.; Kim, S.G.; Lee, Y.M.; Giri, S.S.; Kang, J.W.; Lee, S.B.; Jung, W.J.; Hwang, M.H.; Park, J.; Cheng, C.; et al. Evaluation of the antimicrobial potential and characterization of novel T7-Like Erwinia bacteriophages. Biology 2023, 12, 180. [Google Scholar] [CrossRef]
- Abreu, G.; Garcia, E.; Oliveira, A.; Oliveira, H. Genome sequence of Erwinia amylovora bacteriophage Omen. Microbiol. Resour. Announc. 2024, 13, e0012224. [Google Scholar] [CrossRef]
- Born, Y.; Fieseler, L.; Marazzi, J.; Lurz, R.; Duffy, B.; Loessner, M.J. Novel virulent and broad-host-range Erwinia amylovora bacteriophages reveal a high degree of mosaicism and a relationship to Enterobacteriaceae phages. Appl. Environ. Microbiol. 2011, 77, 5945–5954. [Google Scholar] [CrossRef]
- Buttimer, C.; Born, Y.; Lucid, A.; Loessner, M.J.; Fieseler, L.; Coffey, A. Erwinia amylovora phage vB_EamM_Y3 represents another lineage of hairy Myovirus. Res. Microbiol. 2018, 169, 505–514. [Google Scholar] [CrossRef]
- Knecht, L.E.; Born, Y.; Pelludat, C.; Pothier, J.F.; Smits, T.H.M.; Loessner, M.J.; Fieseler, L. Spontaneous resistance of Erwinia amylovora against bacteriophage Y2 affects infectivity of multiple phages. Front. Microbiol. 2022, 13, 908346. [Google Scholar] [CrossRef] [PubMed]
- Knecht, L.E.; Born, Y.; Pothier, J.F.; Loessner, M.J.; Fieseler, L. Complete genome sequences of Erwinia amylovora phages vB_EamP-S2 and vB_EamM-Bue1. Microbiol. Resour. Announc. 2018, 7, e00891-18. [Google Scholar] [CrossRef] [PubMed]
- Faidiuk, I.V.; Boyko, A.A.; Muchnyk, F.V.; Tovkach, F.I. Morphology and structural organization of polyvalent bacteriophages TT10-27 and KEY. Mikrobiol. Z. 2015, 77, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Buttimer, C.; McAuliffe, O.; Ross, R.P.; Hill, C.; O’Mahony, J.; Coffey, A. Bacteriophages and bacterial plant diseases. Front. Microbiol. 2017, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Lehman, S.M.; Kropinski, A.M.; Castle, A.J.; Svircev, A.M. Complete genome of the broad-host-range Erwinia amylovora phage phiEa21-4 and its relationship to Salmonella phage Felix O1. Appl. Environ. Microbiol. 2009, 75, 2139–2147. [Google Scholar] [CrossRef] [PubMed]
- Gayder, S.; Parcey, M.; Nesbitt, D.; Castle, A.J.; Svircev, A.M. Population dynamics between Erwinia amylovora, Pantoea agglomerans and bacteriophages: Exploiting synergy and competition to improve phage cocktail efficacy. Microorganisms 2020, 8, 1449. [Google Scholar] [CrossRef]
- Ritchie, D.; Klos, E. Isolation of Erwinia amylovora bacteriophage from aerial parts of apple trees. Phytopathology 1977, 67, 101–104. [Google Scholar] [CrossRef]
- Ritchie, D.; Klos, E. Some properties of Erwinia amylovora bacteriophages. Phytopathology 1979, 69, 1078–1083. [Google Scholar] [CrossRef]
- Roach, D.R.; Sjaarda, D.R.; Castle, A.J.; Svircev, A.M. Host exopolysaccharide quantity and composition impact Erwinia amylovora bacteriophage pathogenesis. Appl. Environ. Microbiol. 2013, 79, 3249–3256. [Google Scholar] [CrossRef]
- Gayder, S.; Parcey, M.; Castle, A.J.; Svircev, A.M. Host range of bacteriophages against a world-wide collection of Erwinia amylovora determined using a quantitative PCR assay. Viruses 2019, 11, 910. [Google Scholar] [CrossRef]
- Gross, M.; Geier, G.; Rudolph, K.; Geider, K. Levan and levansucrase synthesized by the fireblight pathogen Erwinia amylovora. Physiol. Mol. Plant Pathol. 1992, 40, 371–381. [Google Scholar] [CrossRef]
- Bogs, J.; Geider, K. Molecular analysis of sucrose metabolism of Erwinia amylovora and influence on bacterial virulence. J. Bacteriol. 2000, 182, 5351–5358. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.H.; Yang, S.; Liu, Z.; Zhang, Y.; Wang, X.; Xu, Z.; Wang, J.; Li, S.C. PhageScope: A well-annotated bacteriophage database with automatic analyses and visualizations. Nucl. Acids Res. 2024, 52, D756–D761. [Google Scholar] [CrossRef] [PubMed]
- Wojtus, J.K.; Frampton, R.A.; Warring, S.; Hendrickson, H.; Fineran, P.C. Genome sequence of a jumbo bacteriophage that infects the kiwifruit phytopathogen Pseudomonas syringae pv. actinidiae. Microbiol. Resour. Announc. 2019, 8, e00224-19. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Gao, M. Jumbo bacteriophages: An overview. Front. Microbiol. 2017, 8, 403. [Google Scholar] [CrossRef] [PubMed]
- Born, Y.; Fieseler, L.; Klumpp, J.; Eugster, M.R.; Zurfluh, K.; Duffy, B.; Loessner, M.J. The tail-associated depolymerase of Erwinia amylovora phage L1 mediates host cell adsorption and enzymatic capsule removal, which can enhance infection by other phage. Environ. Microbiol. 2014, 16, 2168–2180. [Google Scholar] [CrossRef]
- Correa, A.M.S.; Howard-Varona, C.; Coy, S.R.; Buchan, A.; Sullivan, M.B.; Weitz, J.S. Revisiting the rules of life for viruses of microorganisms. Nat. Rev. Microbiol. 2021, 19, 501–513. [Google Scholar] [CrossRef]
- Kim, B.; Lee, S.Y.; Park, J.; Song, S.; Kim, K.P.; Roh, E. Bacteriophage cocktail comprising Fifi044 and Fifi318 for biocontrol of Erwinia amylovora. Plant Pathol. J. 2024, 40, 160–170. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, Y.; Chen, J.; Hong, X.; Xu, X.; Wu, Z.; Ahmed, T.; Loh, B.; Leptihn, S.; Hassan, S.; et al. Identification and characterization of a new type of holin-endolysin lysis cassette in Acidovorax oryzae phage AP1. Viruses 2022, 14, 167. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, Y.; Xu, X.; Ahmed, T.; Yang, Y.; Loh, B.; Leptihn, S.; Yan, C.; Chen, J.; Li, B. The holin-endolysin lysis system of the OP2-like phage X2 infecting Xanthomonas oryzae pv. oryzae. Viruses 2021, 13, 1949. [Google Scholar] [CrossRef]
- Zhang, M.; Qian, J.; Xu, X.; Ahmed, T.; Yang, Y.; Yan, C.; Elsharkawy, M.M.; Hassan, M.M.; Alorabi, J.A.; Chen, J.; et al. Resistance of Xanthomonas oryzae pv. oryzae to lytic phage X2 by spontaneous mutation of lipopolysaccharide synthesis-related glycosyltransferase. Viruses 2022, 14, 1088. [Google Scholar] [PubMed]
- Howard-Varona, C.; Hargreaves, K.R.; Abedon, S.T.; Sullivan, M.B. Lysogeny in nature: Mechanisms, impact and ecology of temperate phages. The ISME J. 2017, 11, 1511–1520. [Google Scholar] [CrossRef] [PubMed]
- Farooq, T.; Hussain, M.D.; Shakeel, M.T.; Tariqjaveed, M.; Aslam, M.N.; Naqvi, S.A.H.; Amjad, R.; Tang, Y.; She, X.; He, Z. Deploying viruses against phytobacteria: Potential use of phage cocktails as a multifaceted approach to combat resistant bacterial plant pathogens. Vireses 2022, 14, 171. [Google Scholar] [CrossRef] [PubMed]
- Koskella, B.; Lin, D.M.; Buckling, A.; Thompson, J.N. The costs of evolving resistance in heterogeneous parasite environments. Proc. Roy. Soc. B Biol. Sci. 2012, 279, 1896–1903. [Google Scholar] [CrossRef] [PubMed]
- Hyman, P.; Abedon, S.T. Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 2010, 70, 217–248. [Google Scholar] [PubMed]
- Knecht, L.E.; Heinrich, N.; Born, Y.; Felder, K.; Pelludat, C.; Loessner, M.J.; Fieseler, L. Bacteriophage S6 requires bacterial cellulose for Erwinia amylovora infection. Environ. Microbiol. 2022, 24, 3436–3450. [Google Scholar] [CrossRef]
- Schmerer, M.; Molineux, I.J.; Bull, J.J. Synergy as a rationale for phage therapy using phage cocktails. PeerJ 2014, 2, e590. [Google Scholar] [CrossRef]
- Jończyk-Matysiak, E.; Łodej, N.; Kula, D.; Owczarek, B.; Orwat, F.; Międzybrodzki, R.; Neuberg, J.; Bagińska, N.; Weber-Dąbrowska, B.; Górski, A. Factors determining phage stability/activity: Challenges in practical phage application. Expert Rev. Anti-infe. 2019, 17, 583–606. [Google Scholar] [CrossRef]
- Borkotoky, S.; Murali, A. A computational assessment of pH-dependent differential interaction of T7 lysozyme with T7 RNA polymerase. BMC Struct. Biol. 2018, 17, 7. [Google Scholar]
- Wdowiak, M.; Paczesny, J.; Raza, S. Enhancing the stability of bacteriophages using physical, chemical, and nano-based approaches: A review. Pharmaceutics 2022, 14, 1936. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, M.; Hu, R.; Bai, J.; He, X.; Jin, Y. Isolation of the novel phage PHB09 and its potential use against the plant pathogen Pseudomonas syringae pv. actinidiae. Viruses 2021, 13, 2275. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, L.A.M.; Pereira, C.; Frazão, C.; Balcão, V.M.; Almeida, A. Efficiency of phage Φ6 for biocontrol of Pseudomonas syringae pv. syringae: An in vitro preliminary study. Microorganisms 2019, 7, 286. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, T.; Xu, X.; Noman, M.; Wang, Q.; Li, B. Phage-guided nanocarriers: A precision strategy against bacterial pathogens. Trends Biotechnol. 2024. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.B.; Vallad, G.E.; Iriarte, F.B.; Obradović, A.; Wernsing, M.H.; Jackson, L.E.; Balogh, B.; Hong, J.C.; Momol, M.T. Considerations for using bacteriophages for plant disease control. Bacteriophage 2012, 2, e23857. [Google Scholar] [CrossRef] [PubMed]
- Malik, D.J.; Sokolov, I.J.; Vinner, G.K.; Mancuso, F.; Cinquerrui, S.; Vladisavljevic, G.T.; Clokie, M.R.J.; Garton, N.J.; Stapley, A.G.F.; Kirpichnikova, A. Formulation, stabilisation and encapsulation of bacteriophage for phage therapy. Adv. Colloid Interface Sci. 2017, 249, 100–133. [Google Scholar] [CrossRef]
- Jończyk, E.; Kłak, M.; Międzybrodzki, R.; Górski, A. The influence of external factors on bacteriophages—Review. Folia Microbiol. 2011, 56, 191–200. [Google Scholar] [CrossRef]
- Yin, Y.; Ni, P.; Deng, B.; Wang, S.; Xu, W.; Wang, D. Isolation and characterisation of phages against Pseudomonas syringae pv. actinidiae. Acta Agric. Scand. B-Soil Plant Sci. 2019, 69, 199–208. [Google Scholar]
- Pereira, C.; Costa, P.; Pinheiro, L.; Balcão, V.M.; Almeida, A. Kiwifruit bacterial canker: An integrative view focused on biocontrol strategies. Planta 2021, 253, 49. [Google Scholar] [CrossRef]
- Born, Y.; Bosshard, L.; Duffy, B.; Loessner, M.J.; Fieseler, L. Protection of Erwinia amylovora bacteriophage Y2 from UV-induced damage by natural compounds. Bacteriophage 2015, 5, e1074330. [Google Scholar] [CrossRef]
- Gdanetz, K.; Dobbins, M.R.; Villani, S.M.; Outwater, C.A.; Slack, S.M.; Nesbitt, D.; Svircev, A.M.; Lauwers, E.M.; Zeng, Q.; Cox, K.D.; et al. Multisite field evaluation of bacteriophages for fire blight management: Incorporation of ultraviolet radiation protectants and impact on the apple flower microbiome. Phytopathology 2024, 114, 1028–1038. [Google Scholar] [CrossRef]
- Jo, S.J.; Kim, S.G.; Park, J.; Lee, Y.M.; Giri, S.S.; Lee, S.B.; Jung, W.J.; Hwang, M.H.; Park, J.H.; Roh, E.; et al. Optimizing the formulation of Erwinia bacteriophages for improved UV stability and adsorption on apple leaves. Heliyon 2023, 9, e22034. [Google Scholar] [CrossRef] [PubMed]
- Van Houte, S.; Buckling, A.; Westra, E.R. Evolutionary ecology of prokaryotic immune mechanisms. Microbiol. Mol. Biol. Rev. 2016, 80, 745–763. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.S.; Salm, H.; Geider, K. Expression of bacteriophage phiEa1h lysozyme in Escherichia coli and its activity in growth inhibition of Erwinia amylovora. Microbiology 2004, 150, 2707–2714. [Google Scholar] [CrossRef] [PubMed]
- Schnabel, E.; Fernando, W.; Meyer, M.; Jones, A.; Jackson, L. Bacteriophage of Erwinia amylovora and their potential for biocontrol. Acta Horticult. 1999, 489, 649–653. [Google Scholar] [CrossRef]
- Born, Y.; Fieseler, L.; Thony, V.; Leimer, N.; Duffy, B.; Loessner, M.J. Engineering of bacteriophages Y2::dpoL1-C and Y2::luxAB for efficient control and rapid detection of the fire blight pathogen, Erwinia amylovora. Appl. Environ. Microbiol. 2017, 83, e00341-17. [Google Scholar] [CrossRef]
- Ahern, S.J.; Das, M.; Bhowmick, T.S.; Young, R.; Gonzalez, C.F. Characterization of novel virulent broad-host-range phages of Xylella Fastidiosa and Xanthomonas. J. Bacteriol. 2014, 196, 459–471. [Google Scholar] [CrossRef]
- Das, M.; Bhowmick, T.S.; Ahern, S.J.; Young, R.; Gonzalez, C.F. Control of Pierce’s disease by phage. PLoS ONE 2015, 10, e0128902. [Google Scholar] [CrossRef]
- Wei, C.; Liu, J.; Maina, A.N.; Mwaura, F.B.; Yu, J.; Yan, C.; Zhang, R.; Wei, H. Developing a bacteriophage cocktail for biocontrol of potato bacterial wilt. Virol. Sin. 2017, 32, 476–484. [Google Scholar] [CrossRef]
- Grace, E.R.; Rabiey, M.; Friman, V.P.; Jackson, R.W. Seeing the forest for the trees: Use of phages to treat bacterial tree diseases. Plant Pathol. 2021, 70, 1987–2004. [Google Scholar] [CrossRef]
- Choi, O.; Kang, B.; Lee, Y.; Lee, Y.; Kim, J. Pantoea ananatis carotenoid production confers toxoflavin tolerance and is regulated by Hfq-controlled quorum sensing. Microbiologyopen 2021, 10, e1143. [Google Scholar] [CrossRef]
- Ibrahim, N.; Nesbitt, D.; Guo, Q.; Lin, J.; Svircev, A.; Wang, Q.; Weadge, J.T.; Anany, H. Improved viability of spray-dried Pantoea agglomerans for phage-carrier mediated control of fire blight. Viruses 2024, 16, 257. [Google Scholar] [CrossRef] [PubMed]
- McCullor, K.; Postoak, B.; Rahman, M.; King, C.; McShan, W.M. Genomic sequencing of high-efficiency transducing Streptococcal bacteriophage A25: Consequences of escape from lysogeny. J. Bacteriol. 2018, 200, e00358-18. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.S.; Karmakar, S.; Kumar, P.; Mishra, V. Application of filamentous phages in environment: A tectonic shift in the science and practice of ecorestoration. Ecol. Evol. 2019, 9, 2263–2304. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, T.; Luo, J.; Noman, M.; Ijaz, M.; Wang, X.; Masood, H.A.; Manzoor, N.; Wang, Y.; Li, B. Microbe-mediated nanoparticle intervention for the management of plant diseases. Crop Health 2023, 1, 3. [Google Scholar] [CrossRef]
- Pratama, A.A.; Terpstra, J.; de Oliveria, A.L.M.; Salles, J.F. The role of rhizosphere bacteriophages in plant health. Trends Microbiol. 2020, 28, 709–718. [Google Scholar] [CrossRef]
- Nagy, J.K.; Schwarczinger, I.; Kuenstler, A.; Pogany, M.; Kiraly, L. Penetration and translocation of Erwinia amylovora-specific bacteriophages in apple—A possibility of enhanced control of fire blight. Eur. J. Plant Pathol. 2015, 142, 815–827. [Google Scholar] [CrossRef]
- Kimmelshue, C.; Goggi, A.S.; Cademartiri, R. The use of biological seed coatings based on bacteriophages and polymers against Clavibacter michiganensis subsp. nebraskensis in maize seeds. Sci. Rep. 2019, 9, 17950. [Google Scholar] [CrossRef]
- Korniienko, N.; Kharina, A.; Budzanivska, I.; Burketova, L.; Kalachova, T. Phages of phytopathogenic bacteria: High potential, but challenging application. Plant Protect. Sci. 2022, 58, 81–91. [Google Scholar] [CrossRef]
- Tom, E.F.; Molineux, I.J.; Paff, M.L.; Bull, J.J. Experimental evolution of UV resistance in a phage. PeerJ 2018, 6, e5190. [Google Scholar] [CrossRef]
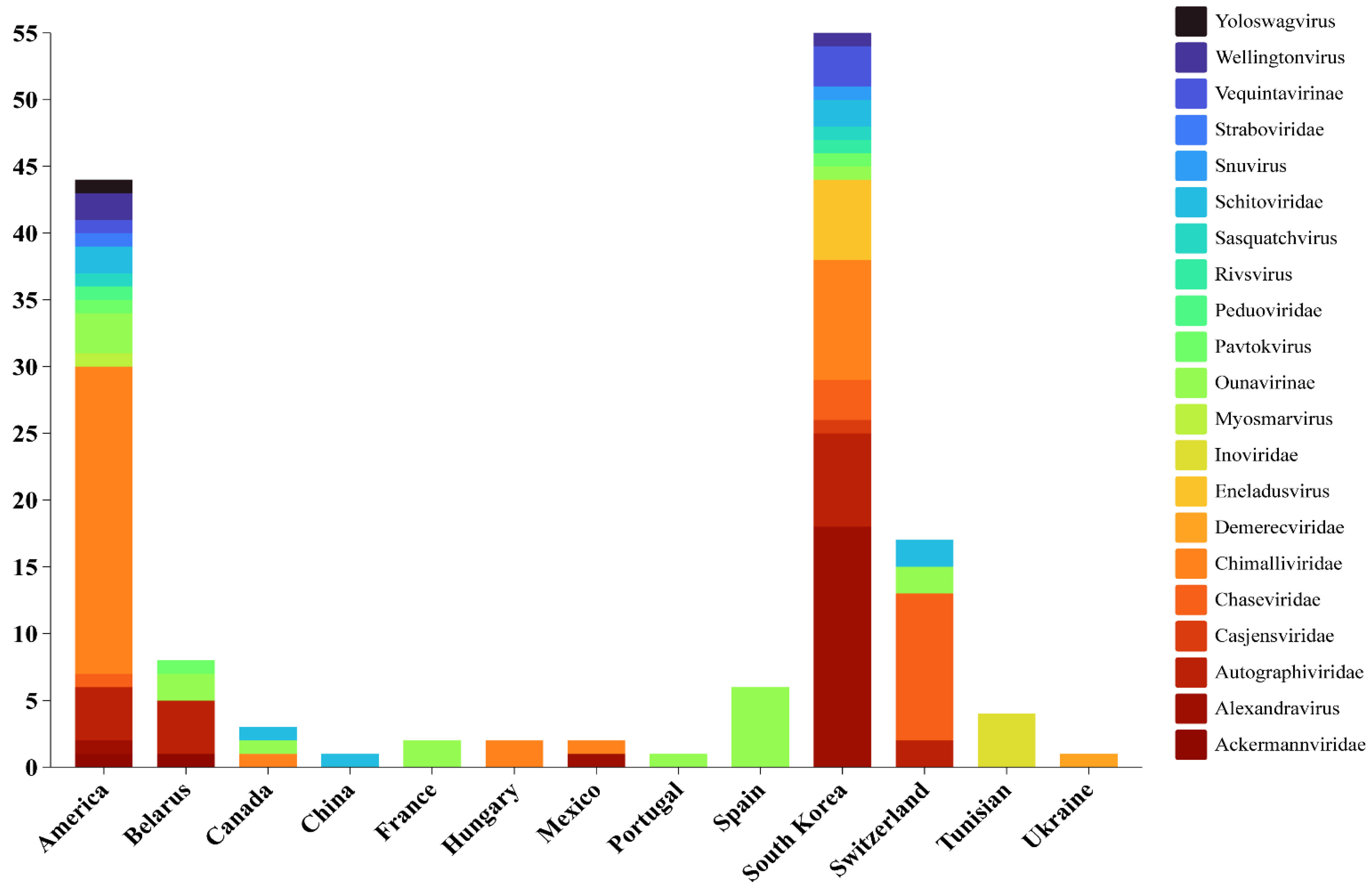

| Phage Name | Region | Morphology | Head Size (nm) | Tail Size (nm) | References |
|---|---|---|---|---|---|
| phiEa104 | USA | Myovirus | 71.56 ± 2.20 | 114.42 ± 2.51 | [36] |
| phiEa116 | USA | Myovirus | 73.36 ± 1.89 | 114.62 ± 2.28 | [36] |
| vB_EamM_RAY | USA | Myovirus | 128 ± 5.96 | 159 ± 11 | [24] |
| vB_EamM_Simmy50 | USA | Myovirus | - | - | [24] |
| vB_EamM_Special G | USA | Myovirus | - | - | [24] |
| vB_EamM_Deimos-Minion | USA | Myovirus | - | - | [24] |
| vB_EamM_Bosolaphorus | USA | Myovirus | - | - | [24] |
| vB_EamM_Desertfox | USA | Myovirus | - | - | [37] |
| vB_EamM_MadMel | USA | Myovirus | - | - | [37] |
| vB_EamM_Asesino | USA | Myovirus | - | - | [37] |
| vB_EamM_Alexandra | USA | Myovirus | - | - | [37] |
| vB_EamM_Mortimer | USA | Myovirus | - | - | [37] |
| vB_EamM_SunLIRen | USA | Myovirus | - | - | [37] |
| vB_EamM_Wellington | USA | Myovirus | - | - | [37] |
| vB_EamM_RisingSun | USA | Myovirus | - | - | [38] |
| vB_EamM_Joad | USA | Myovirus | - | - | [38] |
| vB_EamM_Caitlin | USA | Myovirus | - | - | [38] |
| vB_EamM_ChrisDB | USA | Myovirus | - | - | [38] |
| vB_EamM_EarlPhillipIV | USA | Myovirus | - | - | [38] |
| vB_EamM_Huxley | USA | Myovirus | - | - | [38] |
| vB_EamM_Kwan | USA | Myovirus | - | - | [38] |
| vB_EamM_Machina | USA | Myovirus | - | - | [38] |
| vB_EamM_Parshik | USA | Myovirus | - | - | [38] |
| vB_EamM_Phobos | USA | Myovirus | - | - | [38] |
| vB_EamM_Stratton | USA | Myovirus | - | - | [38] |
| vB_EamM_Yoloswag | USA | Myovirus | - | - | [38] |
| phiEa100 | USA | Podovirus | 61.42 ± 2.14 | - | [39] |
| Era103 | USA | Podovirus | - | - | [39] |
| vB_EamP_Pavtok | USA | Podovirus | - | - | [38] |
| vB_EamP_Frozen | USA | Podovirus | - | - | [38] |
| vB_EamP_Gutmeister | USA | Podovirus | - | - | [38] |
| vB_EamP_Rexella | USA | Podovirus | - | - | [38] |
| Micant | Belarus | Podovirus | 56.39 ± 2.69 | - | [40] |
| Loshitsa2 | Belarus | Podovirus | 59.80 ± 2.60 | - | [40] |
| VyarbaL | Belarus | Podovirus | - | - | [41] |
| phiEa2809 | Belarus | Myovirus | [42] | ||
| Hena1 | Belarus | Myovirus | 72.36 ± 5.38 | 126.28 ± 5.27 | [7] |
| Hena2 | Belarus | Myovirus | - | - | [41] |
| phiEa21-4 | Canada | Myovirus | - | - | [5] |
| vB_EamM_Ea35-70 | Canada | Myovirus | [5] | ||
| Kuerle | China | Podovirus | [43] | ||
| Ea1594-24 | Colombia | Myovirus | 48 | 74 | [21] |
| Ea21-4g | Colombia | Myovirus | 50 | 74 | [21] |
| Ea2345-6 | Colombia | Myovirus | 54 | 77 | [21] |
| Ea1615-26 | Colombia | Myovirus | 62 | 105 | [21] |
| Ea2345-19 | Colombia | Myovirus | 65 | 113 | [21] |
| Ea1594-26 | Colombia | Myovirus | 72 | 97 | [21] |
| Ea1598-6 | Colombia | Myovirus | 80 | 136 | [21] |
| Ea1337-26 | Colombia | Podovirus | 53 | 14 | [21] |
| Ea1598-19 | Colombia | Podovirus | 60 | 20 | [21] |
| phiEa1H | Germany | Podovirus | 59.89 ± 1.49 | - | [36] |
| ΦEaH2B | Hungary | Myovirus | 57 ± 7 | 60 ± 39 | [23] |
| ΦEaH2A | Hungary | Myovirus | 69 ± 7 | 107 ± 11 | [23] |
| ΦEaH1A | Hungary | Myovirus | 70 ± 3 | 117 ± 4 | [23] |
| ΦEaH4B | Hungary | Myovirus | 70 ± 9 | 98 ± 18 | [23] |
| ΦEaH7A | Hungary | Myovirus | 71 ± 8 | 99 ± 7 | [23] |
| ΦEaH12B | Hungary | Myovirus | 72 ± 4 | 103 ± 4 | [23] |
| ΦEaH5K | Hungary | Myovirus | 73 ± 4 | 107 ± 9 | [23] |
| ΦEaH5B | Hungary | Myovirus | 74 ± 5 | 104 ± 9 | [23] |
| ΦEaH7B | Hungary | Myovirus | 77 ± 5 | 108 ± 6 | [23] |
| ΦEaH4A | Hungary | Myovirus | 78 ± 5 | 108 ± 10 | [23] |
| ΦEaH5K | Hungary | Myovirus | - | - | [23] |
| ΦEaH11 | Hungary | Podovirus | 55 ± 2 | 13 ± 2 | [23] |
| ΦEaH9B | Hungary | Podovirus | 61 ± 7 | 9 ± 3 | [23] |
| PhiEaH1 | Hungary | Siphovirus | - | - | [26] |
| PhiEaH2 | Hungary | Siphovirus | - | - | [25] |
| EP-IT22 | Italy | Myovirus | 90 ± 5 | 100 ± 10 | [44] |
| pEa_SNUABM_47 | South Korea | Myovirus | 127 ± 6 | 127 ±3 | [45] |
| pEa_SNUABM_12 | South Korea | Myovirus | 130 ± 5.9 | 126.7 ± 2.6 | [45] |
| pEa_SNUABM_32 | South Korea | Myovirus | 130 ± 6 | 169 ±7 | [46] |
| pEa_SNUABM_31 | South Korea | Myovirus | 139 ± 5 | 196 ± 11 | [46] |
| pEa_SNUABM_48 | South Korea | Myovirus | 140 ± 2 | 150 ± 17 | [46] |
| pEa_SNUABM_27 | South Korea | Myovirus | 69 ± 3 | 115 ± 2 | [46] |
| pEp_SNUABM_01 | South Korea | Myovirus | 78.29 ± 0.91 | - | [47] |
| Fifi106 | South Korea | Myovirus | 79.8 ± 4.3 | 114.1 ± 5.2 | [48] |
| pEa_SNUABM_55 | South Korea | Myovirus | 81.88 ± 2.20 | 6 | [47] |
| pEa_SNUABM_50 | South Korea | Myovirus | - | - | [45] |
| Ea46-1-A1 | South Korea | Podovirus | - | - | [48] |
| phiEaP-8 | South Korea | Podovirus | 75 | - | [49] |
| pEp_SNUABM_04 | South Korea | Podovirus | 55 ± 3 | 16 ± 2 | [50] |
| pEp_SNUABM_03 | South Korea | Podovirus | 56 ± 2 | 17 ± 2 | [50] |
| pEp_SNUABM_11 | South Korea | Podovirus | 56 ± 3 | 18 ± 1 | [50] |
| pEp_SNUABM_12 | South Korea | Podovirus | 63 ± 2 | 17 ± 1 | [50] |
| pEp_SNUABM_08 | South Korea | Siphovirus | 62 ± 4 | 190 ± 12 | [31] |
| Omen | Portugal | Myovirus | 2 ± 5 | 112 ± 9 | [51] |
| vEam_PM_21 | Spain | Myovirus | 59.82 ± 3.98 | 94.56 ± 7.45 | [15] |
| vEam_PM_6 | Spain | Myovirus | 61.11 ± 5.06 | 93.02 ± 3.31 | [15] |
| vEam_PM_27 | Spain | Myovirus | 63.46 ± 4.62 | 101.92 ± 4.62 | [15] |
| vEam_W_25 | Spain | Myovirus | 63.85 ± 5.00 | 94.23 ± 9.23 | [15] |
| vEam_S_24 | Spain | Myovirus | 69.28 ± 5.32 | 94.22 ± 4.53 | [15] |
| vEam_W_28 | Spain | Myovirus | 78.63 ± 7.41 | 102.77 ± 5.4 | [15] |
| vB_EamP_Y2 | Switzerland | Myovirus | 67 | 124 | [52] |
| vB_EamP_M7 | Switzerland | Myovirus | 77 | 116 | [52] |
| vB_EamM_Y3 | Switzerland | Myovirus | 129 ± 4 | 192 ± 12 | [53] |
| vB_EamM-Bue1 | Switzerland | Myovirus | 79 ± 2 | 126 ± 7 | [54] |
| vB_EamP_L1 | Switzerland | Podovirus | 58 | - | [52] |
| vB_EamP_S6 | Switzerland | Podovirus | 66 | - | [52] |
| vB_EamP-S2 | Switzerland | Podovirus | 64 ± 5 | - | [55] |
| PEar1 | Tunisia | Inovirus | - | - | [10] |
| PEar2 | Tunisia | Inovirus | - | - | [10] |
| PEar4 | Tunisia | Inovirus | - | - | [10] |
| PEar6 | Tunisia | Inovirus | - | - | [10] |
| TT10-27 | Ukraine | Podovirus | 71.3 | 22 | [56] |
| KEY | Ukraine | Siphovirus | 80 ± 1 | 169 ± 10 | [56] |
| Phage | Unknown | Integration | Immune | Regulation | Lysis | Packaging | Replication | Assembly | Infection | Hypothetical |
|---|---|---|---|---|---|---|---|---|---|---|
| Ackermannviridae | ||||||||||
| vB_EamM-Bue1 | 95 | 2 | 1 | 6 | 9 | 9 | 21 | 18 | 14 | 49 |
| phiEa2809 | 93 | 2 | 1 | 6 | 9 | 7 | 21 | 18 | 14 | 50 |
| Alexandravirus | ||||||||||
| pEa_SNUABM_1 | 254 | 2 | 3 | 3 | 4 | 10 | 15 | 8 | 23 | 11 |
| pEa_SNUABM_16 | 262 | 2 | 3 | 3 | 4 | 9 | 15 | 8 | 23 | 10 |
| vB_EamM_Alexandra | 255 | 3 | 3 | 2 | 7 | 9 | 17 | 6 | 25 | 12 |
| pEa_SNUABM_3 | 253 | 3 | 3 | 2 | 3 | 10 | 16 | 9 | 24 | 11 |
| pEa_SNUABM_32 | 252 | 2 | 4 | 3 | 5 | 10 | 15 | 8 | 22 | 10 |
| pEa_SNUABM_2 | 253 | 2 | 2 | 2 | 4 | 9 | 16 | 9 | 24 | 11 |
| pEa_SNUABM_22 | 260 | 3 | 3 | 3 | 4 | 9 | 15 | 8 | 23 | 11 |
| pEa_SNUABM_17 | 242 | 3 | 3 | 3 | 4 | 10 | 16 | 8 | 24 | 10 |
| pEa_SNUABM_33 | 251 | 2 | 3 | 4 | 4 | 11 | 16 | 10 | 24 | 10 |
| pEa_SNUABM_35 | 253 | 2 | 3 | 3 | 4 | 9 | 16 | 8 | 24 | 11 |
| pEa_SNUABM_30 | 244 | 2 | 3 | 4 | 4 | 11 | 17 | 9 | 24 | 12 |
| pEa_SNUABM_28 | 264 | 2 | 3 | 3 | 4 | 9 | 15 | 8 | 23 | 11 |
| pEa_SNUABM_40 | 253 | 3 | 3 | 2 | 3 | 11 | 17 | 9 | 24 | 10 |
| pEa_SNUABM_18 | 260 | 2 | 3 | 3 | 4 | 9 | 15 | 8 | 23 | 10 |
| pEa_SNUABM_20 | 252 | 3 | 3 | 2 | 3 | 10 | 16 | 9 | 24 | 10 |
| pEa_SNUABM_23 | 253 | 3 | 3 | 2 | 3 | 10 | 16 | 9 | 24 | 10 |
| pEa_SNUABM_31 | 253 | 3 | 3 | 2 | 3 | 10 | 16 | 9 | 24 | 10 |
| pEa_SNUABM_39 | 253 | 2 | 2 | 2 | 4 | 9 | 16 | 9 | 24 | 11 |
| vB_Ea_2910A | 247 | 2 | 3 | 4 | 4 | 13 | 19 | 9 | 24 | 10 |
| pEa_SNUABM_36 | 253 | 2 | 3 | 3 | 4 | 9 | 16 | 8 | 24 | 11 |
| Autographiviridae | ||||||||||
| phiEa100 | 11 | 0 | 3 | 1 | 5 | 7 | 7 | 5 | 5 | 9 |
| phiEa1H | 11 | 0 | 3 | 1 | 5 | 7 | 7 | 5 | 5 | 9 |
| vB_EamP-S2 | 13 | 0 | 4 | 0 | 4 | 5 | 5 | 5 | 5 | 9 |
| Era103 | 9 | 0 | 3 | 2 | 5 | 7 | 7 | 5 | 5 | 9 |
| Tapenade | 12 | 0 | 4 | 0 | 4 | 5 | 5 | 5 | 5 | 9 |
| VyarbaL | 12 | 0 | 4 | 0 | 4 | 5 | 5 | 5 | 5 | 9 |
| pEp_SNUABM_12 | 17 | 0 | 1 | 0 | 3 | 4 | 8 | 7 | 6 | 4 |
| pEp_SNUABM_03 | 21 | 0 | 0 | 3 | 3 | 4 | 6 | 7 | 6 | 3 |
| pEp_SNUABM_10 | 21 | 0 | 0 | 3 | 3 | 4 | 6 | 7 | 6 | 3 |
| pEp_SNUABM_09 | 20 | 0 | 0 | 3 | 3 | 4 | 6 | 7 | 6 | 3 |
| pEp_SNUABM_04 | 19 | 0 | 0 | 3 | 3 | 4 | 6 | 7 | 6 | 3 |
| pEp_SNUABM_11 | 18 | 0 | 0 | 3 | 3 | 4 | 6 | 7 | 6 | 3 |
| Stepyanka | 13 | 0 | 0 | 4 | 4 | 4 | 7 | 8 | 6 | 4 |
| vB_EamP-L1 | 10 | 0 | 0 | 4 | 3 | 5 | 7 | 8 | 6 | 4 |
| pEa_SNUABM_57 | 11 | 0 | 0 | 2 | 3 | 4 | 7 | 7 | 7 | 5 |
| Loshitsa2 | 21 | 0 | 1 | 0 | 5 | 5 | 5 | 5 | 6 | 4 |
| Micant | 21 | 0 | 1 | 0 | 5 | 5 | 5 | 5 | 6 | 4 |
| Casjensviridae | ||||||||||
| pEp_SNUABM_08 | 36 | 0 | 2 | 1 | 3 | 3 | 3 | 8 | 10 | 13 |
| Chaseviridae | ||||||||||
| vB_EamM-Y2 | 26 | 1 | 0 | 1 | 3 | 4 | 8 | 6 | 13 | 18 |
| Papaline | 29 | 1 | 0 | 1 | 3 | 4 | 8 | 7 | 14 | 16 |
| Calisson | 31 | 2 | 0 | 0 | 3 | 4 | 7 | 6 | 14 | 16 |
| Fougasse | 29 | 1 | 0 | 0 | 3 | 3 | 7 | 6 | 14 | 16 |
| Nougat | 29 | 1 | 0 | 0 | 3 | 3 | 7 | 6 | 14 | 16 |
| Mauresque | 28 | 1 | 0 | 1 | 3 | 2 | 7 | 6 | 14 | 16 |
| Faunus | 29 | 1 | 0 | 1 | 4 | 3 | 7 | 6 | 14 | 14 |
| Aioli | 29 | 1 | 0 | 0 | 3 | 4 | 7 | 6 | 14 | 14 |
| Navette | 27 | 1 | 0 | 0 | 3 | 2 | 7 | 6 | 14 | 16 |
| Farigoule | 27 | 1 | 0 | 0 | 3 | 2 | 7 | 6 | 14 | 16 |
| Orgeat | 29 | 1 | 0 | 0 | 3 | 2 | 7 | 6 | 14 | 16 |
| Fifi440 | 40 | 1 | 0 | 0 | 3 | 3 | 7 | 5 | 4 | 15 |
| Berlingot | 29 | 1 | 0 | 0 | 3 | 2 | 7 | 6 | 14 | 16 |
| Fifi451 | 30 | 1 | 0 | 0 | 3 | 3 | 7 | 5 | 14 | 15 |
| pEa_SNUABM_27 | 28 | 1 | 0 | 0 | 3 | 2 | 7 | 5 | 14 | 14 |
| Chimalliviridae | ||||||||||
| Derbicus | 107 | 3 | 3 | 0 | 7 | 4 | 19 | 35 | 32 | 24 |
| vB_EamM_Earl | 107 | 3 | 3 | 0 | 7 | 4 | 19 | 35 | 32 | 24 |
| vB_EamM_Phobos | 107 | 2 | 4 | 0 | 8 | 4 | 20 | 39 | 36 | 22 |
| vB_EamM_MadMel | 180 | 3 | 1 | 4 | 8 | 6 | 26 | 25 | 30 | 37 |
| vB_EamM_Mortimer | 187 | 3 | 1 | 4 | 8 | 4 | 27 | 25 | 29 | 35 |
| Rebecca | 3 | 1 | 3 | 8 | 4 | 25 | 26 | 31 | 36 | |
| vB_EamM_Deimos-Minion | 183 | 3 | 1 | 3 | 8 | 5 | 25 | 25 | 30 | 35 |
| vB_EamM_SpecialG | 182 | 3 | 1 | 4 | 8 | 5 | 27 | 24 | 28 | 37 |
| vB_EamM_Desertfox | 180 | 3 | 1 | 4 | 8 | 4 | 26 | 25 | 30 | 36 |
| vB_EamM_Bosolaphorus | 183 | 3 | 1 | 3 | 8 | 4- | 26 | 25 | 30 | 35 |
| vB_EamM_RAY | 181 | 3 | 1 | 3 | 8 | 4 | 25 | 24 | 28 | 36 |
| vB_EamM_Simmy50 | 181 | 3 | 1 | 4 | 8 | 4 | 26 | 24 | 28 | 36 |
| Ea35-70 | 185 | 3 | 1 | 3 | 8 | 4 | 25 | 24 | 27 | 35 |
| vB_EamM_Earl | 107 | 3 | 3 | 0 | 7 | 4 | 19 | 35 | 32 | 24 |
| pEa_SNUABM_38 | 106 | 3 | 2 | 0 | 7 | 4 | 18 | 37 | 35 | 24 |
| vB_EamM_Asesino | 137 | 2 | 3 | 0 | 6 | 3 | 22 | 37 | 33 | 23 |
| vB_EamM_Stratton | 128 | 3 | 4 | 2 | 7 | 4 | 21 | 38 | 33 | 23 |
| phiEaH2 | 131 | 3 | 4 | 2 | 6 | 4 | 21 | 37 | 32 | 22 |
| PhiEaH1 | 128 | 3 | 1 | 1 | 7 | 4 | 15 | 30 | 29 | 28 |
| vB_Ea277G | 127 | 3 | 1 | 1 | 7 | 4 | 15 | 30 | 29 | 28 |
| vB_EamM_Machina | 125 | 4 | 5 | 0 | 8 | 4 | 23 | 36 | 33 | 24 |
| vB_EamM_Huxley | 126 | 4 | 5 | 0 | 7 | 4 | 23 | 35 | 32 | 25 |
| pEa_SNUABM_8 | 127 | 3 | 5 | 0 | 7 | 4 | 20 | 43 | 41 | 26 |
| pEa_SNUABM_4 | 127 | 3 | 5 | 0 | 7 | 4 | 20 | 43 | 41 | 26 |
| pEa_SNUABM_9 | 122 | 3 | 5 | 1 | 8 | 4 | 21 | 41 | 39 | 25 |
| vB_EamM_Chris | 117 | 3 | 5 | 1 | 8 | 4 | 23 | 40 | 37 | 26 |
| pEa_SNUABM_43 | 121 | 4 | 5 | 1 | 8 | 4 | 23 | 39 | 37 | 23 |
| pEa_SNUABM_42 | 121 | 4 | 5 | 1 | 8 | 4 | 23 | 39 | 37 | 23 |
| vB_EamM_Caitlin | 126 | 3 | 5 | 1 | 8 | 4 | 22 | 36 | 33 | 26 |
| vB_EamM_Parshik | 126 | 4 | 5 | 0 | 7 | 4 | 23 | 35 | 32 | 25 |
| pEa_SNUABM_6 | 126 | 4 | 5 | 0 | 8 | 5 | 22 | 37 | 35 | 23 |
| pEa_SNUABM_10 | 132 | 4 | 5 | 0 | 8 | 4 | 21 | 36 | 34 | 23 |
| vB_EamM_Joad | 126 | 2 | 3 | 2 | 2 | 5 | 18 | 30 | 32 | 20 |
| vB_EamM_RisingSun | 129 | 2 | 3 | 1 | 2 | 5 | 17 | 30 | 32 | 21 |
| pEa_SNUABM_29 | 121 | 3 | 5 | 1 | 9 | 4 | 22 | 47 | 43 | 25 |
| pEa_SNUABM_11 | 143 | 3 | 7 | 2 | 6 | 5 | 20 | 40 | 36 | 22 |
| vB_EamM_Kwan | 146 | 2 | 6 | 2 | 6 | 6 | 22 | 33 | 30 | 21 |
| Wellington | 144 | 2 | 5 | 3 | 7 | 6 | 22 | 33 | 30 | 21 |
| Demerecviridae | ||||||||||
| KEY | 76 | 0 | 1 | 2 | 9 | 7 | 19 | 9 | 19 | 38 |
| Eneladusvirus | ||||||||||
| pEa_SNUABM_12 | 234 | 4 | 11 | 8 | 8 | 10 | 28 | 74 | 63 | 106 |
| pEa_SNUABM_49 | 228 | 3 | 10 | 11 | 8 | 13 | 27 | 74 | 63 | 106 |
| pEa_SNUABM_50 | 234 | 4 | 11 | 8 | 8 | 8 | 27 | 74 | 63 | 103 |
| pEa_SNUABM_44 | 232 | 4 | 11 | 7 | 8 | 7 | 27 | 74 | 63 | 108 |
| pEa_SNUABM_47 | 231 | 4 | 11 | 8 | 8 | 7 | 27 | 74 | 63 | 105 |
| pEa_SNUABM_19 | 231 | 4 | 11 | 8 | 8 | 7 | 27 | 74 | 63 | 106 |
| Inoviridae | ||||||||||
| PEar6 | 0 | 2 | 0 | 1 | 0 | 0 | 2 | 5 | 1 | 0 |
| PEar4 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 5 | 1 | 0 |
| PEar2 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 5 | 1 | 0 |
| PEar1 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 5 | 1 | 0 |
| Myosmarvirus | ||||||||||
| vB_EamM_TropicalSun | 60 | 0 | 1 | 1 | 1 | 1 | 7 | 5 | 14 | 11 |
| Ounavirinae | ||||||||||
| Omen | 38 | 0 | 1 | 5 | 10 | 1 | 14 | 10 | 11 | 26 |
| Tian | 40 | 0 | 1 | 4 | 10 | 1 | 14 | 10 | 11 | 26 |
| Roscha1 | 42 | 0 | 1 | 3 | 10 | 2 | 13 | 10 | 11 | 25 |
| vB_EamM-M7 | 39 | 0 | 1 | 4 | 10 | 2 | 14 | 10 | 11 | 24 |
| vEam_W_28 | 40 | 0 | 1 | 3 | 10 | 1 | 13 | 10 | 11 | 25 |
| vEam_S_24 | 40 | 0 | 1 | 3 | 10 | 1 | 13 | 10 | 11 | 25 |
| vEam_PM_21 | 40 | 0 | 1 | 3 | 10 | 1 | 13 | 10 | 11 | 25 |
| vEam_PM_27 | 40 | 0 | 1 | 3 | 10 | 1 | 13 | 10 | 11 | 25 |
| vEam_PM_6 | 40 | 0 | 1 | 3 | 10 | 1 | 13 | 10 | 11 | 25 |
| vEam_W_25 | 40 | 0 | 1 | 3 | 10 | 1 | 13 | 10 | 11 | 25 |
| Panisse | 39 | 0 | 1 | 3 | 10 | 1 | 14 | 11 | 11 | 26 |
| phiEa21-4 | 42 | 0 | 1 | 3 | 10 | 1 | 13 | 10 | 11 | 25 |
| phiEa104 | 39 | 0 | 1 | 4 | 11 | 1 | 13 | 10 | 11 | 26 |
| SunLIRen | 40 | 0 | 1 | 3 | 10 | 1 | 13 | 10 | 11 | 26 |
| Pistou | 41 | 0 | 1 | 3 | 10 | 1 | 13 | 10 | 11 | 25 |
| FIfi106 | 39 | 0 | 1 | 4 | 11 | 1 | 13 | 10 | 11 | 25 |
| Rouille | 40 | 0 | 1 | 3 | 10 | 1 | 13 | 10 | 11 | 25 |
| Hena2 | 40 | 0 | 1 | 3 | 10 | 1 | 13 | 10 | 11 | 25 |
| Pavtokvirus | ||||||||||
| Pavtok | 22 | 2 | 2 | 3 | 3 | 3 | 2 | 8 | 6 | 9 |
| PEp14 | 27 | 2 | 2 | 2 | 2 | 3 | 3 | 7 | 6 | 9 |
| Stean | 22 | 3 | 3 | 2 | 3 | 3 | 3 | 7 | 6 | 10 |
| Peduoviridae | ||||||||||
| ENT90 | 4 | 2 | 0 | 4 | 3 | 2 | 1 | 8 | 14 | 0 |
| Rivsvirus | ||||||||||
| pEa_SNUABM_5 | 273 | 2 | 2 | 2 | 4 | 11 | 16 | 7 | 20 | 9 |
| Sasquatchvirus | ||||||||||
| vB_EamM_Y3 | 255 | 3 | 2 | 3 | 3 | 10 | 16 | 8 | 22 | 11 |
| pEp_SNUABM_52 | 257 | 3 | 2 | 3 | 2 | 10 | 15 | 8 | 21 | 12 |
| Schitoviridae | ||||||||||
| phiEaP8 | 30 | 1 | 1 | 2 | 6 | 3 | 9 | 7 | 3 | 17 |
| Kuerle | 35 | 1 | 1 | 2 | 6 | 3 | 9 | 7 | 3 | 20 |
| Ea9-2 | 40 | 2 | 1 | 2 | 6 | 3 | 9 | 7 | 3 | 19 |
| vB_EamP_Rexella | 38 | 2 | 1 | 2 | 6 | 4 | 9 | 7 | 3 | 18 |
| Fifi067 | 35 | 1 | 1 | 2 | 6 | 3 | 9 | 7 | 3 | 19 |
| vB_EamP-S6 | 67 | 0 | 0 | 0 | 5 | 2 | 8 | 8 | 3 | 13 |
| Pastis | 76 | 1 | 0 | 0 | 4 | 2 | 8 | 7 | 3 | 11 |
| vB_EamP_Gutmeister | 33 | 1 | 1 | 2 | 6 | 2 | 9 | 7 | 3 | 18 |
| Snuvirus | ||||||||||
| pEa_SNUABM_7 | 267 | 2 | 3 | 2 | 3 | 9 | 15 | 9 | 23 | 8 |
| Straboviridae | ||||||||||
| Cronus | 137 | 4 | 4 | 9 | 14 | 11 | 26 | 35 | 21 | 44 |
| Vequintavirinae | ||||||||||
| Hena1 | 92 | 2 | 4 | 6 | 7 | 6 | 21 | 18 | 21 | 67 |
| pEa_SNUABM_56 | 92 | 3 | 4 | 6 | 7 | 7 | 18 | 16 | 17 | 76 |
| pEp_SNUABM_01 | 91 | 3 | 4 | 5 | 7 | 7 | 18 | 16 | 18 | 76 |
| pEa_SNUABM_55 | 88 | 3 | 4 | 6 | 7 | 7 | 20 | 16 | 17 | 77 |
| Yoloswagvirus | ||||||||||
| vB_EamM_Yoloswag | 248 | 3 | 3 | 3 | 6 | 11 | 16 | 9 | 23 | 7 |
| Unknown | ||||||||||
| pEa_SNUABM_48 | 208 | 2 | 0 | 1 | 5 | 4 | 22 | 36 | 39 | 32 |
| pEa_SNUABM_37 | 211 | 3 | 0 | 1 | 6 | 4 | 23 | 33 | 36 | 32 |
| pEa_SNUABM_54 | 188 | 2 | 2 | 1 | 3 | 5 | 19 | 30 | 31 | 35 |
| pEa_SNUABM_21 | 264 | 3 | 3 | 3 | 3 | 9 | 15 | 8 | 23 | 9 |
| pEa_SNUABM_13 | 262 | 2 | 3 | 3 | 3 | 8 | 15 | 9 | 24 | 10 |
| pEa_SNUABM_34 | 264 | 3 | 3 | 2 | 3 | 9 | 15 | 8 | 22 | 10 |
| pEa_SNUABM_14 | 264 | 2 | 3 | 3 | 3 | 9 | 15 | 9 | 23 | 9 |
| pEa_SNUABM_45 | 264 | 3 | 3 | 3 | 3 | 9 | 15 | 8 | 22 | 10 |
| pEa_SNUABM_46 | 264 | 3 | 3 | 3 | 3 | 9 | 15 | 8 | 22 | 10 |
| pEa_SNUABM_25 | 262 | 2 | 3 | 3 | 3 | 9 | 15 | 9 | 23 | 9 |
| phiEt88 | 39 | 0 | 1 | 1 | 5 | 4 | 4 | 5 | 9 | 12 |
| Phage | Complete Genome Acc No. | Size (bp) | GC Content (%) | ORFs (tRNA) |
|---|---|---|---|---|
| pEp_SNUABM_01 | MN184887 | 147,321 | 48.7 | 249 (26) |
| pEa_SNUABM_55 | OP480062 | 146,979 | 48.8 | 247 (25) |
| pEp_SNUABM_03 | MT822284.1 | 39,879 | 52.1 | 52 |
| pEp_SNUABM_04 | MT822285.1 | 39,649 | 52.2 | 52 |
| pEp_SNUABM_11 | MT822287.1 | 39,626 | 52.1 | 49 |
| pEp_SNUABM_12 | MT822288.1 | 39,980 | 51.2 | 50 |
| pEp_SNUABM_08 | MN184886 | 62,715 | 57.2 | 79 (0) |
| pEa_SNUABM_12 | MT939486 | 358,115 | 34.4 | 546 (32) |
| pEa_SNUABM_47 | MT939487 | 355,376 | 34.5 | 540 (35) |
| pEa_SNUABM_50 | MT939488 | 356,948 | 34.4 | 540 (34) |
| PEar1 | MT901797 | 6646 | 41.7 | 10 |
| PEar2 | MT901798 | 6651 | 41.7 | 10 |
| PEar4 | MT901799 | 6801 | 41.7 | 10 |
| PEar6 | MT901800 | 6608 | 41.7 | 11 |
| Kuerle | OQ181210.1 | 75,599 | 48.0 | 85 |
| RH-42-1 | PP099880 | 14,942 | 48.2 | 28 (0) |
| Fifi106 | OR284297.1 | 84,405 | 43.4 | 114 (26) |
| vB_EamM_RAY | KU886224 | 271,182 | 49.9 | 317 |
| vB_EamM_Special G | KU886222 | 273,224 | 49.8 | 321 |
| key | MZ616364 | 115,651 | 39.0 | 182 (27) |
| Omen | PP278848 | 85,304 | 43.7 | 133 |
| vB_EamM_Simmy50 | NC_041974.1 | 271,088 | 49.9 | 322 |
| vB_EamM_Y3 | KY984068 | 261,365 | 47.2 | 333 (0) |
| phiEaP-8 | MH160392 | 75,929 | 46.8 | 785 (5) |
| vB_EamP_Pavtok | MH426726 | 61,401 | 62.0 | 62 (0) |
| vB_EamM_SunLIRen | MH426725 | 84,559 | 43.8 | 141 (22) |
| vB_EamM_Wellington | MH426724 | 244,950 | 52.1 | 295 (8) |
| vB_EamM_Asesino | KX397364 | 246,290 | 51.2 | 289 (12) |
| vB_EamM_Alexandra | MH248138 | 266,532 | 50.1 | 349 (0) |
| vB_EamM_Bosolaphorus | MG655267 | 272,228 | 49.8 | 321 (1) |
| vB_EamM_Desertfox | MG655268 | 272,458 | 49.9 | 320 (0) |
| vB_EamM_Mortimer | MG655270 | 273,914 | 49.8 | 325 (1) |
| vB_EamM_MadMel | MG655269 | 275,000 | 49.7 | 321 (0) |
| vB_EamP-S2 | NC_047917.1 | 45,495 | 49.8 | 49 (0) |
| vB_EamM-Bue1 | NC_048702.1 | 164,037 | 50.2 | 175 (1) |
| PhiEa2809 | KP037007 | 162,160 | 50.3 | 145 (1) |
| PhiEaH1 | KF623294 | 218,339 | 52.3 | 241 |
| PhiEaH2 | JX316028 | 243,050 | 51.3 | 262 |
| phiEa1H | FQ482084 | 45,522 | 49.7 | 50 (0) |
| phiEa100 | FQ482086 | 45,554 | 49.7 | 50 (0) |
| phiEa104 | FQ482083 | 84,564 | 43.8 | 118 (24) |
| vB_EamP_S6 | HQ728266 | 74,669 | 52.1 | 115 |
| vB_EamP_Y2 | HQ728264 | 56,621 | 44.2 | 90 |
| vB_EamP_M7 | HQ728263 | 84,694 | 43.4 | 117 |
| vB_EamP_L1 | HQ728265 | 39,282 | 51.9 | 49 |
| vB_EamP_Cutmeister | KX098391 | 71,173 | 46.9 | 84 (8) |
| vB_EamP_Frozen | KX098389 | 75,147 | 46.9 | 92 (8) |
| vB_EamP_Rexella | KX098390 | 75,448 | 46.9 | 92 (7) |
| vB_EamM_Deimos-Minion | KU886225 | 273,501 | 49.9 | 326 (0) |
| vB_EamM_Caitlin | KX397365 | 241,147 | 52.2 | 271 (7) |
| vB_EamM_ChrisDB | KX397366 | 244,840 | 49.4 | 277 (11) |
| vB_EamM_EarlPhilliplv | KX397367 | 223,935 | 50.6 | 241 (0) |
| vB_EamM_Huxley | KX397368 | 240,761 | 51.1 | 271 (9) |
| vB_EamM_Kwan | KX397369 | 246,390 | 52.1 | 285 (8) |
| vB_EamM_Machina | KX397370 | 241,654 | 51.0 | 272 (9) |
| vB_EamM_Parshik | KX397371 | 241,050 | 51.0 | 271 (10) |
| vB_EamM_Phobos | KX397372 | 229,501 | 49.1 | 247 (0) |
| vB_EamM_Stratton | KX397373 | 243,953 | 51.3 | 276 (12) |
| vB_EamM_Yoloswag | KY448244 | 259,700 | 46.9 | 334 (0) |
| vB_EamM_RisingSun | MF459646 | 235,108 | 48.32 | 243 (0) |
| vB_EamM_Joad | MF459647 | 235,374 | 48.29 | 245 (0) |
| phiEa21-4 | EU710883.1 | 84,576 | 43.8 | 117 (26) |
| Ea35-70 | KF806589 | 271,084 | 49.9 | 318 (1) |
| Phages | Lytic/Lysogenic | Latent Period (min) | Burst Size (PFU/Host Cell) | References |
|---|---|---|---|---|
| pEp_SNUABM_01 | Lytic | 20 | 67 | [47] |
| pEp_SNUABM_55 | Lytic | 20 | 65 | [47] |
| pEp_SNUABM_03 | Lytic | 10 | 76 | [50] |
| pEp_SNUABM_08 | Lytic | 40 | 20 | [31] |
| pEa_SNUABM_12 | Lytic | 40 | 17.51 ± 1.48 | [45] |
| pEa_SNUABM_47 | Lytic | 40 | 19.94 ± 3.31 | [45] |
| pEa_SNUABM_50 | Lytic | 40 | 15.51 ± 1.46 | [45] |
| PEar1 | Lysogenic | 10 | 280 | [10] |
| PEar2 | Lysogenic | 10 | 280 | [10] |
| PEar4 | Lysogenic | 10 | 280 | [10] |
| PEar6 | Lysogenic | 10 | 280 | [10] |
| Kuerle | Lytic | 50 | 240 | [43] |
| RH-42-1 | Lytic | 10 | 207 | [9] |
| vB_EamM_Deimos-Minion | Lytic | 180–240 | 4.6–4.9 | [24] |
| Fifi106 | Lytic | 20 | 310 ± 30 | [48] |
| Fifi044 | Lytic | 40 | 20 | [71] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ke, D.; Luo, J.; Liu, P.; Shou, L.; Ijaz, M.; Ahmed, T.; Shahid, M.S.; An, Q.; Mustać, I.; Ondrasek, G.; et al. Advancements in Bacteriophages for the Fire Blight Pathogen Erwinia amylovora. Viruses 2024, 16, 1619. https://doi.org/10.3390/v16101619
Ke D, Luo J, Liu P, Shou L, Ijaz M, Ahmed T, Shahid MS, An Q, Mustać I, Ondrasek G, et al. Advancements in Bacteriophages for the Fire Blight Pathogen Erwinia amylovora. Viruses. 2024; 16(10):1619. https://doi.org/10.3390/v16101619
Chicago/Turabian StyleKe, Dufang, Jinyan Luo, Pengfei Liu, Linfei Shou, Munazza Ijaz, Temoor Ahmed, Muhammad Shafiq Shahid, Qianli An, Ivan Mustać, Gabrijel Ondrasek, and et al. 2024. "Advancements in Bacteriophages for the Fire Blight Pathogen Erwinia amylovora" Viruses 16, no. 10: 1619. https://doi.org/10.3390/v16101619
APA StyleKe, D., Luo, J., Liu, P., Shou, L., Ijaz, M., Ahmed, T., Shahid, M. S., An, Q., Mustać, I., Ondrasek, G., Wang, Y., Li, B., & Lou, B. (2024). Advancements in Bacteriophages for the Fire Blight Pathogen Erwinia amylovora. Viruses, 16(10), 1619. https://doi.org/10.3390/v16101619









