Abstract
Alignment of picornavirus proteinase/polymerase sequences reveals this family evolved into five ‘supergroups’. Interestingly, the nature of the 2A region of the picornavirus polyprotein is highly correlated with this phylogeny. Viruses within supergroup 4, the Paavivirinae, have complex 2A regions with many viruses encoding multiple 2ANPGP sequences. In vitro transcription/translation analyses of a synthetic polyprotein comprising green fluorescent protein (GFP) linked to β-glucuronidase (GUS) via individual 2ANPGPs showed two main phenotypes: highly active 2ANPGP sequences—similar to foot-and-mouth disease virus 2ANPGP—and, surprisingly, a novel phenotype of some 2ANPGP sequences which apparently terminate translation at the C-terminus of 2ANPGP without detectable re-initiation of downstream sequences (GUS). Probing databases with the short sequences between 2ANPGPs did not reveal any potential ‘accessory’ functions. The novel, highly active, 2A-like sequences we identified substantially expand the toolbox for biomedical/biotechnological co-expression applications.
1. Introduction
Picornavirus genomes are single-stranded, positive sense RNA of some 7.5 to 8.5 kb in length. Their genomes have a common architecture: (i) a long 5′ untranslated region (UTR) containing an internal ribosome entry site (IRES), conferring an m7G cap-independent mode of the initiation of translation, (ii) a long open reading frame (ORF; ~2200aa ‘polyprotein’), and (iii) a short 3′ UTR preceding a poly(A) tail. In many genera, the N-terminal region of the polyprotein comprises a ‘leader’ sequence. The P1 domain of the polyprotein comprises four different proteins (1A-1D), 60 copies of each of which assemble to form the capsid. Proteins within the P2 (2A, 2B, 2C) and P3 (3A-3D) domains are replication proteins. Early studies showed the 3C protein to be a virus-encoded proteinase (3Cpro), responsible for a ‘primary’ (co-translational) cleavage between the P2 and P3 regions, but also subsequent (‘secondary’) polyprotein processing, with 3Cpro being conserved amongst all picornaviruses (for review see [1]).
In the case of entero- and human rhinoviruses, a second virus-encoded proteinase, 2Apro (~17 kDa), was identified and shown to be responsible for a single primary cleavage between the capsid proteins (P1) and P2 domains of the polyprotein—and, shown latterly, also to cleave key cellular proteins, thereby enhancing virus replication [2,3,4,5]. Simple inspection of the relatively modest number of picornavirus genome sequences available at that time—such as entero- and rhinoviruses, the foot-and-mouth disease virus (FMDV; aphthovirus), the encephalomyocarditis virus (EMCV; cardiovirus), and the hepatitis A virus (HAV; hepatovirus)—showed their 2A regions to be quite different from one another; indeed, the 2A protein of FMDV was only 18aa long. Research into polyprotein processing mechanisms of these other non-enterovirus genera showed that the capsid proteins domain was separated from the replication protein domains by either (i) the 3C proteinase cleaving the polyprotein in this region [6,7], or (ii) a proposed ‘ribosome skipping’ mechanism mediated by the 2A oligopeptide sequence proposed to interact with the ribosome exit tunnel, bringing about a discontinuity in the polypeptide backbone at the C-terminus of 2A—not by a proteolytic ‘cleavage’, but by ‘skipping’ the synthesis of a specific peptide backbone bond [7,8,9]. A completely conserved motif at the C-terminus of these 2A sequences (-NPG↓P-; position of discontinuity of the peptide backbone shown with arrow) gives rise to this type of picornavirus 2A being referred to in the literature as 2ANPGP. It should be noted, however, that 2ANPGP (‘2A-like’) sequences are also found in a wide range of non-picornavirus virus families [10,11,12], plus non-LTR retrotransposons [13,14,15] and some cellular sequences (NOD-like receptor proteins—NLRs) [16,17]. Our previous work has shown that the ‘ribosome skipping’ activity resides within a ~25aa tract (the 2A C-terminal delimiter being -NPG↓P) [13,18]. Cardiovirus 2A proteins are ~143aa long; however, the C-terminal region of the cardiovirus-like 2As shares sequence similarity with the short aphthovirus-like 2As, both possessing the same proposed ribosome skipping function [19] outlined in Figure 1A.
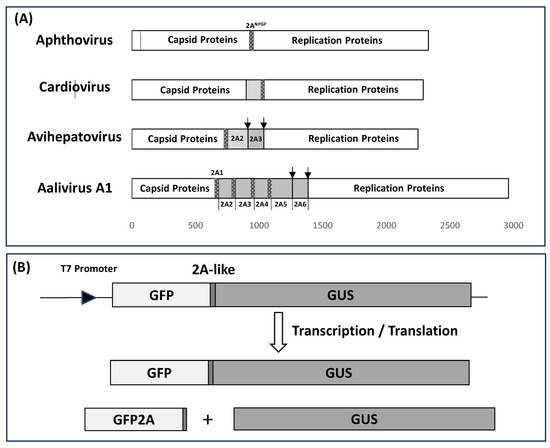
Figure 1.
Schematic (drawn to scale) showing the positions of 2ANPGP sequences (25aa) within the genomes of aphtho-, cardio-, avihepato-, and aalivirus polyproteins (Panel A). The [GFP2AGUS] artificial polyprotein (GFP stop codon removed) was used to test the various 2ANPGP sequences. In the case of FMDV 2ANPGP, three translation products were observed: ‘uncleaved’ [GFP2AGUS] together with the ‘cleavage’ products [GFP2A] and GUS (Panel B).
It has been shown recently, however, that the cardioviruses EMCV and Theilers murine encephalitis virus (TMEV) 2A proteins are bifunctional in that they also stimulate a programmed ribosomal (−1) frameshift by binding an RNA secondary structure formed closely downstream—the first demonstration of protein-stimulated regulation of programmed ribosomal frameshifting. Here, a shift site occurs just 5′ of a stem-loop structure within the region encoding the 2B protein, producing a truncated 2B protein (2B*), effectively terminating translation prior to the translation of downstream replication proteins, which comprises some 50% of the entire polyprotein [20,21,22]. In the case of one genus, the dicipiviruses, however, the P1 region is encoded by an ORF separated from the second ORF (encoding the P2–P3 region) by a second, intergenic IRES that initiates translation of the downstream ORF [23].
Presently, the family Picornaviridae comprises almost 70 genera (Knowles www.picornaviridae.com; [24] accessed on 5 September 2024). Alignment and phylogenetic analyses of the nucleotide sequences corresponding to the uncleaved form ([3CD]) of 3Cpro and the 3D RNA-dependent RNA polymerase (3Dpol) shows five major lineages, or ‘supergroups’ (SG), within the family: the Caphthovirinae (SG1), the Kodimesavirinae (SG2), the Ensavirinae (SG3), the Paavivirinae (SG4), and the Heptrevirinae (SG5) [25].
In the case of supergroups 2 (Kodimesavirinae—22 genera) and 5 (Heptrevirinae—7 genera), the separation between capsid and replication proteins is brought about by 3Cpro cleavage. In the case of supergroup 3 (Ensavirinae—8 genera), this separation is brought about by 2Apro, a proteinase unique to this supergroup. In supergroup 1 (Caphthovirinae—16 genera), this separation is brought about, in the vast majority cases, by a single copy of 2ANPGP. In the case of supergroup 4 (Paavivirinae—13 genera), different species encode either a single 2ANPGP or multiple copies of 2ANPGPs.
In the case of the avihepatoviruses, typified by duck hepatitis virus type 1 (DHV-1), the 2A region comprises three proteins (Figure 1A): (i) 2A1 (20aa) is highly similar to the short aphthovirus 2A sequence—a highly efficient ribosome skipping sequence separating the capsid from replication proteins domains [26], (ii) 2A2 (161aa) has been shown to possess GTPase activity inducing apoptosis [27], and (iii) 2A3 (124aa) possesses similarity with parechovirus 2A, comprising an H-box/NC motif and related to the host-cell H-rev protein family [28], recently shown to promote cell proliferation [29]. In the case of Duck Picornavirus GL/12 (aalivirus A1) the complexity of the 2A region is increased, since it comprises six 2A proteins [30] (Figure 1A). Here, the 2A1 protein (19aa) is aphthovirus-like, 2A2 is 133aa, 2A3 is 150aa, and 2A4 is 131aa; all have C-terminal region (~25aa) similarity with aphthovirus-like 2ANPGP sequences. Protein 2A5 has similarity with the avihepatovirus 2A2 protein, whilst 2A6 has similarity with the avihepatovirus 2A3 (parechovirus-like). Recently, the genome of the duck egg-reducing syndrome virus (DERSV) has been determined [31]. This virus encodes seven 2A proteins with six highly conserved 2ANPGPs.
Since a single iteration of 2ANPGP is sufficient to separate encapsidation from replication proteins (e.g., SG1), the question arises, what function(s) do these ‘additional’ 2ANPGP- type 2As serve? The polypeptide tracts between successive 2ANPGPs are between ~50 and ~150aa long and could represent the acquisition of ‘accessory’ polypeptides. Multiple 2ANPGPs between the capsid and replication polyprotein domains could affect protein biogenesis, a key question being, what is the ribosome skipping activity of each of these sequences? Our previous work showed the activity of 2A/2A-like sequences resides within a ~25aa tract, including the N-terminal proline of the 2B downstream protein. In this study, we inserted a 2A/2A-like sequence (25aa, in-frame) between the green fluorescent protein (GFP: ~27 kDa—stop codon deleted) and β-glucuronidase (GUS: ~70 kDa), creating a single, long ORF [9,13].
It should be noted that certainly not all 2A-like sequences containing the conserved [D-V/I-E-X-NPGP] motif are active in ribosome skipping: the sequence immediately upstream, although not highly conserved, plays an essential role in ‘cleavage’. It is essential, therefore, to perform an assay to determine the activity of each sequence. Here, we used an in vitro transcription/translation system—the ‘skipping’ activity of each test sequence was determined by the incorporation of 35S-methionine into each translation product (Figure 1B). For viruses in supergroup 4, however, amino acid tracts between the 2ANPGPs are generally short; analysing the 2A region as a single tract could produce a significantly more complex mixture of translation products with either (i) poor resolution on SDS gels or (ii) low methionine content proving very difficult to detect, or (iii) producing translation products too small to be detected. We chose, therefore, to use our GFP/GUS system to determine the ‘skipping’ activity of these 2ANPGPs individually, rather than in their native, concatenated forms.
2. Materials and Methods
2.1. Bioinformatic Analyses
Picornavirus genome sequences were downloaded from the National Center for Biotechnology Information (NCBI) via the links provided on https://www.picornaviridae.com (accessed on 5 September 2024). The uncleaved form of the proteinase and polymerase ([3CD]) protein sequences were aligned using CLUSTALX [32]. The dendrogram file from CLUSTALX (.dnd) was opened using FIGTREE (http://tree.bio.ed.ac.uk/software/figtree/, accessed on 5 September 2024) to construct the phylogenetic tree. Polyprotein sequences were searched for occurrences of the ‘signature’ NPGP motif completely conserved amongst all 2A-like sequences to identify 2ANPGPs. Arbitrarily, 25aa tracts were chosen for further analyses (Table 1). Nucleotide and amino acid sequences of tracts between picornavirus 2ANPGPs were submitted to NCBI BlastN/BlastP (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 5 September 2024) for database similarity searches.

Table 1.
2ANPGP amino acid sequences and accession codes.
2.2. Cloning of 2ANPGP Sequences into pSTA1
All plasmids were constructed using standard methods and confirmed by automated nucleotide sequencing (Eurofins Genomics, Ebersberg, Germany). Restriction enzymes were purchased from Promega (Southampton, UK) and New England Biolabs (Hitchin, UK), whilst oligonucleotides were obtained from Eurofins Genomics (Ebersberg, Germany). PCR products encoding each 2A-like sequence were amplified from our pcDNA™ 3.1 mammalian expression vector, encoding ([pGFPF2AGUS]; pSTA1) [9] using the T7 ‘forward’ primer (5′-TAATACGACTCACTATAGGG-3′) and ‘reverse’ oligonucleotide primers listed in Table 2, such that a panel of GFP/2A-like PCR products was generated. Each PCR product was restricted with BamHI and ApaI, gel purified, and cloned into pSTA1, similarly restricted. Sequence identities were confirmed by automated DNA sequencing using an oligonucleotide primer corresponding to that encoding a C-terminal region of GFP (5′-CTGTCCACACAATCTGCCC-3′).

Table 2.
Reverse oligonucleotide primers used to amplify GFP, thereby adding each 2ANPGP 3′ extension; the ApaI restriction site used in cloning is indicated in bold typeface.
2.3. In Vitro Transcription/Translation
Plasmid DNA was linearised with PstI and purified using the Wizard SV system (Promega, Southampton, UK). Purified, linearised DNA (200 ng) was used to program a Wheat Germ Extract coupled transcription/translation system (Promega) supplemented with L-[35S]-methionine (EasyTag™ Perkin Elmer Beaconsfield, UK; 1 μL = 10 μCi) and the amino acid mixture (minus Met). The final reaction volume was adjusted to 25 μL using nuclease-free water. Reactions were incubated at 30 °C for 90 min before the addition of 2x SDS-PAGE loading buffer (Jena Bioscience, Jena, Germany). Translation products were analysed by 4–12% gradient SDS-PAGE (NuPAGE, Invitrogen, Paisley, UK) and the distribution of the radiolabel visualised by autoradiography.
3. Results
3.1. Bioinformatic Analyses
NOTE: Alignments and virus polyprotein sequences showing the positions of 2ANPGPs discussed below are shown in the Supplementary Data. Our alignment of [3CD] amino acid sequences produced the same phylogenetic relationships as the nucleotide sequence alignments reported by Zell and co-workers [25]. Our phylogenetic analyses show that picornaviruses encoding 2ANPGPs fall into two distinct supergroups—the Caphthovirinae (SG1) and the Paavivirinae (SG4), with viruses within the latter encoding either a single or multiple 2ANPGPs; indeed, sometimes this was the case for different species within the same genus (Table 1). Interestingly, there was a high correlation between each supergroup and the nature of the 2A region—the most plastic amongst picornavirus polyproteins (Figure 2). In the case of supergroup 1 (Caphthovirinae—16 genera), viruses encoded a single copy of 2ANPGP. The first exception here is mosavirus B1 which encoded two 2ANPGPs. In this virus, the first copy of 2ANPGP appears to occur within the P1 region, whilst the second copy aligns with the single copy of mosavirus A 2ANPGP, which lies between the P1 and P2 polyprotein domains. The second exception is mupivirus, both A1 and A2, neither of which encode a 2ANPGP.
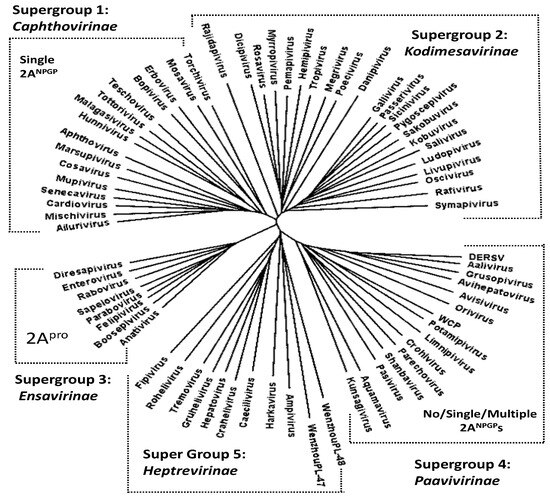
Figure 2.
Dendrogram of aligned 3CD amino acid sequences rendered using FigTree. The supergroups indicated are consistent with those of Zell and co-workers [25]. Viruses encoding a 2A proteinase occurred only within supergroup 3, whilst 2ANPGP sequences were found only within supergroups 1 and 4.
In the case of supergroup 4 (Paavivirinae—13 genera), variability was observed between different species within a genus. Here, a genus may comprise viruses which encode either (i) no 2ANPGP (orivirus A1 and A2: alignment with avisiviruses shows a relative deletion in this region; see Supplementary Data), (ii) a single 2ANPGP (aquamavirus, avihepatovirus, crohivirus, pasivirus, shanbavirus), (iii) multiple copies of 2ANPGPs—aalivirus, limnipivirus, Wuhan carp picornavirus (WCP—unassigned, but clusters within SG4) (NOTE: DERSV is most closely related to aaliviruses and clusters within the Paavivirinae (Figure 2)), (iv) genera comprising viruses encoding either a single or multiple 2ANPGPs (avisivirus, kunsagivirus, potamipivirus), or (v) genera comprising viruses encoding either no or multiple 2ANPGPs (grusopivirus, parechovirus; see Supplementary Data for alignments).
Wenzhou picorna-like virus 48 (WPLV-48; NC_032820) encoded three 2ANPGP sequences but is unique in that 2A1 appears to lie between the P1 capsid and P2 replication protein domains, whilst 2A2 and 2A3 occur in the C-terminal region of the polyprotein—downstream of the WPL-48 sequences that align with the C-terminal 3CD region of all other picornaviruses (see Supplementary Data). In our alignment/phylogenetic analyses, this region was deleted. Although unassigned, WPLV-48 clustered within SG5 (Figure 2), the Heptrevirinae, which do not encode any 2ANPGPs (Figure 2). Interestingly, our 3CD alignment shows Wenzhou picorna-like virus 47 (WPLV-47; NC_033150) also clusters within SG5, although it does not encode a 2A-like sequence. Like many other viruses in this supergroup, WPLV-47 diverged at an early stage in the evolution of this supergroup.
For viruses with multiple 2ANPGPs, peptide sequences lying between each 2ANPGP were used to probe the sequence database; other than similarities with related viruses, no significant matches were detected.
3.2. ‘Cleavage’ Activities of 2A-like Sequences
For all cases, in vitro translation profiles are shown in Figure 3, with estimated ‘cleavage’ activities shown in Table 3.
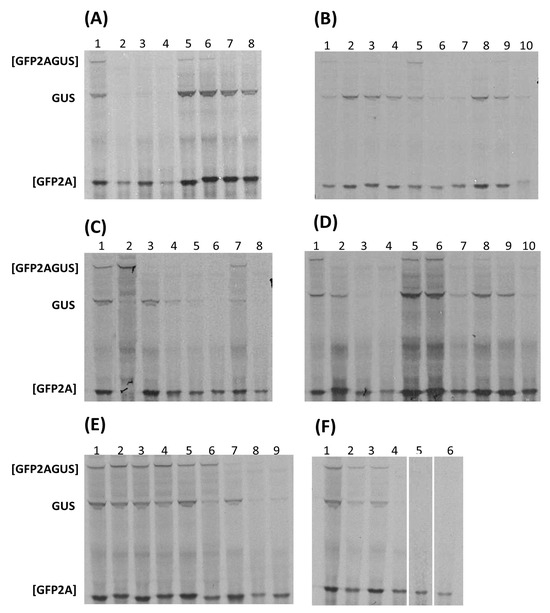
Figure 3.
Translation in vitro. Coupled transcription/translation wheat germ extracts were programmed with the plasmid constructs indicated. Translation products were labelled with [35S] methionine, separated on 4–12% gradient SDS polyacrylamide gels, and detected by autoradiography. Gels are representatives of experiments performed in triplicate. Extended autoradiography did not reveal further visible translation products. Lane 1 panels (A–F) control construct FMDV 25 encoding [GFP-2A(25aa)-GUS]; positions of the uncleaved [GFP-2A-GUS] and cleavage products [GFP-2A] and GUS are indicated. (A) Lanes 2–4, WP-LV48: 2A1–2A3; lanes 5–8, aalivirus-A1: 2A1–2A4. (B) Lanes 2–6, aalivirus-B1: 2A1–2A5; lanes 7 and 8, avisivirus-A1: 2A1/2A2; lanes 9 and 10, avisivirus-B1: 2A1/2A2. (C) Lanes 2 and 3, parechovirus E: 2A1/2A2; lanes 4 and 5, RtPV: 2A1/2A2; lane 6, potamipivirus A1: 2A1; lanes 7 and 8, potamipivirus B1: 2A1/2A2. (D) Lanes 2 and 3, limnipivirus A1: 2A2 and 2A2; lanes 4 and 5, limnipivirus B1: 2A1 and 2A2; lanes 6 and 7, limnipivirus C1: 2A1 and 2A2; lanes 8–10, limnipivirus D1: 2A1, 2A2, and 2A3. (E) Lanes 2–4, grusopivirus-A1: 2A1, 2A2, and 2A3; lanes 5–7, grusopivirus-C: 2A1–2A3; lane 8, YC-4; lane 9, kungsagivirus-C1: 2A1. (F) Lanes 2 and 3, mosavirus B1: 2A1/2A2; lanes 4–6, WCP: 2A1–2A3.

Table 3.
Estimated ‘cleavage’ activities of 2A-like sequences.
Alignments showing relative insertions/deletions in polyproteins within a genus, plus highlights of the position of 2ANPGPs, are provided in the Supplementary Data.
In the single instance of a virus clustering within SG1—mosavirus B1 encoding two 2ANPGPs—the activity of both sequences was directly comparable to FMDV 2A. The first mosavirus B1 2ANPGP occurred within P1, whilst the second aligned with the single 2ANPGP present within other mosavirus sequences and corresponded to the junction between the P1 and P2 domains of the polyprotein.
In the case of viruses encoding 2ANPGPs within SG4, however, a more complex pattern of activity was observed. Aalivirus A1/B1 2ANPGPs showed activities comparable to FMDV 2A, although aalivirus B1 sequences yielded slightly less of the GUS translation product. For both aalivirus A1 and B1 2ANPGPs, 2A1–2A4 (A1) and 2A1–2A5 (B1) produced similar translation profiles. Similarly, in the related grusopiviruses, different viruses encoded different numbers of 2ANPGPs. Grusopiviruses encoded a sequence (-TVIINPGP-) partly resembling 2A within the (predicted) P1 capsid protein domain of the polyprotein, although point mutations within the conserved C-terminal consensus of active 2A/2A-like sequences ([D(V/I)ExNPGP]) strongly suggest these are inactive. Alignment of grusopivirus polyproteins shows relative insertions/deletions within the 2A region are the cause of variable numbers of 2ANPGPs (see Supplementary Data). 2A1–2A3 of grusopivirus A1 and C viruses produced a similar translation profile to FMDV 2A, although GrV-C 2A2 showed a lower translation of the downstream protein, GUS. Again, relative insertions/deletions within the 2A region caused variable numbers of 2ANPGPs—notably virus YC-4, which only encoded one 2ANPGP (see Supplementary Data). In the case of avihepatoviruses, related to aali- and grusopiviruses, all viruses encoded a single 2ANPGP. Avisivirus A1 and B1 viruses encoded two 2ANPGPs, both active but with no detectable ‘uncleaved’ [GFP2AGUS], and somewhat lower levels of GUS in comparison with FMDV 2A. Avisivirus A1 strain USA-IN1 and avisivirus C1, however, encoded a single 2ANPGP, whilst oriviruses did not encode a 2ANPGP—again due to relative insertions/deletions (see Supplementary Data). Wuhan carp picornavirus (WCP) encoded three 2ANPGPs: the translation profile produced by each sequence showed the [GFP2A] translation product but, surprisingly, no detectable ‘uncleaved’ [GFP2AGUS] or downstream GUS translation product. A similar translation profile was observed for the two potamipivirus B1 2ANPGPs, high [GFP2A] with low/no GUS translation, whilst potamipivirus A1 encoded a single 2ANPGP due to relative insertions/deletions. The 2ANPGPs encoded by members of the limnipivirus genus showed a perhaps surprisingly wide range of translation profiles, whilst limnipivirus A1 2A1, type B1 2A2, type C1 2A1, and type D1 2A1–3 gave essentially the same translation profile as FMDV 2A; type A1 2A2 and type B1 2A1 produced the upstream translation product [GFP2A], but the downstream product, GUS, was not detected. Both crohivirus A and B encoded a single 2ANPGP, whilst in the parechoviruses, relative insertions/deletions in this region have produced a variable number of 2ANPGPs: type A has none, types B, C, D, and F have one, whilst types E and RtPV have two. Type E 2A1 only produced a full-length ([GFP2AGUS)] translation product, but type E 2A2 and RtPV 2A1/2A2 produced [GFP2A] as the major translation product with a low level of GUS. Shanba-, pasi-, aquama-, and kunsagiviruses encoded a single 2ANPGP, with the exception of kunsagivirus C, which encoded three 2ANPGPs—again arising from relative insertions/deletions in this genus. In the kunsagivirus polyprotein alignment, kunsagivirus C 2A3 aligned with the single 2ANPGP within types A and B (see Supplementary Data).
Analyses of the ribosome skipping activities showed a mixed pattern (Table 3). Certain genera showed activities similar to the control FMDV 2ANPGP: aaliviruses, avisiviruses, grusopiviruses, mosavirus B1, and parechovirus 2A2. In a number of cases only [GFP2A] could be detected: kunsagivirus C1 2A1, limnipivirus A1 2A2, limnipivirus B1 2A1, potamivirus B1 2A2, WCP 2A1/2A2/2A3, and WP-LV48 2A1/2A2 and 2A3. In the case of parechovirus E, 2A1 was inactive, whilst parechovirus E 2A2, along with RtPV 2A1 and 2A2, was similar to FMDV 2A.
4. Discussion
4.1. 2A and 2A-like Sequences in SG1 and SG4 Viruses
Our phylogenetic analyses were based upon alignment of 3CD amino-acid sequences and showed a surprisingly high, but not complete, correlation with the nature of the 2A region of the polyprotein. It should be noted, however, that in the interpretation of these data, picornavirus mixed infections may show high levels of recombination (reviewed in [33]). Whilst SG1 comprises almost exclusively mammalian viruses, SG4 comprises ‘sub-lineages’ of avian (DERSV, aali-, avihepato-, avisi-, grusopi-, kunsagi-, ori-), fish (WCP, limnipi-, potamipi-), and mammalian (crohi-, parecho-, shanba-, pasi-, aquama-, kunsagi-) viruses. The picornavirus 2A region is highly plastic; viruses within SG4 may encode no 2ANPGPs (e.g., oriviruses), a single 2ANPGP (e.g., aquama- and pasiviruses), or multiple 2ANPGPs. Indeed, viruses within the same genus may encode different numbers of 2ANPGPs (e.g., aali-, avisi-, kunsagi-, limnipi-, parecho-, and potamipiviruses). SG4 polyprotein alignments show that relative insertions/deletions within the 2A region determines the variable numbers of 2ANPGPs (see Supplementary Data).
In the case of SG1 viruses, the single 2ANPGP sequence is responsible for a highly efficient ‘primary’, co-translational cleavage between capsid and replication proteins. Our in vitro transcription/translation analyses showed many 2ANPGP sequences in SG4 to be as active as the FMDV 2A control: aalivirus A1/B1 2A1–2A4, grusopivirus A1/C 2A1–2A3, limnipivirus A1 2A1, B1 2A2, C1 2A1 and D1 2A1–3, and mosavirus B1 2A1, 2A2. We have shown that somewhat less efficient 2ANPGP sequences can also be used for other virus genomes to acquire additional functional ‘modules’. For example, different RNA segments/proteins of certain rota- and totiviruses have acquired a [2A-like/dsRNA-binding protein] ‘module’ [10,34,35]. In the case of SG4 viruses with multiple iterations of 2ANPGP, could sequences between 2ANPGPs comprise ‘accessory’ protein(s)? Protein BLAST was used to probe databases using such sequences. These analyses only produced, however, significant matches against corresponding regions of closely related viruses, providing no indications of accessory functions. As outlined above, it has been shown that cardiovirus 2A proteins, ~143aa long, alongside the ribosome skipping activity, mediate programmed ribosomal frameshifting at a proximal site within protein 2B, such that the frameshifted ribosome quickly encounters a stop codon. Therefore, as the infectious cycle progresses and the level of the 2A protein increases, the expression of (downstream) replication proteins is progressively diminished. Remaining aminoacyl-tRNAs can be devoted, therefore, to the production of capsid proteins, thereby increasing the yield of particles.
4.2. Implications for Our Model of Ribosome Skipping
In our proposed model of 2ANPGP ribosome skipping, the interaction between 2ANPGP and the ribosome exit tunnel leads to the C-terminal portion of 2ANPGP adopting a conformation within the P-site of the peptidyl transferase centre (PTC), such that tRNAPro in the A-site cannot form a peptide bond, since the imino group is sterically restrained, being within a ring structure—the phi angle is constrained. We proposed that the tRNAPro exits the A-site, allowing release factors 1 and 3 (eRF1/3) to enter this site to hydrolyse the bond between the nascent polypeptide and tRNAGly [7,8,9]. eRF1 exit from the A-site is promoted by eRF3 [36]. For the ‘pseudo-reinitiation’ necessary to allow the elongation cycle to continue, (i) the vacant A-site must be re-occupied by tRNAPro and (ii) the tRNAPro must be translocated from the A- to P-site by elongation factor 2 (eEF2), thereby allowing (iii) the next aminoacyl-tRNA to enter the A-site to recommence polypeptide elongation—in this case, the downstream replication proteins. Should any of these steps be inhibited, translation would be terminated at this point without ‘pseudo-reinitiation’—effectively, the phenotype we observed using our reporter system in the case of the kunsagivirus C1 2A1, limnipivirus A1 2A2, limnipivirus B1 2A1, potamivirus B1 2A2, WCP 2A1/2A2/2A3, and WP-LV48 2A1/2A2- and 2A3 2A-like sequences; we observed synthesis of [GFP2A], but no GUS.
Our model proposes that the C-terminal residues of 2ANPGP adopt a conformation within the PTC such that a peptide bond with prolyl-tRNA (A-site) cannot be formed; it is possible that certain subset of 2ANPGP sequences adopt a conformation that also precludes peptidyl-tRNA hydrolysis by eRF1, halting, rather than stalling, elongation. The translation of sequences upstream of 2A (GFP) without detectable translation of sequences downstream of 2A (GUS) has, perhaps, a parallel with ‘no-go decay’. It has been shown that stalled translation complexes can be ‘rescued’ by the activities of mammalian Pelota/Hbs1/ABCE1 [37]. Pelota is a molecular mimic of eRF1, although (i) lacks the -GGQ- motif of eRF1—the peptidyl-tRNA ester linkage is, therefore, not hydrolysed by Pelota, and (ii) binds into a vacant A-site in a stop codon independent manner; Hbs1 is a structural homologue of eRF3 (reviewed in [38,39,40]). It should be noted, however, that Pelota might act in the stop–go process in conjunction with other translational GTPases with which it interacts, e.g., GTPBP2 [41]. ABCE1 promotes dissociation of the ribosome subunits and nascent peptidyl-tRNA is released, with peptidyl-tRNA hydrolase (Pth) subsequently releasing tRNA from peptidyl-tRNA by cleaving the peptide/tRNA ester linkage. Consistent with our observations, this would produce [GFP2A] alone, although, following this analogy, the subsequent decay of cellular mRNA in the no-go decay pathway implies degradation of the virus RNA. The first caveat here is that our analyses of 2A-like sequences from a range of organisms were performed on a ‘synthetic’ reporter construct using an in vitro translation system; how representative is this of virus replication in these disparate hosts? In our previous studies, however, such in vitro analyses have shown that these data have been confirmed by their use in many biotechnological/biomedical applications, including heterologous expression in a wide range of organisms (see Section 4.3 below). The second caveat that arises from our choice of the analysis of 25aa tracts is that our previous work on FMDV 2A indicated that such a tract was highly active and commensurate with the cloning strategy for analysis of larger numbers of 2A-like sequences; it may be the case that other StopGo sequences require additional upstream sequences and/or host-specific tRNAs/translation factor abundances, etc., for maximal efficiencies.
The evidence that 2ANPGP sequences function within the ribosome arises from two main sources. Firstly, construction of artificial polyproteins comprising two proteins, each of which bear N-terminal signal sequences. Here, sequences encoding the p40 and p35 subunits of IL12 were linked via FMDV 2ANPGP to encode [p402Ap35] (p40 stop codon removed). Both subunits were secreted from the cell to form active IL12: the (formerly) N-terminal signal sequence of p35 was recognised by signal recognition particle (SRP) as a nascent N-terminal feature and p35 secreted from the cell, along with p40 [42]. This property has been observed for many other artificial polyprotein systems, e.g., heavy and light chains of monoclonal antibodies, T-cell receptor proteins, etc. [43]. Secondly, the 2ANPGP ‘cleavage’ has been mapped to a 20–30aa tract that can be accommodated within the ribosome exit tunnel—one type amongst a family of ribosome arrest peptides (RAPs) [44]. Indeed, the C-terminal portion of a stalled nascent peptide in the ribosome exit tunnel has been shown to modulate the selectivity of the A-site [45].
Studies of 2ANPGP activity using strains of S. cerevisiae with compromised levels of release factor activity showed in strains with eRF1 depleted and a greater proportion of ribosomes translated through the 2A coding sequence, whilst in strains with impaired eRF3 GTPase activity, many ribosomes failed to ‘pseudo-reinitiate’ and translate sequences downstream of 2ANPGP [46]; these data led us to develop our model of 2ANPGP ‘cleavage’ activity. In contrast, however, studies using reconstituted translation systems in vitro did not show any involvement of eRF1/3 [47]. In conclusion, this sub-set of 2A-like sequences that appear to terminate translation at 2ANPGP poses a conundrum for both (i) our current model of 2ANPGP-mediated ribosome skipping and (ii) the effect of such sequences on the replication of kunsagi-, limnipi-, potami-, WCP, and WP-LV48 viruses.
4.3. Biotechnological and Biomedical Applications
2A/2A-like sequences have been used in a huge range of biotechnological and biomedical applications; these sequences have been shown to be active in all eukaryotic cell types (amoeba, yeast, fungi, algae, plant, animal) but not in prokaryotic cells (see [43]). In this paper, we report a series of 2A-like sequences that are directly comparable—in some cases superior—to those highly active sequences already in use: those encoded by aalivirus A1 2A1–2A4, aalivirus B1 2A1–2A4, avisivirus A1 2A2, avisivirus B1 2A1, grusopivirus A1 2A1–2A3, grusopivirus C 2A1/2A3, limnipivirus A1 2A1, limnipivirus B1 2A2, limnipivirus C1 2A1, limnipivirus D1 2A1/2A2, and mosavirus B1 2A1/2A2.
In the majority of cases, 2ANPGPs from different viruses have been used to co-express multiple different proteins (e.g., components of macromolecular structures, biochemical pathways, etc.) from a single ORF: for example, six different genes were co-expressed using five different 2A-like sequences to create autonomous bioluminescent human cells [48]. Alternatively, multiple iterations of the T2A peptide sequence with different codon usages were used to co-express nine different proteins comprising the carotenoid and violacein pathways in Pichia pastoris [49]. In both cases, however, constructs were designed to minimise the possibility of gene deletion via homologous recombination. In many cases this co-expression technology has replaced the need for time-consuming, costly, sequential transformations by the ability to link multiple genes into a single self-processing construct, thereby introducing a ‘trait’ by a single transformation: e.g., production of pluripotent stem cells, cancer gene therapies, CAR T-cell therapies, golden rice development, co-expression of glyphosate tolerance/BT toxins, introduction of novel biosynthetic pathways, etc. [42]. Here we describe a series of 2A-like sequences which substantially expand the toolbox for biotechnologists.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v16101587/s1, Supplementary Data: The polyprotein sequence alignments plus indications of the positions of 2A-like sequences.
Author Contributions
Conceptualisation, M.D.R. and G.A.L.; Methodology (clone construction, in vitro transcription/translation, etc.), L.S.R. and G.A.L.; Bioinformatic analyses, Y.-T.L., H.-C.W. and M.D.R. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the University of St Andrews.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The picornavirus sequence datasets analyzed in this manuscript were accessed from The National Center for Biotechnology Information (NCBI—https://www.ncbi.nlm.nih.gov/ (accessed on 5 September 2024)). The polyprotein sequence alignments plus indications of the positions of 2A-like sequences are given within the Supplementary Data submitted in this article.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
References
- Martĩnez-Salas, E.; Ryan, M.D. Translation and protein processing. In The Picornaviruses; Ehrenfeld, E., Domingo, E., Roos, R.P., Eds.; ASM Press: Washington, DC, USA, 2010; pp. 141–161. [Google Scholar]
- Yu, S.F.; Benton, P.; Bovee, M.; Sessions, J.; Lloyd, R.E. Defective RNA replication by Poliovirus mutants deficient in 2A protease cleavage activity. J. Virol. 1995, 69, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, H.; Nicklin, M.J.; Murray, M.G.; Anderson, C.W.; Dunn, J.J.; Studier, F.W.; Wimmer, E. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell 1986, 45, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.-Y.; Liu, Y.-N.; Wang, W.; Kao, F.-J.; Kung, S.-H. In vivo dynamics of enterovirus protease revealed by fluorescence resonance emission transfer (FRET) based on a novel FRET pair. Biochem. Biophys. Res. Commun. 2007, 353, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Lina, Y.; Zhao, J.; Yu, J.; Chen, Y.; Lin, M.C.; Kung, H.-F.; He, M.-L. Enterovirus 71 disrupts Interferon signaling by reducing the level of Interferon Receptor 1. J. Virol. 2012, 86, 3767–3776. [Google Scholar] [CrossRef]
- Martin, A.; Escriou, N.; Chao, S.-F.; Girard, M.; Lemon, S.M.; Wychowski, C. Identification and site-directed mutagenesis of the primary (2A/2B) cleavage site of the Hepatitis A Virus polyprotein: Functional impact on the infectivity of HAV RNA transcripts. Virology 1995, 213, 213–222. [Google Scholar] [CrossRef]
- Palmenberg, A.; Neubauer, D.; Skern, T. Genome organization and encoded proteins. In The Picornaviruses; Ehrenfeld, E., Domingo, E., Roos, R.P., Eds.; ASM Press: Washington, DC, USA, 2010; pp. 3–17. [Google Scholar]
- Ryan, M.D.; Donnelly, M.L.L.; Lewis, A.; Mehrotra, A.P.; Wilkie, J.; Gani, D. A model for nonstoichiometric, co-translational protein scission in eukaroytic ribosomes. Bioorg Chem. 1999, 27, 55–79. [Google Scholar] [CrossRef]
- Donnelly, M.L.L.; Luke, G.A.; Mehotra, A.; Li, X.; Hughes, L.E.; Gani, D.; Ryan, M.D. Analysis of the aphthovirus 2A/2B polyprotein “cleavage” mechanism indicates not a proteolytic reaction, but a novel translational effect: A putative ribosomal “skip”. J. Gen. Virol. 2001, 82, 1013–1025. [Google Scholar] [CrossRef] [PubMed]
- Luke, G.A.; de Felipe, P.; Lukashev, A.; Kallioinen, S.E.; Bruno, E.A.; Ryan, M.D. Occurrence, function and evolutionary origins of “2A-like” sequences in virus genomes. J. Gen. Virol. 2008, 89, 1036–1042. [Google Scholar] [CrossRef]
- de Lima, J.G.S.; Teixeira, D.G.; Freitas, T.T.; Lima, J.P.M.S. Evolutionary origin of 2A-like sequences in Totiviridae genomes. Virus Res. 2019, 259, 1–9. [Google Scholar] [CrossRef]
- de Lima, J.G.S.; Lanza, D.C.F. 2A and 2A-like sequences: Distribution in different virus species and applications in biotechnology. Viruses 2021, 13, 2160. [Google Scholar] [CrossRef]
- Donnelly, M.L.L.; Hughes, L.E.; Luke, G.A.; Mendoza, H.; ten Dam, E.; Gani, D.; Ryan, M.D. The “cleavage” activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring “2A-like” sequences. J. Gen. Virol. 2001, 82, 1027–1041. [Google Scholar] [CrossRef] [PubMed]
- Heras, S.R.; Thomas, M.C.; García-Canadas, M.; de Felipe, P.; García-Perez, J.L.; Ryan, M.D.; Lopez, M.C. L1Tc non-LTR retrotransposons from Trypanosoma cruzi contain functional viral-like self-cleaving 2A sequence in frame with active proteins they encode. Cell Mol. Life Sci. 2006, 63, 1449–1460. [Google Scholar] [CrossRef] [PubMed]
- Odon, V.; Luke, G.A.; Roulston, C.; de Felipe, P.; Ruan, L.; Escuin-Ordinas, H.; Brown, J.D.; Ryan, M.D.; Sukhodub, A. APE-type non-LTR retrotransposons of multicellular organisms encode virus-like 2A oligopeptide sequences, which mediate translational recoding during protein synthesis. Mol. Biol. Evol. 2013, 30, 1955–1965. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.D.; Ryan, M.D. Ribosome “Skipping”: “Stop-Carry On” or “Stop-Go” Translation. In Recoding: Expansion of Decoding Rules Enriches Gene Expression; Atkins, J.F., Gesteland, R.F., Eds.; Springer: New York, NY, USA, 2010; pp. 101–121. [Google Scholar]
- Roulston, C.; Luke, G.A.; de Felipe, P.; Ruan, L.; Cope, J.; Nicholson, J.; Sukhodub, A.; Tilsner, J.; Ryan, M.D. “2A-Like” signal sequences mediating translational recoding: A novel form of dual protein targeting. Traffic 2016, 17, 923–939. [Google Scholar] [CrossRef]
- Minskaia, E.; Nicholson, J.; Ryan, M.D. Optimisation of the foot-and-mouth disease virus 2A co-expression system for biomedical applications. BMC Biotechnol. 2013, 13, 67. [Google Scholar] [CrossRef]
- Donnelly, M.L.L.; Gani, D.; Flint, M.; Monaghan, S.; Ryan, M.D. The cleavage activities of aphthovirus and cardiovirus 2A proteins. J. Gen. Virol. 1997, 78, 13–21. [Google Scholar] [CrossRef]
- Loughran, G.; Firth, A.E.; Atkins, J.F. Ribosomal frameshifting into an overlapping gene in the 2B-encoding region of the cardiovirus genome. Proc. Natl. Acad. Sci. USA 2011, 108, E1111–E1119. [Google Scholar] [CrossRef]
- Napthine, S.; Ling, R.; Finch, L.K.; Jones, J.D.; Bell, S.; Brierley, I.; Firth, A.E. Protein-directed ribosomal frameshifting temporally regulates gene expression. Nat. Commun. 2017, 8, 15582. [Google Scholar] [CrossRef]
- Napthine, S.; Bell, S.; Hill, C.H.; Brierley, I.; Firth, A.E. Characterization of the stimulators of protein-directed ribosomal frameshifting in Theiler’s murine encephalomyelitis virus. Nucleic Acids Res. 2019, 47, 8207–8223. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Choi, G.K.Y.; Huang, Y.; Teng, J.L.L.; Tsoi, H.-W.; Tse, H.; Yeung, M.L.; Chan, K.-H.; Jin, D.-Y.; et al. Natural occurrence and characterization of two internal ribosome entry site elements in a novel virus, canine Picodicistrovirus, in the picornavirus-like superfamily. J. Virol. 2012, 86, 2797–2808. [Google Scholar] [CrossRef]
- Zell, R. Picornaviridae—The ever-growing virus family. Arch. Virol. 2018, 163, 299–317. [Google Scholar] [CrossRef] [PubMed]
- Zell, R.; Knowles, N.J.; Simmonds, P. A proposed division of the family Picornaviridae into subfamilies based on phylogenetic relationships and functional genomic organization. Arch. Virol. 2021, 166, 2927–2935. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zeng, Q.; Wang, M.; Cheng, A.; Pan, K.; Zhu, D.; Liu, M.; Jia, R.; Yang, Q.; Wu, Y.; et al. DHAV-1 2A1 peptide—A newly discovered co-expression tool that mediates the ribosomal “Skipping” function. Front. Microbiol. 2018, 9, 2727. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Ou, X.; Zhu, D.; Ma, G.; Cheng, A.; Wang, M.; Chen, S.; Jia, R.; Liu, M.; Sun, K.; et al. The 2A2 protein of Duck hepatitis A virus type 1 induces apoptosis in primary cell culture. Virus Genes 2016, 52, 780–788. [Google Scholar] [CrossRef]
- Hughes, P.J.; Stanway, G. The 2A proteins of three diverse picornaviruses are related to each other and to the H-rev107 family of proteins involved in the control of cell proliferation. J. Gen. Virol. 2000, 81, 201–207. [Google Scholar] [CrossRef]
- Yang, X.; Cheng, A.; Wang, M.; Jia, R.; Sun, K.; Pan, K.; Yang, Q.; Wu, Y.; Zhu, D.; Chen, S.; et al. Structures and corresponding functions of five types of picornaviral 2A proteins. Front. Microbiol. 2017, 8, 1373. [Google Scholar] [CrossRef]
- Wang, X.; Liu, N.; Wang, F.; Ning, K.; Li, Y.; Zhang, D. Genetic characterization of a novel duck-origin picornavirus with six 2A proteins. J. Gen. Virol. 2014, 95, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Shuo, D.; Luo, Y.; Pan, X.; Yan, D.; Li, X.; Lin, W.; Huang, D.; Yang, J.; Yuan, C.; et al. An emerging duck egg-reducing syndrome caused by a novel picornavirus containing seven putative 2A peptides. Viruses 2022, 14, 932. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Lukashev, A.N. Recombination among picornaviruses. Rev. Med. Virol. 2010, 20, 327–337. [Google Scholar] [CrossRef]
- Dantas, M.D.A.; Cavalcante, G.H.O.; Oliveira, R.A.C.; Lanza, D.C.F. New insights about ORF1 coding regions support the proposition of a new genus comprising arthropod viruses in the family Totiviridae. Virus Res. 2016, 211, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Jia, X.; Gao, Y.; Liu, Z.; Zhang, H.; Tan, Q.; Zhang, X.; Zhou, H.; Li, Y.; Wu, D.; et al. Cryo-EM reveals a previously unrecognized structural protein of a dsRNA virus implicated in its extracellular transmission. PLoS Pathog. 2021, 17, e1009396. [Google Scholar] [CrossRef] [PubMed]
- Eyler, D.E.; Wehner, K.A.; Green, R. Eukaryotic release factor 3 is required for multiple turnovers of peptide release catalysis by eukaryotic release factor 1. J. Biol. Chem. 2013, 288, 29530–29538. [Google Scholar] [CrossRef] [PubMed]
- Pisareva, V.P.; Skabkin, M.A.; Hellen, C.U.; Pestova, T.V.; Pisarev, A.V. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 2011, 30, 1804–1817. [Google Scholar] [CrossRef] [PubMed]
- Yip, M.C.J.; Shao, S. Detecting and rescuing stalled ribosomes. Trends Biochem. Sci. 2021, 46, 731–743. [Google Scholar] [CrossRef]
- Powers, K.T.; Szeto, J.A.; Schaffitzel, C. New insights into no-go, non-stop and nonsense-mediated mRNA decay complexes. Curr. Opin. Struct. Biol. 2020, 65, 110–118. [Google Scholar] [CrossRef]
- Harigaya, Y.; Parker, R. No-go decay: A quality control mechanism for RNA in translation. Wiley Interdiscip. Rev. RNA 2010, 1, 132–141. [Google Scholar] [CrossRef]
- Zinoviev, A.; Goyal, A.; Jindal, S.; LaCava, J.; Komar, A.A.; Rodnina, M.V.; Hellen, C.U.T.; Pestova, T.V. Functions of unconventional mammalian translational GTPases GTPBP1 and GTPBP2. Genes Dev. 2018, 32, 1226–1241. [Google Scholar] [CrossRef]
- Chaplin, P.J.; Camon, E.B.; Villarreal-Ramos, B.; Flint, M.; Ryan, M.D.; Collins, R.A. Production of interleukin-12 as a self-processing 2A polypeptide. J. Interferon Cytokine Res. 1999, 19, 235–241. [Google Scholar] [CrossRef]
- Research Papers Using 2A, Plus Reviews Discussing the Use of 2A. Available online: https://www.st-andrews.ac.uk/ryanlab/page10.htm (accessed on 5 September 2024).
- Ito, K.; Chiba, S. Arrest peptides: Cis-acting modulators of translation. Annu. Rev. Biochem. 2013, 82, 171–202. [Google Scholar] [CrossRef]
- Ramu, H.; Vázquez-Laslop, N.; Klepacki, D.; Dai, Q.; Piccirilli, J.; Micura, R.; Mankin, A.S. Nascent peptide in the ribosome exit tunnel affects functional properties of the A-site of the peptidyl transferase center. Mol. Cell 2011, 41, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Doronina, V.A.; Wu, C.; de Felipe, P.; Sachs, M.S.; Ryan, M.D.; Brown, J.D. Site-specific release of nascent chains from ribosomes at a sense codon. Mol. Cell Biol. 2008, 28, 4227–4239. [Google Scholar] [CrossRef] [PubMed]
- Machida, K.; Mikami, S.; Masutani, M.; Mishima, K.; Kobayashi, T.; Imataka, H. A translation system reconstituted with human factors proves that processing of encephalomyocarditis virus proteins 2A and 2B occurs in the elongation phase of translation without eukaryotic release factors. J. Biol. Chem. 2014, 289, 31960–31971. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Ripp, S.; Sayler, G.S.; Close, D.M. Expression of a humanized viral 2A-mediated lux operon efficiently generates autonomous bioluminescence in human cells. PLoS ONE 2014, 9, e96347. [Google Scholar] [CrossRef]
- Geier, M.; Fauland, P.; Vogl, T.; Glieder, A. Compact multi-enzyme pathways in P. pastoris. Chem. Commun. 2015, 51, 1643–1646. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).