The First Pseudomonas Phage vB_PseuGesM_254 Active against Proteolytic Pseudomonas gessardii Strains
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain Isolation, Identification, and Growth Conditions
2.2. Phage Isolation and Propagation
2.3. Biological Properties and Host Range Assay
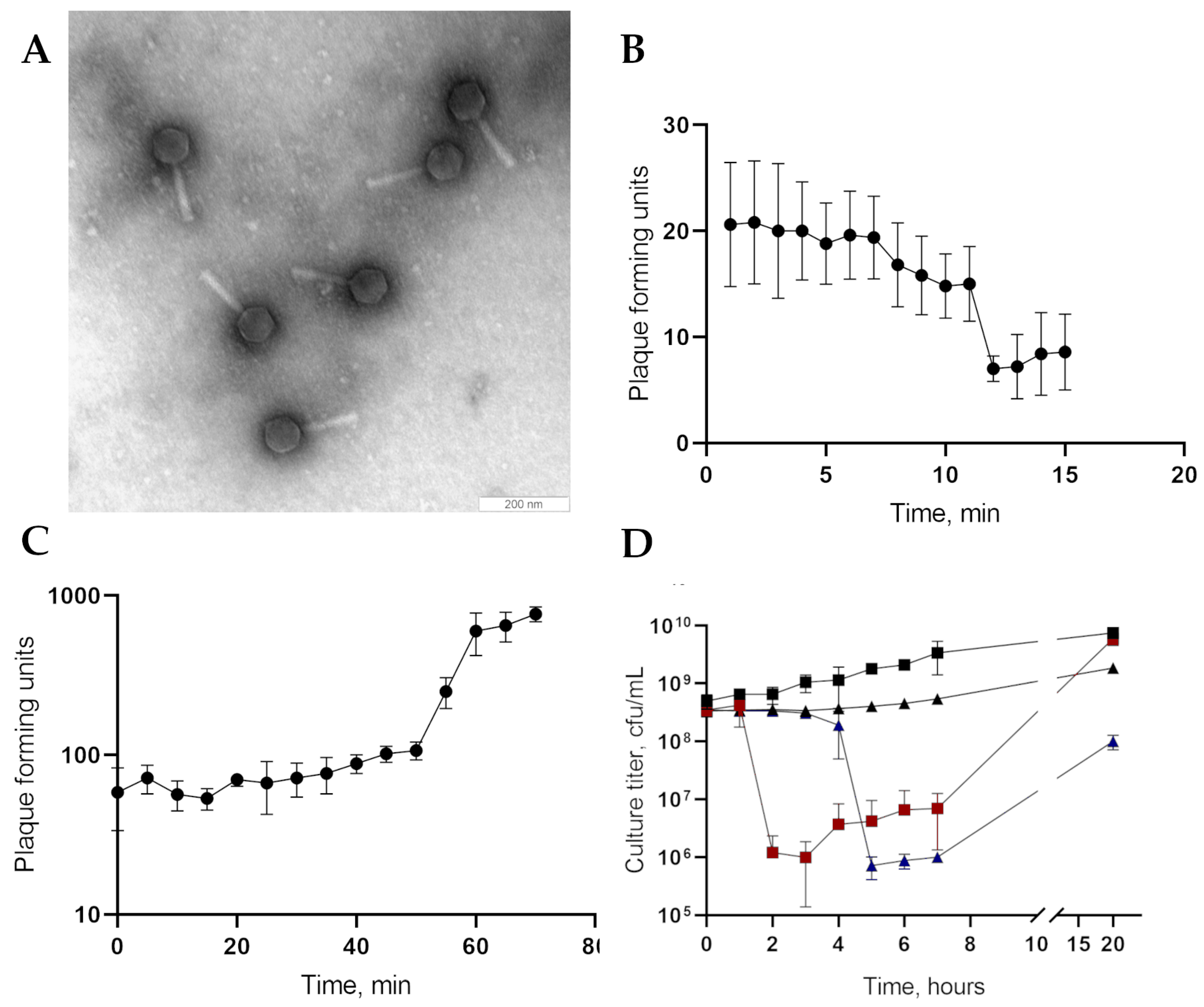
2.4. Phage Plaques and Phage Particles Morphology
2.5. Complete Genome Sequencing and Analysis
3. Results
3.1. Phage Particles Morphology, Biological Properties
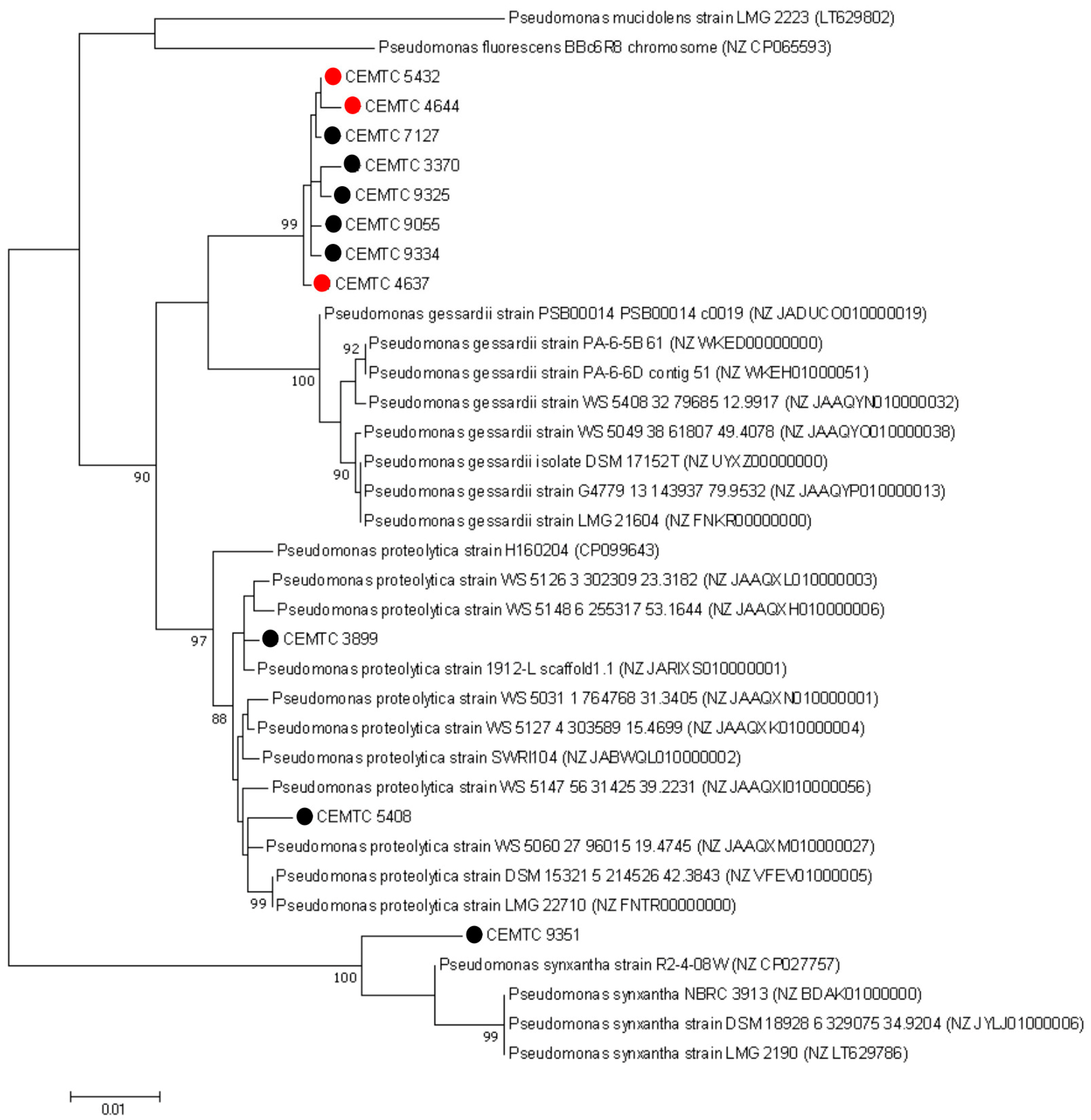
3.2. Bacterial Hosts
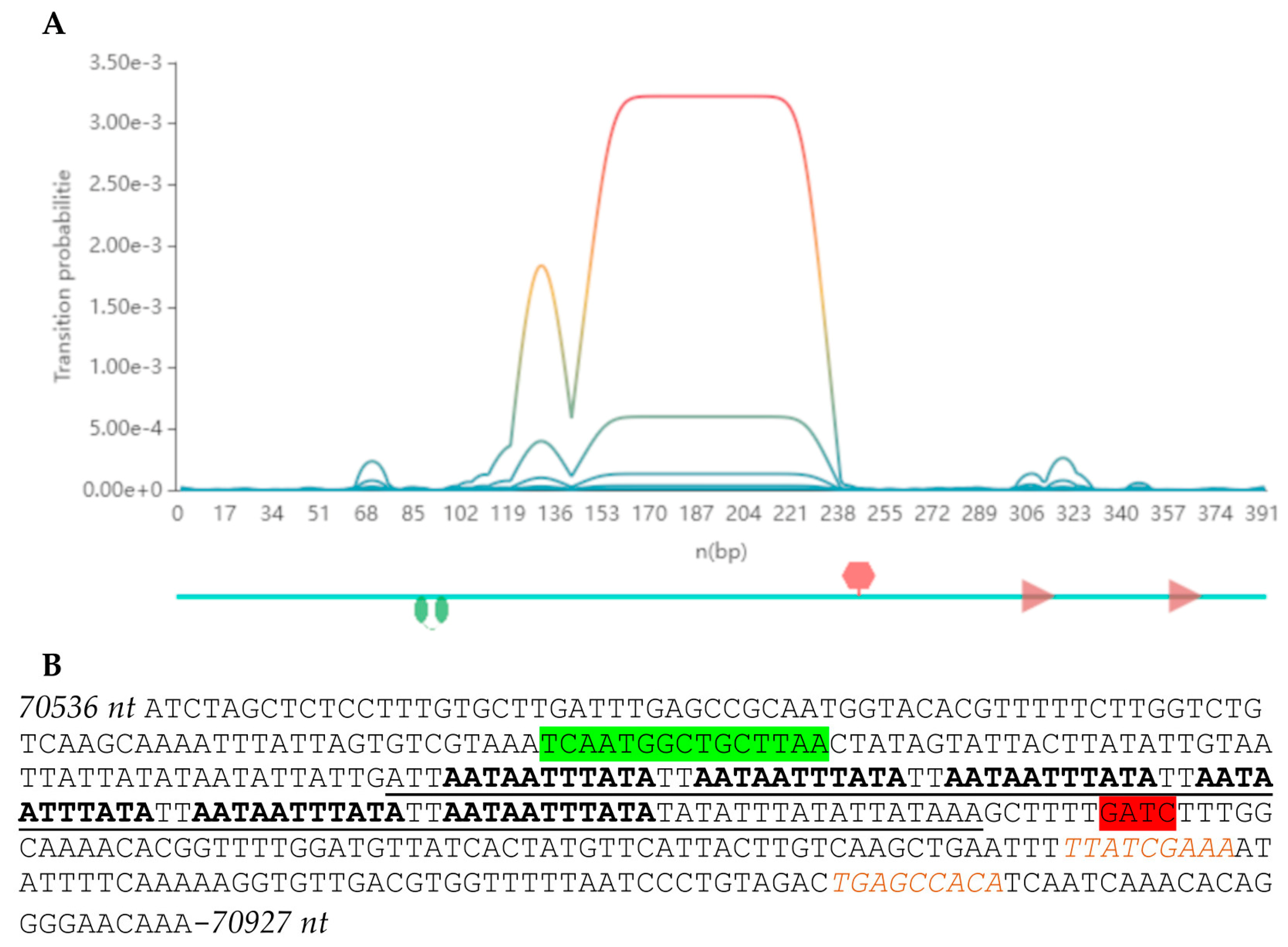
3.3. PseuGes_254 Genome Characteristics
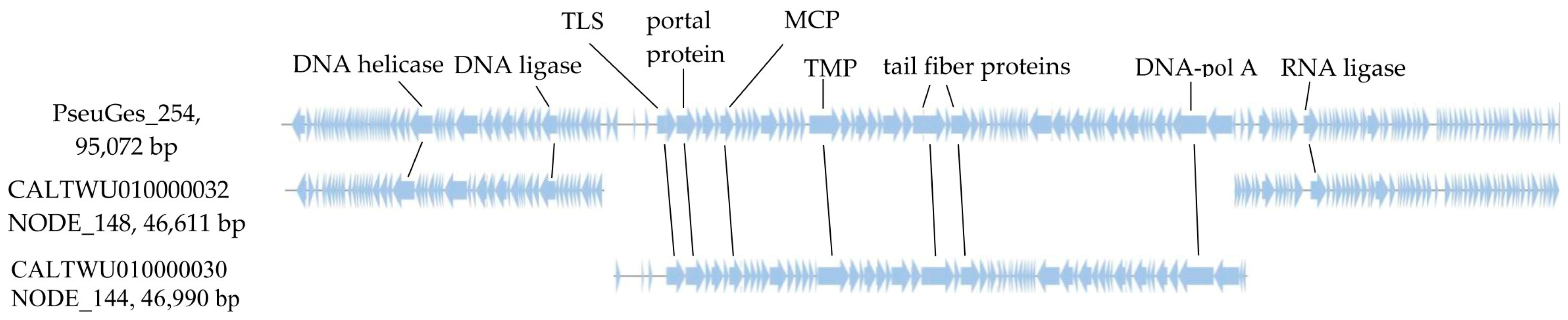
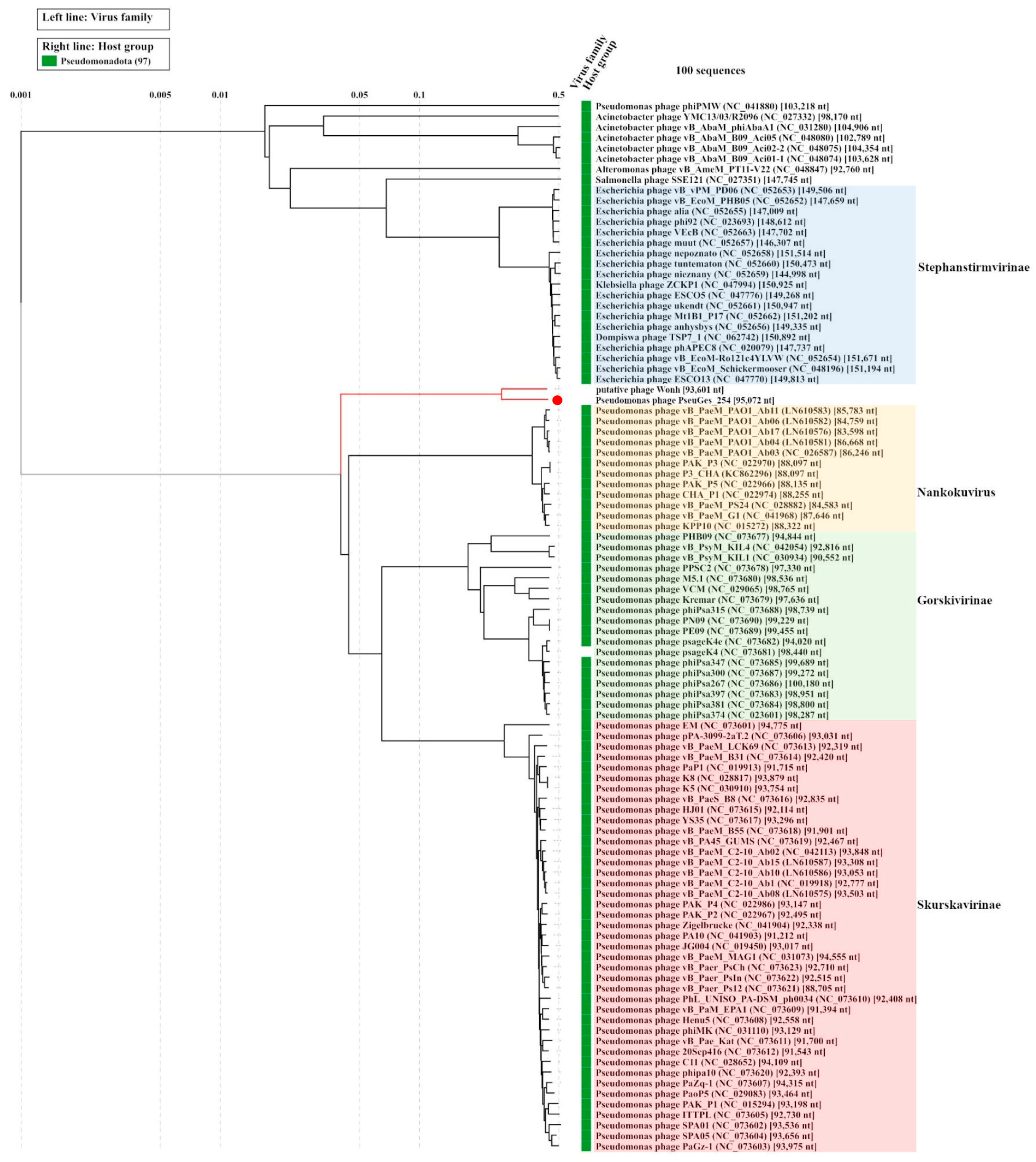
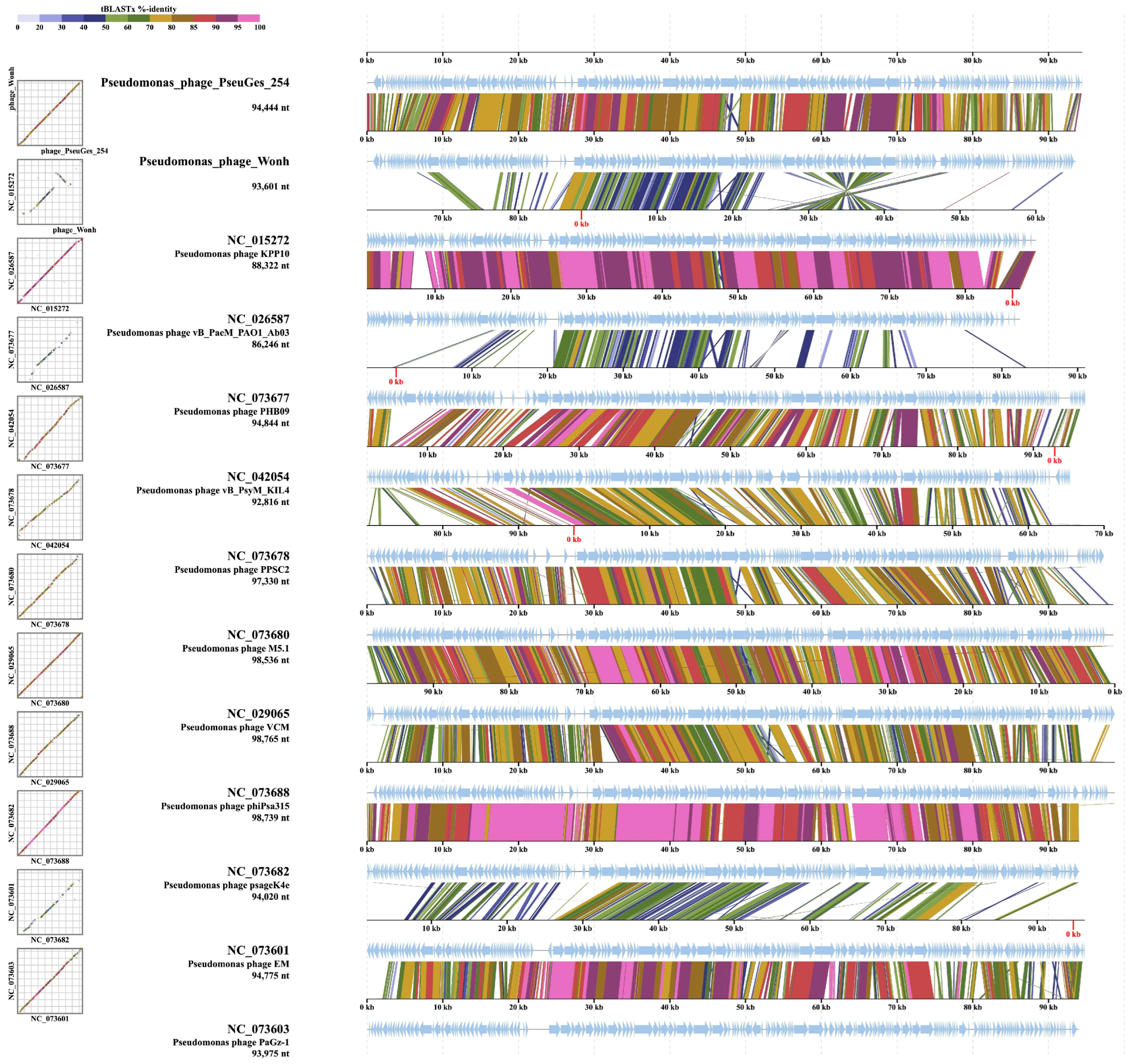
3.4. Comparative Genomic Analysis
3.5. Phylogenetic Analysis of Eessential Phage Proteins
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peix, A.; Ramírez-Bahena, M.H.; Velázquez, E. The current status on the taxonomy of Pseudomonas revisited: An update. Infect. Genet. Evol. 2018, 57, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Lalucat, J.; Mulet, M.; Gomila, M.; García-Valdés, E. Genomics in Bacterial Taxonomy: Impact on the genus Pseudomonas. Genes 2020, 11, 139. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Sanz, D.; Meier-Kolthoff, J.P.; Göker, M.; Martín, M.; Rivilla, R.; Redondo-Nieto, M. Genomic and genetic diversity within the Pseudomonas fluorescens complex. PLoS ONE 2016, 11, e0150183. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Wu, K.; Khan, A.; Jiang, Y.; Ling, Z.; Liu, P.; Chen, Y.; Tao, X.; Li, X. A novel Pseudomonas gessardii strain LZ-E simultaneously degrades naphthalene and reduces hexavalent chromium. Bioresour. Technol. 2016, 207, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Nandy, S.; Arora, U.; Tarar, P.; Viggor, S.; Jõesaar, M.; Kivisaar, M.; Kapley, A. Monitoring the growth, survival and phenol utilization of the fluorescent-tagged Pseudomonas oleovorans immobilized and free cells. Bioresour. Technol. 2021, 338, 125568. [Google Scholar] [CrossRef] [PubMed]
- Viggor, S.; Jõesaar, M.; Soares-Castro, P.; Ilmjärv, T.; Santos, P.M.; Kapley, A.; Kivisaar, M. Microbial metabolic potential of phenol degradation in wastewater treatment plant of crude oil refinery: Analysis of metagenomes and characterization of isolates. Microorganisms 2020, 8, 652. [Google Scholar] [CrossRef]
- Flury, P.; Vesga, P.; Dominguez-Ferreras, A.; Tinguely, C.; Ullrich, C.I.; Kleespies, R.G.; Keel, C.; Maurhofer, M. Persistence of root-colonizing Pseudomonas protegens in herbivorous insects throughout different developmental stages and dispersal to new host plants. ISME J. 2019, 13, 860–872. [Google Scholar] [CrossRef]
- Wood, S.J.; Kuzel, T.M.; Shafikhani, S.H. Pseudomonas aeruginosa: Infections, Animal Modeling, and Therapeutics. Cells 2023, 12, 199. [Google Scholar] [CrossRef]
- Jurado-Martín, I.; Sainz-Mejías, M.; McClean, S. Pseudomonas aeruginosa: An Audacious Pathogen with an Adaptable Arsenal of Virulence Factors. Int. J. Mol. Sci. 2021, 22, 3128. [Google Scholar] [CrossRef]
- Frapolli, M.; Défago, G.; Moënne-Loccoz, Y. Multilocus sequence analysis of biocontrol fluorescent Pseudomonas spp. producing the antifungal compound 2,4-diacetylphloroglucinol. Environ. Microbiol. 2007, 9, 1939–1955. [Google Scholar] [CrossRef]
- Samaddar, S.; Chatterjee, P.; Roy Choudhury, A.; Ahmed, S.; Sa, T. Interactions between Pseudomonas spp. and their role in improving the red pepper plant growth under salinity stress. Microbiol. Res. 2019, 219, 66–73. [Google Scholar] [CrossRef]
- Giugliano, R.; Buonocore, C.; Zannella, C.; Chianese, A.; Palma Esposito, F.; Tedesco, P.; De Filippis, A.; Galdiero, M.; Franci, G.; de Pascale, D. Antiviral Activity of the Rhamnolipids Mixture from the Antarctic Bacterium Pseudomonas gessardii M15 against Herpes Simplex Viruses and Coronaviruses. Pharmaceutics 2021, 13, 2121. [Google Scholar] [CrossRef] [PubMed]
- Kuzina, E.; Mukhamatdyarova, S.; Sharipova, Y.; Makhmutov, A.; Belan, L.; Korshunova, T. Influence of Bacteria of the Genus Pseudomonas on Leguminous Plants and Their Joint Application for Bioremediation of Oil Contaminated Soils. Plants 2022, 11, 3396. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, A.A.; Mullaeva, S.A.; Sazonova, O.I.; Petrikov, K.V.; Vetrova, A.A. Current research on simultaneous oxidation of aliphatic and aromatic hydrocarbons by bacteria of genus Pseudomonas. Folia Microbiol. 2022, 67, 591–604. [Google Scholar] [CrossRef] [PubMed]
- Allen, A.D.; Anderson, W.A.; Ayorinde, F.O.; Eribo, B.E. Biosynthesis and characterization of copolymer poly(3HB-co-3HV) from saponified Jatropha curcas oil by Pseudomonas oleovorans. J. Ind. Microbiol. Biotechnol. 2010, 37, 849–856. [Google Scholar] [CrossRef] [PubMed]
- Ashby, R.D.; Solaiman, D.K.; Foglia, T.A. The synthesis of short- and medium-chain-length poly(hydroxyalkanoate) mixtures from glucose- or alkanoic acid-grown Pseudomonas oleovorans. J. Ind. Microbiol. Biotechnol. 2002, 28, 147–153. [Google Scholar] [CrossRef]
- Ahuja, V.; Bhatt, A.K.; Mehta, S.; Sharma, V.; Rathour, R.K.; Sheetal. Xylitol production by Pseudomonas gessardii VXlt-16 from sugarcane bagasse hydrolysate and cost analysis. Bioprocess. Biosyst. Eng. 2022, 45, 1019–1031. [Google Scholar] [CrossRef]
- Haas, D.; Défago, G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 2005, 3, 307–319. [Google Scholar] [CrossRef]
- Saleem, M.; Asghar, H.N.; Zahir, Z.A.; Shahid, M. Impact of lead tolerant plant growth promoting rhizobacteria on growth, physiology, antioxidant activities, yield and lead content in sunflower in lead contaminated soil. Chemosphere 2018, 195, 606–614. [Google Scholar] [CrossRef]
- Couillerot, O.; Prigent-Combaret, C.; Caballero-Mellado, J.; Moënne-Loccoz, Y. Pseudomonas fluorescens and closely-related fluorescent pseudomonads as biocontrol agents of soil-borne phytopathogens. Lett. Appl. Microbiol. 2009, 48, 505–512. [Google Scholar] [CrossRef]
- Pilipchuk, T.A.; Gerasimovich, A.D.; Ananyeva, I.N.; Kolomiets, E.I.; Popov, F.A.; Novik, G.I. Biopesticide ‘Multiphage’ based on phages of phytopathogenic bacteria P. syringae and P. fluorescens used in agriculture to control plant diseases. Mikrobn. Biotekhnologii 2015, 7, 197–219. (In Russian) [Google Scholar]
- Ramani, K.; Chockalingam, E.; Sekaran, G. Production of a novel extracellular acidic lipase from Pseudomonas gessardii using slaughterhouse waste as a substrate. J. Ind. Microbiol. Biotechnol. 2010, 37, 531–535. [Google Scholar] [CrossRef] [PubMed]
- Zarei, M.; Mohammadpour, H.; Gharibi, D.; Pourmahdi Borujeni, M. Identification of Pseudomonas jessenii and Pseudomonas gessardii as the most proteolytic Pseudomonas isolates in Iranian raw milk and their impact on stability of sterilized milk during storage. J. Dairy Res. 2020, 87, 368–374. [Google Scholar] [CrossRef] [PubMed]
- De Jonghe, V.; Coorevits, A.; Van Hoorde, K.; Messens, W.; Van Landschoot, A.; De Vos, P.; Heyndrickx, M. Influence of storage conditions on the growth of Pseudomonas species in refrigerated raw milk. Appl. Environ. Microbiol. 2011, 77, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Machado, S.G.; Baglinière, F.; Marchand, S.; Van Coillie, E.; Vanetti, M.C.; De Block, J.; Heyndrickx, M. The Biodiversity of the Microbiota Producing Heat-Resistant Enzymes Responsible for Spoilage in Processed Bovine Milk and Dairy Products. Front. Microbiol. 2017, 8, 302. [Google Scholar] [CrossRef]
- Nascimento, E.C.D.; Sabino, M.C.; Corguinha, L.D.R.; Targino, B.N.; Lange, C.C.; Pinto, C.L.O.; Pinto, P.F.; Vidigal, P.M.P.; Sant'Ana, A.S.; Hungaro, H.M. Lytic bacteriophages UFJF_PfDIW6 and UFJF_PfSW6 prevent Pseudomonas fluorescens growth in vitro and the proteolytic-caused spoilage of raw milk during chilled storage. Food Microbiol. 2022, 101, 103892. [Google Scholar] [CrossRef]
- Tayyarcan, E.K.; Boyaci, I.H. Isolation, characterization, and application of bacteriophage cocktails for the biocontrol of Pseudomonas fluorescens group strains in whole and skimmed milk. Braz. J. Microbiol. 2023, 54, 3061–3071. [Google Scholar] [CrossRef]
- Tayyarcan, E.K.; Evran, E.; Guven, K.; Ekiz, E.; Acar Soykut, E.; Boyaci, I.H. Evaluating the efficacy of a phage cocktail against Pseudomonas fluorescens group strains in raw milk: Microbiological, physical, and chemical analyses. Arch. Microbiol. 2024, 206, 283. [Google Scholar] [CrossRef]
- Nowicki, G.; Walkowiak-Nowicka, K.; Zemleduch-Barylska, A.; Mleczko, A.; Frąckowiak, P.; Nowaczyk, N.; Kozdrowska, E.; Barylski, J. Complete genome sequences of two novel autographiviruses infecting a bacterium from the Pseudomonas fluorescens group. Arch. Virol. 2017, 162, 2907–2911. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, Z.; Li, J.; Qin, K.; Wei, Y.; Zhang, Q.; Lin, L.; Ji, X. Complete genome sequence of the lytic cold-active Pseudomonas fluorescens bacteriophage VSW-3 from Napahai plateau wetland. Virus Genes 2017, 53, 146–150. [Google Scholar] [CrossRef]
- Sillankorva, S.; Kropinski, A.M.; Azeredo, J. Genome sequence of the broad-host-range Pseudomonas phage Φ-S1. J. Virol. 2012, 86, 10239. [Google Scholar] [CrossRef] [PubMed]
- Sillankorva, S.; Kluskens, L.D.; Lingohr, E.J.; Kropinski, A.M.; Neubauer, P.; Azeredo, J. Complete genome sequence of the lytic Pseudomonas fluorescens phage ϕIBB-PF7A. Virol. J. 2011, 8, 142. [Google Scholar] [CrossRef] [PubMed]
- Koberg, S.; Gieschler, S.; Brinks, E.; Wenning, M.; Neve, H.; Franz, C.M.A.P. Genome sequence of the novel virulent bacteriophage PMBT14 with lytic activity against Pseudomonas fluorescens DSM 50090R. Arch Virol 2018, 163, 2575–2577. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wu, Y.; Tang, Y.; Gan, B. Genomic characterization and phylogenetic analysis of the novel Pseudomonas phage PPSC2. Arch. Virol. 2018, 163, 1977–1980. [Google Scholar] [CrossRef] [PubMed]
- Sprotte, S.; Brinks, E.; Wagner, N.; Kropinski, A.M.; Neve, H.; Franz, C.M.A.P. Characterization of the first Pseudomonas grimontii bacteriophage, PMBT3. Arch. Virol. 2021, 166, 2887–2894. [Google Scholar] [CrossRef] [PubMed]
- Hungaro, H.M.; Vidigal, P.M.P.; do Nascimento, E.C.; Gomes da Costa Oliveira, F.; Gontijo, M.T.P.; Lopez, M.E.S. Genomic characterisation of UFJF_PfDIW6: A Novel lytic Pseudomonas fluorescens-phage with potential for biocontrol in the dairy industry. Viruses 2022, 14, 629. [Google Scholar] [CrossRef]
- Wang, Y.; Qian, P.Y. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS ONE 2009, 4, e7401. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 8, 3022–3027. [Google Scholar] [CrossRef]
- Bacteriophage λ and its vectors. In Molecular Cloning, 3rd ed.; Sambrook, J., Russell, D., Eds.; Cold Spring Harbour Laboratory Press: New York, NY, USA, 2001; Volume 1, pp. 2.25–2.106. [Google Scholar]
- Kropinski, A.M. Measurement of the rate of attachment of bacteriophage to cells. In Bacteriophages: Methods and Protocols; Clokie, M.R.J., Kropinski, A.M., Eds.; Humana Press: New York, NY, USA, 2009; pp. 151–155. [Google Scholar]
- Pajunen, M.; Kiljunen, S.; Skurnik, M. Bacteriophage phiYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J. Bacteriol. 2000, 182, 5114–5120. [Google Scholar] [CrossRef]
- Heo, Y.J.; Lee, Y.R.; Jung, H.H.; Lee, J.; Ko, G.; Cho, Y.H. Antibacterial Efficacy of phages against Pseudomonas aeruginosa infections in mice and Drosophila melanogaster. Antimicrob. Agents Chemother. 2009, 53, 2469–2474. [Google Scholar] [CrossRef]
- Kutter, E. Phage host range and efficiency of plating. In Bacteriophages: Methods and Protocols; Clokie, M.R.J., Kropinski, A.M., Eds.; Humana Press: New York, NY, USA, 2009; Volume 1, pp. 141–149. [Google Scholar] [CrossRef]
- O’Flaherty, S.; Coffey, A.; Edwards, R.; Meaney, W.; Fitzgerald, G.F.; Ross, R.P. Genome of staphylococcal phage K: A new lineage of Myoviridae infecting gram-positive bacteria with a low GC content. J. Bacteriol. 2004, 186, 2862–2871. [Google Scholar] [CrossRef] [PubMed]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes de novo assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef] [PubMed]
- Garneau, J.R.; Depardieu, F.; Fortier, L.C.; Bikard, D.; Monot, M. PhageTerm: A tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci. Rep. 2017, 7, 8292. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef]
- Söding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, W244–W248. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Dong, M.J.; Luo, H.; Gao, F. Ori-Finder 2022: A Comprehensive Web server for prediction and analysis of bacterial replication origins. Genom. Proteom. Bioinform. 2022, 20, 1207–1213. [Google Scholar] [CrossRef]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. ViPTree: The viral proteomic tree server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef]
- Moraru, C.; Varsani, A.; Kropinski, A.M. VIRIDIC—A novel tool to calculate the intergenomic similarities of prokaryote-infecting viruses. Viruses 2020, 12, 1268. [Google Scholar] [CrossRef] [PubMed]
- Verhille, S.; Baïda, N.; Dabboussi, F.; Hamze, M.; Izard, D.; Leclerc, H. Pseudomonas gessardii sp. nov. and Pseudomonas migulae sp. nov., two new species isolated from natural mineral waters. Int. J. Syst. Bacteriol. 1999, 49 Pt 4, 1559–1572. [Google Scholar] [CrossRef] [PubMed]
- Rajewska, M.; Wegrzyn, K.; Konieczny, I. AT-rich region and repeated sequences—The essential elements of replication origins of bacterial replicons. FEMS Microbiol. Rev. 2012, 36, 408–434. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.; Kropinski, A.M.; Adriaenssens, E.M. A Roadmap for genome-based phage taxonomy. Viruses 2021, 13, 506. [Google Scholar] [CrossRef]


| No. | Species (16S rRNA Gene (GenBank ID) | CEMTC Number | Isolation Source | Date of Isolation | Relative Efficiency of Plating (EOP) 1/Phage Titer, BOE/mL |
|---|---|---|---|---|---|
| 1 | P. gessardii (ON838145) | 4637 * | Chemal river, Altai Republic | 26 January 2022 | Not applicable/3.7 × 1010 |
| 2 | P. gessardii (OQ834588) | 4644 | Chemal river, Altai Republic | 29 January 2022 | Low/8 × 106 |
| 3 | P. gessardii (PP348772) | 5432 | Upper Inegen river, Altai Republic | 7 May 2022 | Low/1.4 × 107 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morozova, V.; Babkin, I.; Mogileva, A.; Kozlova, Y.; Tikunov, A.; Bardasheva, A.; Fedorets, V.; Zhirakovskaya, E.; Ushakova, T.; Tikunova, N. The First Pseudomonas Phage vB_PseuGesM_254 Active against Proteolytic Pseudomonas gessardii Strains. Viruses 2024, 16, 1561. https://doi.org/10.3390/v16101561
Morozova V, Babkin I, Mogileva A, Kozlova Y, Tikunov A, Bardasheva A, Fedorets V, Zhirakovskaya E, Ushakova T, Tikunova N. The First Pseudomonas Phage vB_PseuGesM_254 Active against Proteolytic Pseudomonas gessardii Strains. Viruses. 2024; 16(10):1561. https://doi.org/10.3390/v16101561
Chicago/Turabian StyleMorozova, Vera, Igor Babkin, Alina Mogileva, Yuliya Kozlova, Artem Tikunov, Alevtina Bardasheva, Valeria Fedorets, Elena Zhirakovskaya, Tatiana Ushakova, and Nina Tikunova. 2024. "The First Pseudomonas Phage vB_PseuGesM_254 Active against Proteolytic Pseudomonas gessardii Strains" Viruses 16, no. 10: 1561. https://doi.org/10.3390/v16101561
APA StyleMorozova, V., Babkin, I., Mogileva, A., Kozlova, Y., Tikunov, A., Bardasheva, A., Fedorets, V., Zhirakovskaya, E., Ushakova, T., & Tikunova, N. (2024). The First Pseudomonas Phage vB_PseuGesM_254 Active against Proteolytic Pseudomonas gessardii Strains. Viruses, 16(10), 1561. https://doi.org/10.3390/v16101561







