Urine: A Pitfall for Molecular Detection of Toscana Virus? An Analytical Proof-of-Concept Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Virus Stock Preparation
2.2. Spiking of Clinical Specimens
2.3. RNA Extraction and Viral Genome Detection
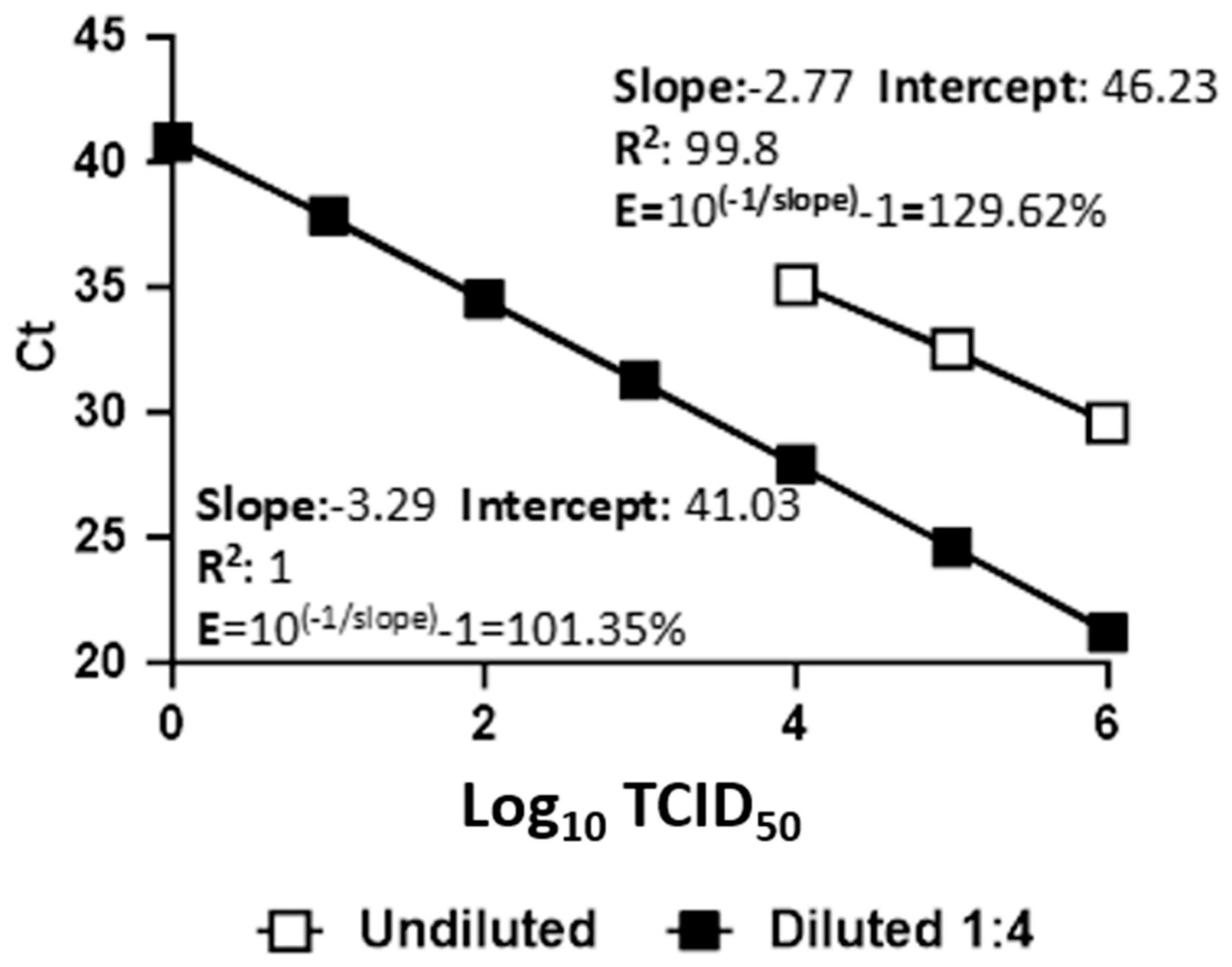
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ayhan, N.; Charrel, R.N. An update on Toscana virus distribution, genetics, medical and diagnostic aspects. Clin. Microbiol. Infect. 2020, 26, 1017–1023. [Google Scholar] [CrossRef] [PubMed]
- Cusi, M.G.; Savellini, G.G.; Zanelli, G. Toscana virus epidemiology: From Italy to beyond. Open Virol. J. 2010, 4, 22–28. [Google Scholar] [CrossRef]
- Cusi, M.G.; Savellini, G.G. Diagnostic tools for Toscana virus infection. Expert. Rev. Anti Infect. Ther. 2011, 9, 799–805. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.M.; Mulkey, S.B.; Campos, J.M.; DeBiasi, R.L. Laboratory diagnosis of CNS infections in children due to emerging and re-emerging neurotropic viruses. Pediatr. Res. 2023. [Google Scholar] [CrossRef] [PubMed]
- Charrel, R.N. Chapter 8—Toscana Virus Infection. In Emerging Infectious Diseases; Ergönül, Ö., Can, F., Madoff, L., Akova, M., Eds.; Academic Press: Amsterdam, The Netherlands, 2014; pp. 111–119. [Google Scholar]
- Li, M.; Wang, B.; Li, L.; Wong, G.; Liu, Y.; Ma, J.; Li, J.; Lu, H.; Liang, M.; Li, A.; et al. Rift Valley Fever Virus and Yellow Fever Virus in Urine: A Potential Source of Infection. Virol. Sin. 2019, 34, 342–345. [Google Scholar] [CrossRef] [PubMed]
- Ergunay, K.; Kaplan, B.; Okar, S.; Akkutay-Yoldar, Z.; Kurne, A.; Arsava, E.M.; Ozkul, A. Urinary detection of toscana virus nucleic acids in neuroinvasive infections. J. Clin. Virol. 2015, 70, 89–92. [Google Scholar] [CrossRef]
- Mahony, J.; Chong, S.; Jang, D.; Luinstra, K.; Faught, M.; Dalby, D.; Sellors, J.; Chernesky, M. Urine specimens from pregnant and nonpregnant women inhibitory to amplification of Chlamydia trachomatis nucleic acid by PCR, ligase chain reaction, and transcription-mediated amplification: Identification of urinary substances associated with inhibition and removal of inhibitory activity. J. Clin. Microbiol. 1998, 36, 3122–3126. [Google Scholar]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors—Occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef]
- Thirion, L.; Pezzi, L.; Pedrosa-Corral, I.; Sanbonmatsu-Gamez, S.; de Lamballerie, X.; Falchi, A.; Perez-Ruiz, M.; Charrel, R.N. Evaluation of a Trio Toscana Virus Real-Time RT-PCR Assay Targeting Three Genomic Regions within Nucleoprotein Gene. Pathogens 2021, 10, 254–264. [Google Scholar] [CrossRef]
- Brisbarre, N.; Plumet, S.; Cotteaux-Lautard, C.; Emonet, S.F.; Pages, F.; Leparc-Goffart, I. A rapid and specific real time RT-PCR assay for diagnosis of Toscana virus infection. J. Clin. Virol. 2015, 66, 107–111. [Google Scholar] [CrossRef]
- Weidmann, M.; Sanchez-Seco, M.P.; Sall, A.A.; Ly, P.O.; Thiongane, Y.; Lô, M.M.; Schley, H.; Hufert, F.T. Rapid detection of important human pathogenic Phleboviruses. J. Clin. Virol. 2008, 41, 138–142. [Google Scholar] [CrossRef]
- Pérez-Ruiz, M.; Collao, X.; Navarro-Marí, J.M.; Tenorio, A. Reverse transcription, real-time PCR assay for detection of Toscana virus. J. Clin. Virol. 2007, 39, 276–281. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty percent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Toye, B.; Woods, W.; Bobrowska, M.; Ramotar, K. Inhibition of PCR in genital and urine specimens submitted for Chlamydia trachomatis testing. J. Clin. Microbiol. 1998, 36, 2356–2358. [Google Scholar] [CrossRef] [PubMed]
- Biel, S.S.; Held, T.K.; Landt, O.; Niedrig, M.; Gelderblom, H.R.; Siegert, W.; Nitsche, A. Rapid quantification and differentiation of human polyomavirus DNA in undiluted urine from patients after bone marrow transplantation. J. Clin. Microbiol. 2000, 38, 3689–3695. [Google Scholar] [CrossRef] [PubMed]
- Merck: RT-PCR/RT-qPCR Troubleshooting. Available online: https://www.sigmaaldrich.cn/CN/zh/technical-documents/technical-article/genomics/pcr/troubleshooting (accessed on 5 November 2023).
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Nolan, T.; Hands, R.; Bustin, S. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef] [PubMed]
- Tyler, K.L. Emerging viral infections of the central nervous system: Part 1. Arch. Neurol. 2009, 66, 939–948. [Google Scholar]
- Gori Savellini, G.; Gandolfo, C.; Cusi, M.G. Epidemiology of Toscana virus in South Tuscany over the years 2011–2019. J. Clin. Virol. 2020, 128, 104452–104456. [Google Scholar] [CrossRef]
- Riccò, M.; Peruzzi, S. Epidemiology of Toscana Virus in Italy (2018–2020), a summary of available evidences. Acta Biomed. 2021, 92, e2021230. [Google Scholar]
- Braito, A.; Corbisiero, R.; Corradini, S.; Marchi, B.; Sancasciani, N.; Fiorentini, C.; Ciufolini, M.G. Evidence of Toscana virus infections without central nervous system involvement: A serological study. Eur. J. Epidemiol. 1997, 13, 761–764. [Google Scholar] [CrossRef]
- Magurano, F.; Baggieri, M.; Gattuso, G.; Fortuna, C.; Remoli, M.E.; Vaccari, G.; Zaccaria, G.; Marchi, A.; Bucci, P.; Benedetti, E.; et al. Toscana virus genome stability: Data from a meningoencephalitis case in Mantua, Italy. Vector Borne Zoonotic Dis. 2014, 14, 866–869. [Google Scholar] [CrossRef]
- Muñoz, C.; Ayhan, N.; Ortuño, M.; Ortiz, J.; Gould, E.A.; Maia, C.; Berriatua, E.; Charrel, R.N. Experimental Infection of Dogs with Toscana Virus and Sandfly Fever Sicilian Virus to Determine Their Potential as Possible Vertebrate Hosts. Microorganisms 2020, 8, 596–605. [Google Scholar] [CrossRef] [PubMed]
- Vilibic-Cavlek, T.; Zidovec-Lepej, S.; Ledina, D.; Knezevic, S.; Savic, V.; Tabain, I.; Ivic, I.; Slavuljica, I.; Bogdanic, M.; Grgic, I.; et al. Clinical, Virological, and Immunological Findings in Patients with Toscana Neuroinvasive Disease in Croatia: Report of Three Cases. Trop. Med. Infect. Dis. 2020, 5, 144–150. [Google Scholar] [CrossRef]
- Okar, S.V.; Bekircan-Kurt, C.E.; Hacıoğlu, S.; Erdem-Özdamar, S.; Özkul, A.; Ergünay, K. Toscana virus associated with Guillain-Barré syndrome: A case-control study. Acta Neurol. Belg. 2021, 121, 661–668. [Google Scholar] [CrossRef]
- Jonsson, N.; Gullberg, M.; Lindberg, A.M. Real-time polymerase chain reaction as a rapid and efficient alternative to estimation of picornavirus titers by tissue culture infectious dose 50% or plaque forming units. Microbiol. Immunol. 2009, 53, 149–154. [Google Scholar]
- Liu, B.; Forman, M.; Valsamakis, A. Optimization and evaluation of a novel real-time RT-PCR test for detection of parechovirus in cerebrospinal fluid. J. Virol. Methods 2019, 272, 113690. [Google Scholar] [CrossRef] [PubMed]
- Regione Veneto, Boll. Sorveglianza Arbovirosi. Available online: https://www.regione.veneto.it/documents/10793/12217504/2023_n12_Bollettino.pdf/63c87ad6-84b0-4581-b436-ef9bf3c06297 (accessed on 27 December 2023).
- Matusali, G.; D’Abramo, A.; Terrosi, C.; Carletti, F.; Colavita, F.; Vairo, F.; Savellini, G.G.; Gandolfo, C.; Anichini, G.; Lalle, E.; et al. Infectious Toscana Virus in Seminal Fluid of Young Man Returning from Elba Island, Italy. Emerg. Infect. Dis. 2022, 28, 865–869. [Google Scholar] [CrossRef]
| TCID50 | Undiluted Urine (Mean Ct ± SD 3) | Undiluted Urine Hit Score | Diluted Urine (Mean Ct ± SD 3) | Diluted Urine Hit Score | Undiluted-Diluted Mean Ct (ΔCt) |
|---|---|---|---|---|---|
| 106 | 29.57 (±0.25) | 4/4 | 21.20 (±0.09) | 4/4 | 8.37 |
| 105 | 32.54 (±0.35) | 4/4 | 24.59 (±0.04) | 4/4 | 7.95 |
| 104 | 35.10 1 (±0.36) | 4/4 | 27.93 (±0.20) | 4/4 | 7.17 |
| 103 | Nd 2 | 0/4 | 31.28 (±0.11) | 4/4 | |
| 102 | nd | 0/4 | 34.54 (±0.20) | 4/4 | |
| 101 | nd | 0/4 | 37.87 1 (±0.69) | 4/4 | |
| 1 | nd | 0/4 | 40.81 (±1.01) | 3/4 | |
| 0.1 | nd | 0/4 | nd | 0/4 | |
| 0.01 | nd | 0/4 | nd | 0/4 | |
| 0.001 | nd | 0/4 | nd | 0/4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mori, A.; Matucci, A.; Pomari, E.; Accordini, S.; Piubelli, C.; Donini, A.; Nicolini, L.; Castilletti, C. Urine: A Pitfall for Molecular Detection of Toscana Virus? An Analytical Proof-of-Concept Study. Viruses 2024, 16, 98. https://doi.org/10.3390/v16010098
Mori A, Matucci A, Pomari E, Accordini S, Piubelli C, Donini A, Nicolini L, Castilletti C. Urine: A Pitfall for Molecular Detection of Toscana Virus? An Analytical Proof-of-Concept Study. Viruses. 2024; 16(1):98. https://doi.org/10.3390/v16010098
Chicago/Turabian StyleMori, Antonio, Andrea Matucci, Elena Pomari, Silvia Accordini, Chiara Piubelli, Annalisa Donini, Lavinia Nicolini, and Concetta Castilletti. 2024. "Urine: A Pitfall for Molecular Detection of Toscana Virus? An Analytical Proof-of-Concept Study" Viruses 16, no. 1: 98. https://doi.org/10.3390/v16010098
APA StyleMori, A., Matucci, A., Pomari, E., Accordini, S., Piubelli, C., Donini, A., Nicolini, L., & Castilletti, C. (2024). Urine: A Pitfall for Molecular Detection of Toscana Virus? An Analytical Proof-of-Concept Study. Viruses, 16(1), 98. https://doi.org/10.3390/v16010098







