microRNA-185 Inhibits SARS-CoV-2 Infection through the Modulation of the Host’s Lipid Microenvironment
Abstract
:1. Introduction
2. Results
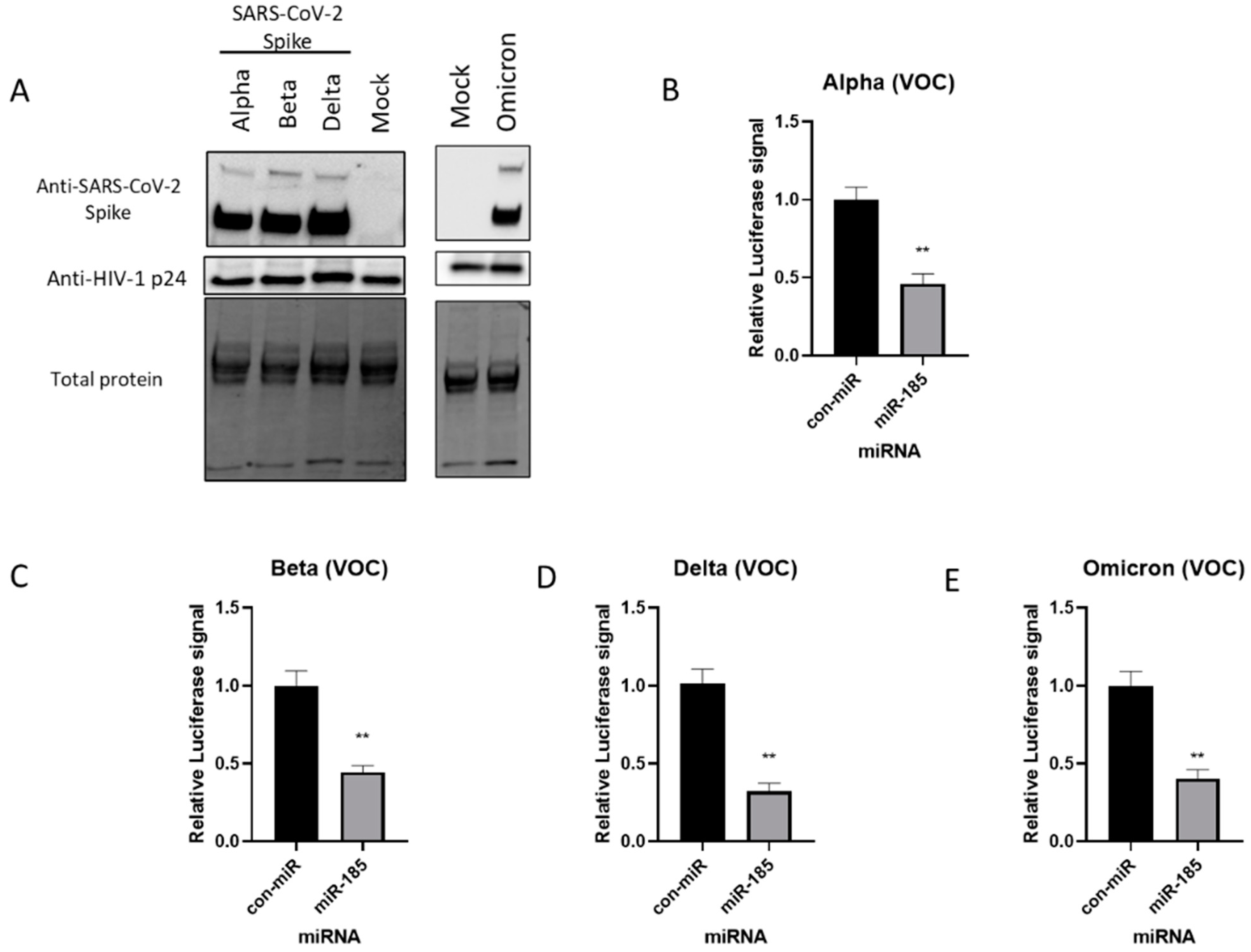
2.1. Expression of SARS-CoV-2 Spike Protein and Pseudovirus Incorporation of SARS-CoV-2 Spike (S) Protein
2.2. miR-185 Antagonizes SARS-CoV-2 S Protein Pseudotyped Virus Entry in Huh7 and Calu-3 Cell Lines
2.3. Inhibition of SREBP2-Modulated Signaling Antagonizes SARS-CoV-2 Spike Pseudotyped Viral Entry
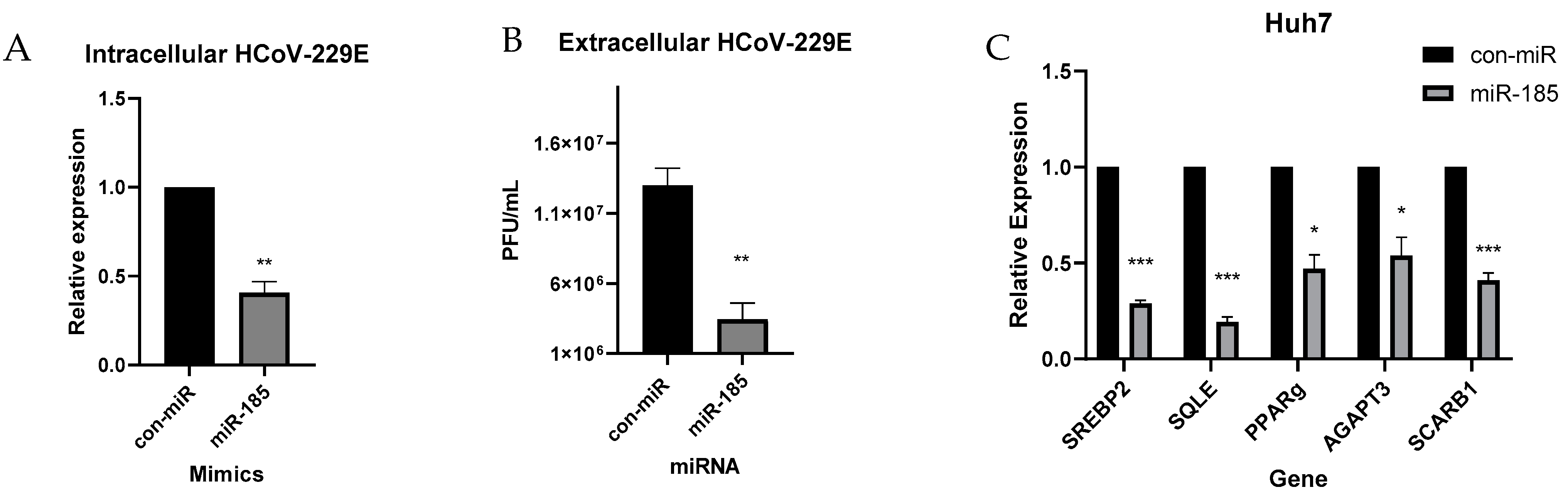
2.4. miR-185 Inhibits SARS-CoV-2 Entry by Modulating Lipid Metabolism and Repression of ACE2 Expression in Calu-3 Cells
2.5. miR-185 Inhibits HCoV-229E Replication and Infectivity
2.6. miR-185 Overexpression Inhibits SARS-CoV-2 Pathogenesis in Calu-3 Cells
3. Discussion
4. Methods
4.1. Reagents and Cell Culture
4.2. Generation of D614G and N501Y Mutants
4.3. Production of Pseudotyped Viral Particles
4.4. Detection of S Protein of SARS-CoV-2 by Western Blot
4.5. Entry Assays
4.6. Fluvastatin and 25-Hydroxycholesterol Treatments
4.7. Transfections and Infections
4.8. HCoV-229E Plaque Assay
4.9. Quantitative Real-Time PCR
4.10. Transfections of miRNAs for SARS-CoV-2 Inoculation
4.11. RNA Extraction from SARS-CoV-2 Infected Cells and Quantitative Real-Time PCR (qRT-PCR)
5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Awadasseid, A.; Wu, Y.; Tanaka, Y.; Zhang, W. Effective drugs used to combat SARS-CoV-2 infection and the current status of vaccines. Biomed. Pharmacother. 2021, 137, 111330. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, M.; Malehi, A.S.; Rahim, F. COVID-19 patients: A systematic review and meta-analysis of laboratory findings, comorbidities, and clinical outcomes comparing medical staff versus the general population. Osong Public Health Res. Perspect. 2020, 11, 269–279. [Google Scholar] [CrossRef]
- Liu, C.H.; Lu, C.H.; Wong, S.H.; Lin, L.T. Update on Antiviral Strategies against COVID-19: Unmet Needs and Prospects. Front. Immunol. 2021, 11, 616595. [Google Scholar] [CrossRef]
- Kyriakidis, N.C.; López-Cortés, A.; González, E.V.; Grimaldos, A.B.; Prado, E.O. SARS-CoV-2 vaccines strategies: A comprehensive review of phase 3 candidates. npj Vaccines 2021, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.-L.; Cheng, J.-S.-Y.; Gong, H.-L.; Yuan, M.-D.; Zhao, X.-H.; Li, Z.; Wei, D.-X. Advances in the design and development of SARS-CoV-2 vaccines. Mil. Med. Res. 2021, 8, 67. [Google Scholar] [CrossRef]
- Barone, P.; DeSimone, R.A. Convalescent plasma to treat coronavirus disease 2019 (COVID-19): Considerations for clinical trial design. Transfusion 2020, 60, 1123–1127. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Liu, L.; Iketani, S.; Guo, Y.; Chan, J.F.-W.; Wang, M.; Liu, L.; Luo, Y.; Chu, H.; Huang, Y.; Nair, M.S.; et al. Striking Antibody Evasion Manifested by the Omicron Variant of SARS-CoV-2. Nature 2021, 202, 676–681. [Google Scholar] [CrossRef]
- Arora, P.; Rocha, C.; Kempf, A.; Nehlmeier, I.; Graichen, L.; Winkler, M.S.; Lier, M.; Schulz, S.; Jäck, H.M.; Cossmann, A.; et al. The spike protein of SARS-CoV-2 variant A.30 is heavily mutated and evades vaccine-induced antibodies with high efficiency. Cell. Mol. Immunol. 2021, 18, 2673–2675. [Google Scholar] [CrossRef]
- Chen, R.E.; Zhang, X.; Case, J.B.; Winkler, E.S.; Liu, Y.; VanBlargan, L.A.; Liu, J.; Errico, J.M.; Xie, X.; Suryadevara, N.; et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021, 27, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, D.X.; Tam, J.P. Lipid rafts are involved in SARS-CoV entry into Vero E6 cells. Biochem. Biophys. Res. Commun. 2008, 369, 344–349. [Google Scholar] [CrossRef]
- Radenkovic, D.; Chawla, S.; Pirro, M.; Sahebkar, A.; Banach, M. Cholesterol in relation to COVID-19: Should we care about it? J. Clin. Med. 2020, 9, 1909. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, S.; Nordqvist, H.; Ambikan, A.T.; Gupta, S.; Sperk, M.; Svensson-Akusjärvi, S.; Mikaeloff, F.; Benfeitas, R.; Saccon, E.; Ponnan, S.M.; et al. Metabolic perturbation associated with COVID-19 disease severity and SARS-CoV-2 replication. Mol. Cell. Proteomics 2021, 20, 100159. [Google Scholar] [CrossRef] [PubMed]
- Mohr, A.M.; Mott, J.L. Overview of microRNA biology. Semin. Liver Dis. 2015, 35, 3–11. [Google Scholar] [CrossRef]
- Singaravelu, R.; O’Hara, S.; Jones, D.M.; Chen, R.; Taylor, N.G.; Srinivasan, P.; Quan, C.; Roy, D.G.; Steenbergen, R.H.; Kumar, A.; et al. MicroRNAs regulate the immunometabolic response to viral infection in the liver. Nat. Chem. Biol. 2015, 11, 988–993. [Google Scholar] [CrossRef]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef]
- Deng, H.K.; Liu, R.; Ellmeier, W.; Choe, S.; Unutmaz, D.; Burkhart, M.; Di Marzio, P.; Marmon, S.; Sutton, R.E.; Mark Hill, C.; et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature 1996, 381, 661–666. [Google Scholar] [CrossRef]
- Sherman, E.J.; Emmer, B.T. ACE2 protein expression within isogenic cell lines is heterogeneous and associated with distinct transcriptomes. Sci. Rep. 2021, 11, 15900. [Google Scholar] [CrossRef]
- Hou, Y.J.; Chiba, S.; Halfmann, P.; Ehre, C.; Kuroda, M.; Dinnon, K.H.; Leist, S.R.; Schäfer, A.; Nakajima, N.; Takahashi, K.; et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 2020, 370, 1464–1468. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Plante, K.S.; Plante, J.A.; Xie, X.; Zhang, X.; Ku, Z.; An, Z.; Scharton, D.; Schindewolf, C.; et al. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature 2021, 602, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Zang, R.; Case, J.B.; Yutuc, E.; Ma, X.; Shen, S.; Castro, M.F.G.; Liu, Z.; Zeng, Q.; Zhao, H.; Son, J.; et al. Cholesterol 25-hydroxylase suppresses SARS-CoV-2 replication by blocking membrane fusion. Proc. Natl. Acad. Sci. USA 2020, 117, 32105–32113. [Google Scholar] [CrossRef]
- Zu, S.; Deng, Y.Q.; Zhou, C.; Li, J.; Li, L.; Chen, Q.; Li, X.F.; Zhao, H.; Gold, S.; He, J.; et al. 25-Hydroxycholesterol is a potent SARS-CoV-2 inhibitor. Cell Res. 2020, 30, 1043–1045. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Deng, Y.Q.; Wang, S.; Ma, F.; Aliyari, R.; Huang, X.Y.; Zhang, N.N.; Watanabe, M.; Dong, H.L.; Liu, P.; et al. 25-Hydroxycholesterol Protects Host against Zika Virus Infection and Its Associated Microcephaly in a Mouse Model. Immunity 2017, 46, 446–456. [Google Scholar] [CrossRef]
- Anggakusuma; Romero-Brey, I.; Berger, C.; Colpitts, C.C.; Boldanova, T.; Engelmann, M.; Todt, D.; Perin, P.M.; Behrendt, P.; Vondran, F.W.R.; et al. Interferon-inducible cholesterol-25-hydroxylase restricts hepatitis C virus replication through blockage of membranous web formation. Hepatology 2015, 62, 702–714. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.Y.; Sanchez, D.J.; Aliyari, R.; Lu, S.; Cheng, G. Systematic identification of type I and type II interferon-induced antiviral factors. Proc. Natl. Acad. Sci. USA 2012, 109, 4239–4244. [Google Scholar] [CrossRef]
- Gomes, B.; Gonçalves, S.; Disalvo, A.; Hollmann, A.; Santos, N.C. Effect of 25-hydroxycholesterol in viral membrane fusion: Insights on HIV inhibition. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1171–1178. [Google Scholar] [CrossRef]
- Liu, S.Y.; Aliyari, R.; Chikere, K.; Li, G.; Marsden, M.D.; Smith, J.K.; Pernet, O.; Guo, H.; Nusbaum, R.; Zack, J.A.; et al. Interferon-Inducible Cholesterol-25-Hydroxylase Broadly Inhibits Viral Entry by Production of 25-Hydroxycholesterol. Immunity 2013, 38, 92–105. [Google Scholar] [CrossRef]
- Adams, C.M.; Reitz, J.; De Brabander, J.K.; Feramisco, J.D.; Li, L.; Brown, M.S.; Goldstein, J.L. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and insigs. J. Biol. Chem. 2004, 279, 52772–52780. [Google Scholar] [CrossRef]
- Ahn, A.; Gibbons, D.L.; Kielian, M. The Fusion Peptide of Semliki Forest Virus Associates with Sterol-Rich Membrane Domains. J. Virol. 2002, 76, 3267–3275. [Google Scholar] [CrossRef] [PubMed]
- Li, G.M.; Li, Y.G.; Yamate, M.; Li, S.M.; Ikuta, K. Lipid rafts play an important role in the early stage of severe acute respiratory syndrome-coronavirus life cycle. Microbes Infect. 2007, 9, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Sanders, D.W.; Jumper, C.C.; Ackerman, P.J.; Bracha, D.; Donlic, A.; Kim, H.; Kenney, D.; Castello-Serrano, I.; Suzuki, S.; Tamura, T.; et al. SARS-CoV-2 requires cholesterol for viral entry and pathological syncytia formation. eLife 2021, 10, e65962. [Google Scholar] [CrossRef]
- Glende, J.; Schwegmann-Wessels, C.; Al-Falah, M.; Pfefferle, S.; Qu, X.; Deng, H.; Drosten, C.; Naim, H.Y.; Herrler, G. Importance of cholesterol-rich membrane microdomains in the interaction of the S protein of SARS-coronavirus with the cellular receptor angiotensin-converting enzyme 2. Virology 2008, 381, 215–221. [Google Scholar] [CrossRef]
- Madison, B.B. Srebp2: A master regulator of sterol and fatty acid synthesis1. J. Lipid Res. 2016, 57, 333–335. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Wan, L.; Yan, Q.; Wang, X.; Zhang, J.; Yang, X.; Zhang, Y.; Fan, C.; Li, D.; Deng, Y.; et al. HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nat. Metab. 2020, 2, 1391–1400. [Google Scholar] [CrossRef]
- Teissier, É.; Pécheur, E.I. Lipids as modulators of membrane fusion mediated by viral fusion proteins. Eur. Biophys. J. 2007, 36, 887–899. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho, M.V.; Gonçalves-De-albuquerque, C.F.; Silva, A.R. PPAR gamma: From definition to molecular targets and therapy of lung diseases. Int. J. Mol. Sci. 2021, 22, 805. [Google Scholar] [CrossRef] [PubMed]
- Ebert, M.S.; Sharp, P.A. Roles for MicroRNAs in conferring robustness to biological processes. Cell 2012, 149, 515–524. [Google Scholar] [CrossRef]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. (Lausanne) 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Tambyah, P.A.; Ching, C.S.; Sepramaniam, S.; Ali, J.M.; Armugam, A.; Jeyaseelan, K. microRNA expression in blood of dengue patients. Ann. Clin. Biochem. 2015, 53, 466–476. [Google Scholar] [CrossRef]
- Ouyang, X.; Jiang, X.; Gu, D.; Zhang, Y.; Kong, S.K.; Jiang, C.; Xie, W. Dysregulated serum miRNA profile and promising biomarkers in dengue-infected patients. Int. J. Med. Sci. 2016, 13, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C.L.; Yi, M.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of Hepatitis C Virus RNA Abundance by a Liver-specific MicroRNA. Science 2005, 1, 1577–1581. [Google Scholar] [CrossRef] [PubMed]
- Singaravelu, R.; Ahmed, N.; Quan, C.; Srinivasan, P.; Christopher, J.; Roy, D.G.; Pezacki, J.P. A conserved miRNA-183 cluster regulates the innate antiviral response. J. Biol. Chem. 2019, 294, 19785–19794. [Google Scholar] [CrossRef]
- Van Der Ree, M.H.; Van Der Meer, A.J.; Van Nuenen, A.C.; De Bruijne, J.; Ottosen, S.; Janssen, H.L.; Kootstra, N.A.; Reesink, H.W. Miravirsen dosing in chronic hepatitis C patients results in decreased microRNA-122 levels without affecting other microRNAs in plasma. Aliment. Pharmacol. Ther. 2016, 43, 102–113. [Google Scholar] [CrossRef]
- van der Ree, M.H.; de Vree, J.M.; Stelma, F.; Willemse, S.; van der Valk, M.; Rietdijk, S.; Molenkamp, R.; Schinkel, J.; van Nuenen, A.C.; Beuers, U.; et al. Safety, tolerability, and antiviral effect of RG-101 in patients with chronic hepatitis C: A phase 1B, double-blind, randomised controlled trial. Lancet 2017, 389, 709–717. [Google Scholar] [CrossRef]
- Cagno, V.; Civra, A.; Rossin, D.; Calfapietra, S.; Caccia, C.; Leoni, V.; Dorma, N.; Biasi, F.; Poli, G.; Lembo, D. Inhibition of herpes simplex-1 virus replication by 25-hydroxycholesterol and 27-hydroxycholesterol. Redox Biol. 2017, 12, 522–527. [Google Scholar] [CrossRef]
- Liu, Y.; Wei, Z.; Zhang, Y.; Ma, X.; Chen, Y.; Yu, M.; Ma, C.; Li, X.; Cao, Y.; Liu, J.; et al. Activation of liver X receptor plays a central role in antiviral actions of 25-hydroxycholesterol. J. Lipid Res. 2018, 59, 2287–2296. [Google Scholar] [CrossRef]
- Wang, S.; Li, W.; Hui, H.; Tiwari, S.K.; Zhang, Q.; Croker, B.A.; Rawlings, S.; Smith, D.; Carlin, A.F.; Rana, T.M. Cholesterol 25-Hydroxylase inhibits SARS -CoV-2 and other coronaviruses by depleting membrane cholesterol. EMBO J. 2020, 39, e106057. [Google Scholar] [CrossRef]
- Yan, B.; Chu, H.; Yang, D.; Sze, K.H.; Lai, P.M.; Yuan, S.; Shuai, H.; Wang, Y.; Kao, R.Y.T.; Chan, J.F.W.; et al. Characterization of the lipidomic profile of human coronavirus-infected cells: Implications for lipid metabolism remodeling upon coronavirus replication. Viruses 2019, 11, 73. [Google Scholar] [CrossRef] [PubMed]
- Nardacci, R.; Colavita, F.; Castilletti, C.; Lapa, D.; Matusali, G.; Meschi, S.; Del Nonno, F.; Colombo, D.; Capobianchi, M.R.; Zumla, A.; et al. Evidences for lipid involvement in SARS-CoV-2 cytopathogenesis. Cell Death Dis. 2021, 12, 263. [Google Scholar] [CrossRef] [PubMed]
- Sagan, S.M.; Rouleau, Y.; Leggiadro, C.; Supekova, L.; Schultz, P.G.; Su, A.I.; Pezacki, J.P. The influence of cholesterol and lipid metabolism on host cell structure and hepatitis C virus replication. Biochem. Cell Biol. 2006, 84, 67–79. [Google Scholar] [CrossRef]
- Spann, N.J.; Glass, C.K. Sterols and oxysterols in immune cell function. Nat. Immunol. 2013, 14, 893–900. [Google Scholar] [CrossRef] [PubMed]
- Nomura, R.; Kiyota, A.; Suzaki, E.; Kataoka, K.; Ohe, Y.; Miyamoto, K.; Senda, T.; Fujimoto, T. Human Coronavirus 229E Binds to CD13 in Rafts and Enters the Cell through Caveolae. J. Virol. 2004, 78, 8701–8708. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Huang, M.; Yuan, Q.; Wei, Y.; Gao, Y.; Mao, L.; Gu, L.; Tan, Y.W.; Zhong, Y.; Liu, D.; et al. The important role of lipid raft-mediated attachment in the infection of cultured cells by coronavirus infectious bronchitis virus beaudette strain. PLoS ONE 2017, 12, e0170123. [Google Scholar] [CrossRef]
- Li, X.; Zhu, W.; Fan, M.; Zhang, J.; Peng, Y.; Huang, F.; Wang, N.; He, L.; Zhang, L.; Holmdahl, R.; et al. Dependence of SARS-CoV-2 infection on cholesterol-rich lipid raft and endosomal acidification. Comput. Struct. Biotechnol. J. 2021, 19, 1933–1943. [Google Scholar] [CrossRef]
- Marchand-Senécal, X.; Kozak, R.; Mubareka, S.; Salt, N.; Gubbay, J.B.; Eshaghi, A.; Allen, V.; Li, Y.; Bastien, N.; Gilmour, M.; et al. Diagnosis and Management of First Case of COVID-19 in Canada: Lessons Applied from SARS-CoV-1. Clin. Infect. Dis. 2020, 71, 2207–2210. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Paquette, S.G.; Banner, D.; Huang, S.S.H.; Almansa, R.; Leon, A.; Xu, L.; Bartoszko, J.; Kelvin, D.J.; Kelvin, A.A. Influenza Transmission in the Mother-Infant Dyad Leads to Severe Disease, Mammary Gland Infection, and Pathogenesis by Regulating Host Responses. PLoS Pathog. 2015, 11, e1005173. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.W.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, N.; Francis, M.E.; Ahmed, N.; Kelvin, A.A.; Pezacki, J.P. microRNA-185 Inhibits SARS-CoV-2 Infection through the Modulation of the Host’s Lipid Microenvironment. Viruses 2023, 15, 1921. https://doi.org/10.3390/v15091921
Ahmed N, Francis ME, Ahmed N, Kelvin AA, Pezacki JP. microRNA-185 Inhibits SARS-CoV-2 Infection through the Modulation of the Host’s Lipid Microenvironment. Viruses. 2023; 15(9):1921. https://doi.org/10.3390/v15091921
Chicago/Turabian StyleAhmed, Nadine, Magen E. Francis, Noreen Ahmed, Alyson A. Kelvin, and John Paul Pezacki. 2023. "microRNA-185 Inhibits SARS-CoV-2 Infection through the Modulation of the Host’s Lipid Microenvironment" Viruses 15, no. 9: 1921. https://doi.org/10.3390/v15091921
APA StyleAhmed, N., Francis, M. E., Ahmed, N., Kelvin, A. A., & Pezacki, J. P. (2023). microRNA-185 Inhibits SARS-CoV-2 Infection through the Modulation of the Host’s Lipid Microenvironment. Viruses, 15(9), 1921. https://doi.org/10.3390/v15091921




