Different Outcomes of Chicken Infection with UK-Origin H5N1-2020 and H5N8-2020 High-Pathogenicity Avian Influenza Viruses (Clade 2.3.4.4b)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Virus Origin and Propagation
2.2. Ethics and Safety Statement
2.3. Experimental Design: Minimum Infectious Dose (MID50), Intra-Species Transmission, and Pathogenesis of H5N8-2020 and H5N1-2020 in Chickens
2.4. Collection of Clinical and Environmental Samples followed by RNA Extraction
2.5. AIV Reverse Transcription Real-Time PCR (RRT-PCR)
2.6. Serology
2.7. Immunohistochemistry
2.8. Whole-Genome Sequencing (WGS) of Progeny Viruses
3. Results
3.1. Determination of the MID50 and Onward Transmission among Chickens Infected with H5N8-2020 and H5N1-2020
3.2. Morbidity and Mortality of Chickens Infected with H5N8-2020 and H5N1-2020
3.3. Pathogenesis Investigation: Systemic Viral Distribution in Chickens after Infection with H5N8-2020 or H5N1-2020
3.4. Environmental Testing
3.5. Viral Genetic Polymorphisms from Chickens Infected with H5N8-2020 and H5N1-2020
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alexander, D.J. An overview of the epidemiology of avian influenza. Vaccine 2007, 25, 5637–5644. [Google Scholar] [CrossRef] [PubMed]
- Patapiou, P.A.; Slomka, M.J.; Seekings, A.H.; James, J.; Thomas, S.S.; Reid, S.M.; Hansen, R.D.E.; Lewis, N.S.; Banyard, A.C. JMM Profile: Avian influenza: A veterinary pathogen with zoonotic potential. J. Med. Microbiol. 2022, 71, 001491. [Google Scholar] [CrossRef]
- Brown, I.H.; Banks, J.; Manvell, R.J.; Essen, S.C.; Shell, W.; Slomka, M.; Londt, B.; Alexander, D.J. Recent epidemiology and ecology of influenza A viruses in avian species in Europe and the Middle East. Dev. Biol. 2006, 124, 45–50. [Google Scholar]
- FAO. Approaches to Controlling, Preventing and Eliminating H5N1 Highly Pathogenic Avian Influenza in Endemic Countries. In Animal Production and Health Paper No.171; FAO: Rome, Italy, 2011; Available online: https://www.fao.org/3/i2150e/i2150e00.htm (accessed on 17 July 2023).
- Smith, G.J.; Donis, R.O.; World Health Organization/World Organisation for Animal Health; Food Agriculture Organization; H5 Evolution Working Group. Nomenclature updates resulting from the evolution of avian influenza A(H5) virus clades 2.1.3.2a, 2.2.1, and 2.3.4 during 2013–2014. Influenza Other Respir Viruses 2015, 9, 271–276. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization Global Influenza Program Surveillance Network. Evolution of H5N1 avian influenza viruses in Asia. Emerg. Infect. Dis. 2005, 11, 1515–1521. [Google Scholar] [CrossRef]
- Global Consortium for H5N8 and Related Influenza Viruses. Role for migratory wild birds in the global spread of avian influenza H5N8. Science 2016, 354, 213–217. [Google Scholar] [CrossRef]
- Alarcon, P.; Brouwer, A.; Venkatesh, D.; Duncan, D.; Dovas, C.I.; Georgiades, G.; Monne, I.; Fusaro, A.; Dan, A.; Smietanka, K.; et al. Comparison of 2016-17 and Previous Epizootics of Highly Pathogenic Avian Influenza H5 Guangdong Lineage in Europe. Emerg. Infect. Dis. 2018, 24, 2270–2283. [Google Scholar] [CrossRef]
- EFSA; European Food Safety Authority; European Centre for Disease Prevention and Control and European Union Reference Laboratory for Avian Influenza; Adlhoch, C.; Fusaro, A.; Gonzales, J.L.; Kuiken, T.; Marangon, S.; Niqueux, E.; Staubach, C.; et al. Avian Influenza overview August-December 2020. EFSA J. 2020, 18, 6379. [Google Scholar] [CrossRef]
- Poen, M.J.; Venkatesh, D.; Bestebroer, T.M.; Vuong, O.; Scheuer, R.D.; Oude Munnink, B.B.; de Meulder, D.; Richard, M.; Kuiken, T.; Koopmans, M.P.G.; et al. Co-circulation of genetically distinct highly pathogenic avian influenza A clade 2.3.4.4 (H5N6) viruses in wild waterfowl and poultry in Europe and East Asia, 2017–2018. Virus Evol. 2019, 5, vez004. [Google Scholar] [CrossRef]
- Lewis, N.S.; Banyard, A.C.; Whittard, E.; Karibayev, T.; Al Kafagi, T.; Chvala, I.; Byrne, A.; Meruyert Akberovna, S.; King, J.; Harder, T.; et al. Emergence and spread of novel H5N8, H5N5 and H5N1 clade 2.3.4.4 highly pathogenic avian influenza in 2020. Emerg. Microbes Infect. 2021, 10, 148–151. [Google Scholar] [CrossRef]
- Pacey, A.C.; Coxon, C.; Gale, P.; Gauntlett, F.; Wild, C. Updated Outbreak Assessment #16, 15 February 2021. Highly pathogenic avian influenza (HPAI) in the UK, and Europe. Department for Environment, Food and Rural Affairs; Animal & Plant Health Agency Advice Services Team—International Disease Monitoring. London, UK. 2021. Available online: https://webarchive.nationalarchives.gov.uk/ukgwa/20210802175516/https://www.gov.uk/government/publications/avian-influenza-bird-flu-in-europe (accessed on 17 July 2023).
- Puranik, A.; Slomka, M.J.; Warren, C.J.; Thomas, S.S.; Mahmood, S.; Byrne, A.M.P.; Ramsay, A.M.; Skinner, P.; Watson, S.; Everett, H.E.; et al. Transmission dynamics between infected waterfowl and terrestrial poultry: Differences between the transmission and tropism of H5N8 highly pathogenic avian influenza virus (clade 2.3.4.4a) among ducks, chickens and turkeys. Virology 2020, 541, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Seekings, A.H.; Warren, C.J.; Thomas, S.S.; Mahmood, S.; James, J.; Byrne, A.M.P.; Watson, S.; Bianco, C.; Nunez, A.; Brown, I.H.; et al. Highly pathogenic avian influenza virus H5N6 (clade 2.3.4.4b) has a preferable host tropism for waterfowl reflected in its inefficient transmission to terrestrial poultry. Virology 2021, 559, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Slomka, M.J.; Puranik, A.; Mahmood, S.; Thomas, S.S.; Seekings, A.H.; Byrne, A.M.P.; Nunez, A.; Bianco, C.; Mollett, B.C.; Watson, S.; et al. Ducks Are Susceptible to Infection with a Range of Doses of H5N8 Highly Pathogenic Avian Influenza Virus (2016, Clade 2.3.4.4b) and Are Largely Resistant to Virus-Specific Mortality, but Efficiently Transmit Infection to Contact Turkeys. Avian Dis. 2019, 63, 172–180. [Google Scholar] [CrossRef] [PubMed]
- WOAH. Avian Influenza (Including Infection with High Pathogenicity Avian Influenza Viruses): Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, Chapter 3.3.4. 2021. Available online: https://www.woah.org/fileadmin/Home/eng/Health_standards/tahm/3.03.04_AI.pdf (accessed on 17 July 2023).
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Home Office. Guidance on the Operation of the Animals (Scientific Procedures) Act 1986. March 2020 update. Available online: https://www.gov.uk/guidance/guidance-on-the-operation-of-the-animals-scientific-procedures-act-1986 (accessed on 17 July 2023).
- Slomka, M.J.; Seekings, A.H.; Mahmood, S.; Thomas, S.; Puranik, A.; Watson, S.; Byrne, A.M.P.; Hicks, D.; Nunez, A.; Brown, I.H.; et al. Unexpected infection outcomes of China-origin H7N9 low pathogenicity avian influenza virus in turkeys. Sci. Rep. 2018, 8, 7322. [Google Scholar] [CrossRef]
- Slomka, M.J.; Pavlidis, T.; Coward, V.J.; Voermans, J.; Koch, G.; Hanna, A.; Banks, J.; Brown, I.H. Validated RealTime reverse transcriptase PCR methods for the diagnosis and pathotyping of Eurasian H7 avian influenza viruses. Influenza Other Respir. Viruses 2009, 3, 151–164. [Google Scholar] [CrossRef]
- Nagy, A.; Vostinakova, V.; Pirchanova, Z.; Cernikova, L.; Dirbakova, Z.; Mojzis, M.; Jirincova, H.; Havlickova, M.; Dan, A.; Ursu, K.; et al. Development and evaluation of a one-step real-time RT-PCR assay for universal detection of influenza A viruses from avian and mammal species. Arch. Virol. 2010, 155, 665–673. [Google Scholar] [CrossRef]
- Londt, B.Z.; Nunez, A.; Banks, J.; Nili, H.; Johnson, L.K.; Alexander, D.J. Pathogenesis of highly pathogenic avian influenza A/turkey/Turkey/1/2005 H5N1 in Pekin ducks (Anas platyrhynchos) infected experimentally. Avian Pathol. 2008, 37, 619–627. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- James, J.; Billington, E.; Warren, C.J.; Sliva, D.D.; Genova, C.D.; Airey, M.; Meyer, S.M.; Lewis, T.; Peers-Dent, J.; Thomas, S.S.; et al. Clade 2.3.4.4b H5N1 high pathogenicity avian influenza virus (HPAIV) from the 2021/22 epizootic is highly duck adapted and poorly adapted to chickens. J. Gen. Virol. 2023, 104, 001852. [Google Scholar] [CrossRef] [PubMed]
- Leyson, C.; Youk, S.S.; Smith, D.; Dimitrov, K.; Lee, D.H.; Larsen, L.E.; Swayne, D.E.; Pantin-Jackwood, M.J. Pathogenicity and genomic changes of a 2016 European H5N8 highly pathogenic avian influenza virus (clade 2.3.4.4) in experimentally infected mallards and chickens. Virology 2019, 537, 172–185. [Google Scholar] [CrossRef]
- DeJesus, E.; Costa-Hurtado, M.; Smith, D.; Lee, D.H.; Spackman, E.; Kapczynski, D.R.; Torchetti, M.K.; Killian, M.L.; Suarez, D.L.; Swayne, D.E.; et al. Changes in adaptation of H5N2 highly pathogenic avian influenza H5 clade 2.3.4.4 viruses in chickens and mallards. Virology 2016, 499, 52–64. [Google Scholar] [CrossRef]
- Bertran, K.; Swayne, D.E.; Pantin-Jackwood, M.J.; Kapczynski, D.R.; Spackman, E.; Suarez, D.L. Lack of chicken adaptation of newly emergent Eurasian H5N8 and reassortant H5N2 high pathogenicity avian influenza viruses in the U.S. is consistent with restricted poultry outbreaks in the Pacific flyway during 2014–2015. Virology 2016, 494, 190–197. [Google Scholar] [CrossRef]
- Pohlmann, A.; King, J.; Fusaro, A.; Zecchin, B.; Banyard, A.C.; Brown, I.H.; Byrne, A.M.P.; Beerens, N.; Liang, Y.; Heutink, R.; et al. Has Epizootic Become Enzootic? Evidence for a Fundamental Change in the Infection Dynamics of Highly Pathogenic Avian Influenza in Europe, 2021. mBio 2022, 13, e0060922. [Google Scholar] [CrossRef] [PubMed]
- Caliendo, V.; Lewis, N.S.; Pohlmann, A.; Baillie, S.R.; Banyard, A.C.; Beer, M.; Brown, I.H.; Fouchier, R.A.M.; Hansen, R.D.E.; Lameris, T.K.; et al. Transatlantic spread of highly pathogenic avian influenza H5N1 by wild birds from Europe to North America in 2021. Sci. Rep. 2022, 12, 11729. [Google Scholar] [CrossRef] [PubMed]
- Pan American Health Organization (PAHO). PAHO Issues Alert on Outbreaks of Avian Influenza in Birds in Ten Countries of the Americas (17 January 2023). Available online: https://www.paho.org/en/news/17-1-2023-paho-issues-alert-outbreaks-avian-influenza-birds-ten-countries-americas (accessed on 17 July 2023).
- EFSA (European Food Safety Authority); ECDC (European Centre for Disease Prevention and Control); EURL (European Reference Laboratory for Avian Influenza); Adlhoch, C.; Fusaro, A.; Gonzales, J.L.; Kuiken, T.M.S.; Niqueux, É.; Staubach, C.; Terregino, C.; et al. Scientific report: Avian influenza overview December 2020–February 2021. EFSA J. 2021, 19, e06497. [Google Scholar] [CrossRef]
- Byrne, A.M.P.; James, J.; Mollett, B.C.; Meyer, S.M.; Lewis, T.; Czepiel, M.; Seekings, A.H.; Mahmood, S.; Thomas, S.S.; Ross, C.S.; et al. Investigating the Genetic Diversity of H5 Avian Influenza Viruses in the United Kingdom from 2020–2022. Microbiol. Spectr. 2023, 11, e04776-22. [Google Scholar] [CrossRef]
- Freath, L.; Pacey, T.; Gale, P.; Perrin, L. Updated Outbreak Assessment #10, 17 January 2022. Highly Pathogenic Avian Influenza (HPAI) in the UK and Europe. 2022. Available online: https://www.gov.uk/government/publications/avian-influenza-bird-flu-in-europe (accessed on 29 August 2023).
- Slomka, M.J.; Reid, S.M.; Byrne, A.M.P.; Coward, V.J.; Seekings, J.; Cooper, J.L.; Peers-Dent, J.; Agyeman-Dua, E.; de Silva, D.; Hansen, R.D.E.; et al. Efficient and Informative Laboratory Testing for Rapid Confirmation of H5N1 (Clade 2.3.4.4) High-Pathogenicity Avian Influenza Outbreaks in the United Kingdom. Viruses 2023, 15, 1344. [Google Scholar] [CrossRef]
- Liang, Y.; Hjulsager, C.K.; Seekings, A.H.; Warren, C.J.; Lean, F.Z.X.; Nunez, A.; James, J.; Thomas, S.S.; Banyard, A.C.; Slomka, M.J.; et al. Pathogenesis and infection dynamics of high pathogenicity avian influenza virus (HPAIV) H5N6 (clade 2.3.4.4b) in pheasants and onward transmission to chickens. Virology 2022, 577, 138–148. [Google Scholar] [CrossRef]
- Palya, V.; Tatar-Kis, T.; Walkone Kovacs, E.; Kiss, I.; Homonnay, Z.; Gardin, Y.; Kertesz, K.; Dan, A. Efficacy of a Recombinant Turkey Herpesvirus AI (H5) Vaccine in Preventing Transmission of Heterologous Highly Pathogenic H5N8 Clade 2.3.4.4b Challenge Virus in Commercial Broilers and Layer Pullets. J. Immunol. Res. 2018, 2018, 3143189. [Google Scholar] [CrossRef] [PubMed]
- James, J.; Warren, C.J.; De Silva, D.; Lewis, T.; Grace, K.; Reid, S.M.; Falchieri, M.; Brown, I.H.; Banyard, A.C. The Role of Airborne Particles in the Epidemiology of Clade 2.3.4.4b H5N1 High Pathogenicity Avian Influenza Virus in Commercial Poultry Production Units. Viruses 2023, 15, 1002. [Google Scholar] [CrossRef] [PubMed]
- Parker, C.D.; Irvine, R.M.; Slomka, M.J.; Pavlidis, T.; Hesterberg, U.; Essen, S.; Cox, B.; Ceeraz, V.; Alexander, D.J.; Manvell, R.; et al. Outbreak of Eurasian lineage H5N1 highly pathogenic avian influenza in turkeys in Great Britain in November 2007. Vet. Rec. 2014, 175, 282. [Google Scholar] [CrossRef]
- Grund, C.; Hoffmann, D.; Ulrich, R.; Naguib, M.; Schinkothe, J.; Hoffmann, B.; Harder, T.; Saenger, S.; Zscheppang, K.; Tonnies, M.; et al. A novel European H5N8 influenza A virus has increased virulence in ducks but low zoonotic potential. Emerg. Microbes Infect. 2018, 7, 132. [Google Scholar] [CrossRef] [PubMed]
- Swayne, D.E.; Halvorson, D.A. Influenza. In Diseases of Poultry, 12th ed.; Saif, Y.M., Fadly, A.M., Glisson, J.R., McDougald, L.R., Nolan, L.K., Eds.; Blackwell Publishing: Ames, IA, USA, 2008; pp. 153–184. [Google Scholar]
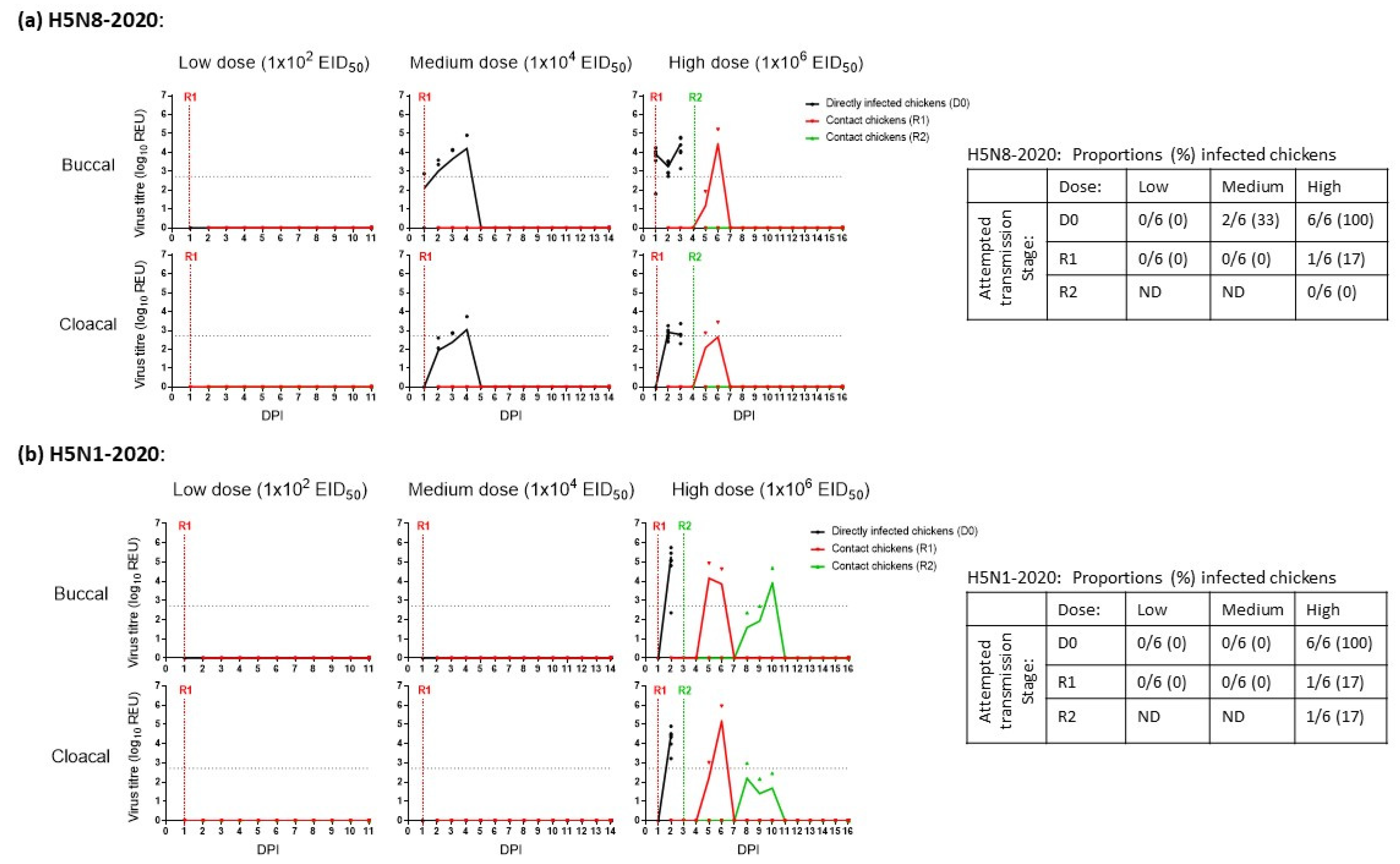
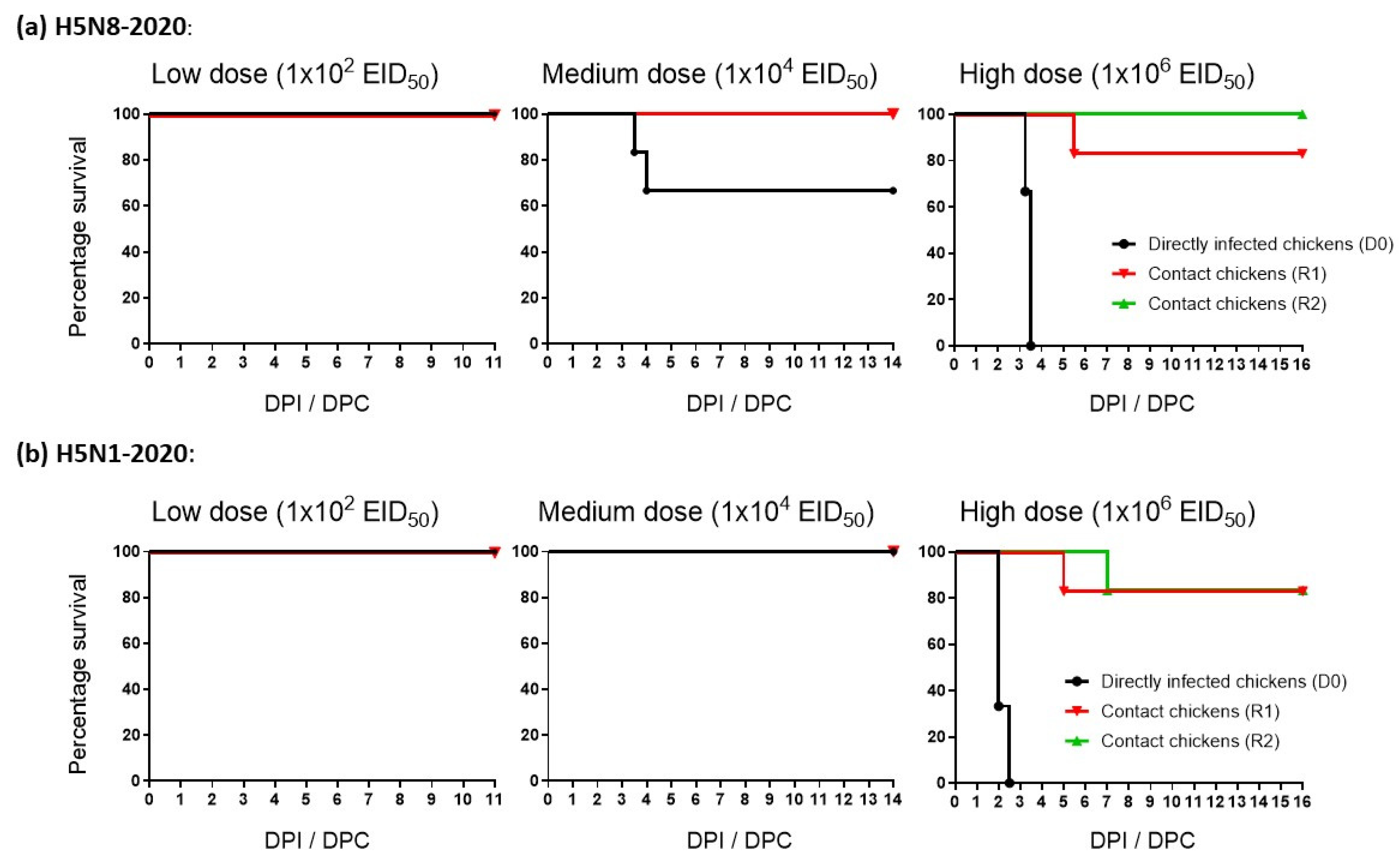
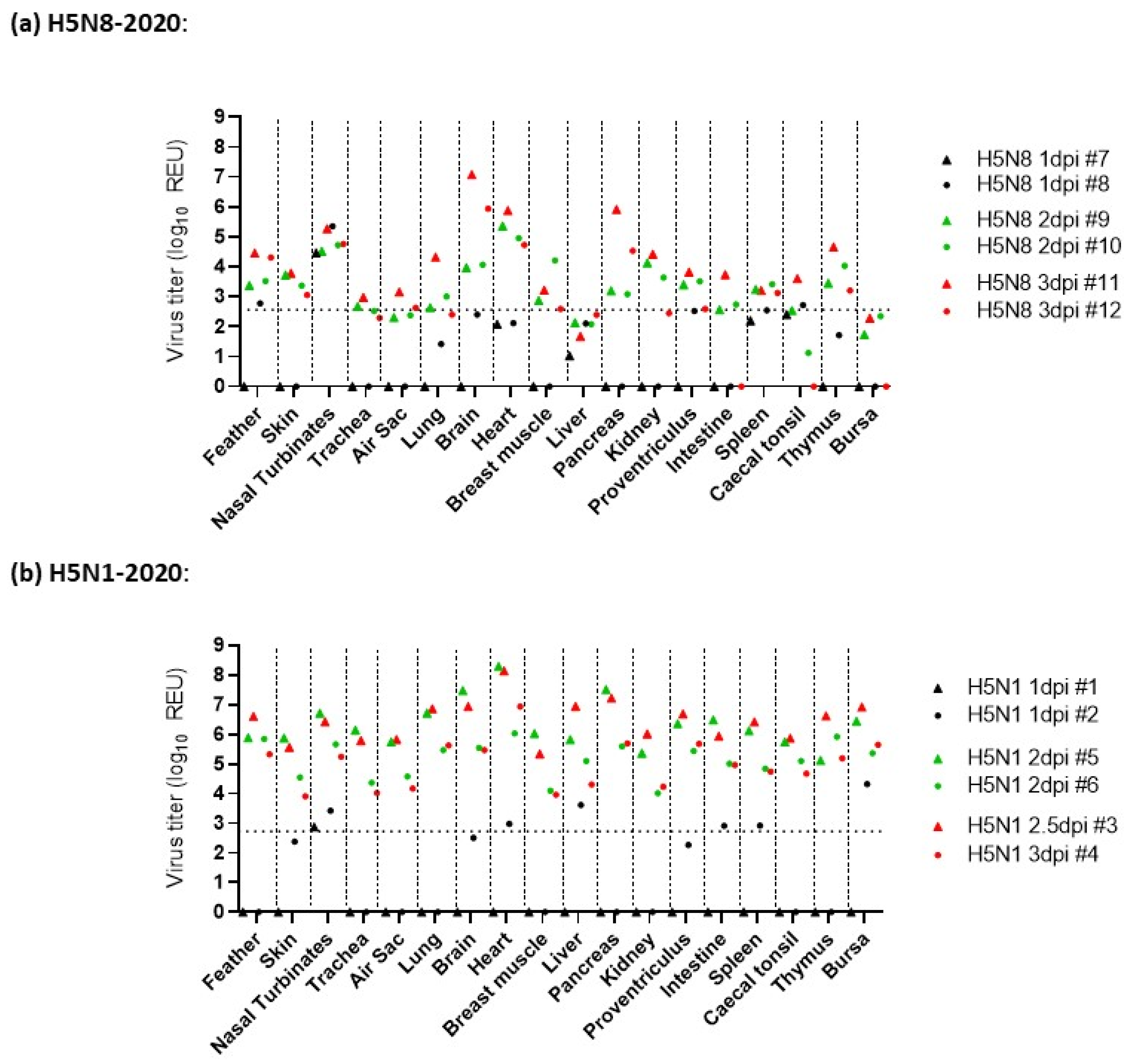
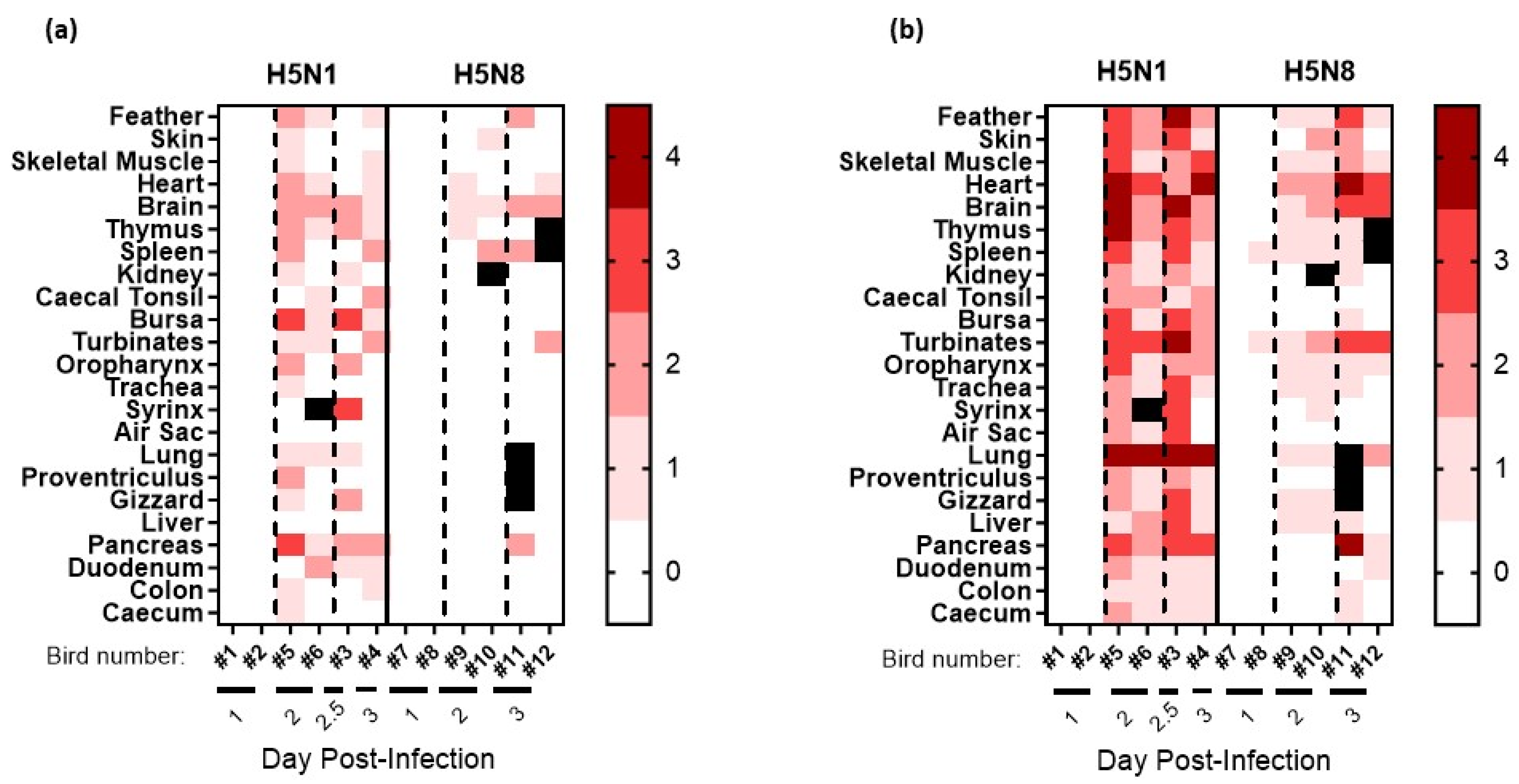

| H5Nx Clade 2.3.4.4 HPAIV | Infectivity: (MID50) 1 | Reference | ||
|---|---|---|---|---|
| Subtype-Year | Isolate Name | Layer Chickens | Ducks 4 | |
| H5N8-2020 | A/chicken/England/030786/2020 | 4.3 2 | ND | This manuscript |
| H5N1-2020 | A/mute swan/England/SA14-234255/2020 | 5.0 2 | ND | |
| H5N1-2021 | A/chicken/England/053052/2021 | 4.7 2 | <3.0 | [25] |
| H5N6-2017 | A/mute swan/England/AVP18-001986/2017 | ND | 3.0 | [14] |
| H5N8-2016 | A/tufted-duck/Denmark/11470/ LWPL/2016 | 5.0 3 | 3.0 5 | [26] |
| H5N2-2015 | A/turkey/Arkansas/7791/2015 | 5.1 3 | ND | [27] |
| H5N2-2015 | A/turkey/Minnesota/12582/2015 | 3.6 3 | <2.0 5 | |
| H5N2-2015 | A/turkey/South Dakota/12511/2015 | 3.2 3 | ND | |
| H5N2-2015 | A/chicken/Iowa/13388/2015 | 3.5 3 | ND | |
| H5N2-2014 | A/Northern pintail/Washington/ 40964/2014 | 5.7 3 | <2.0 5 | Chickens: [28] Ducks: [27] |
| H5N8-2014 | A/gyrfalcon/Washington/40188–6/2014 | 4.4 3 | <2.0 5 | |
| H5N8-2014 | A/duck/England/1279/2014 | ND | <4.0 | [13] |
| Chicken Group (Transmission Stage) and Individual Identifier | Clinical Specimen | Ct Value | Genetic Polymorphisms | |
|---|---|---|---|---|
| D0 medium dose | #71 | brain | 19.23 | PB2: M/V 202V PB1: E/G 178G NS1: D/G 209D |
| #72 | brain | 19.55 | PB2: M/V 202M PB1: E/G 178E NS1: D/G 209D | |
| D0 high dose | #73 | brain | 20.19 | PB1: E/G 178G |
| #77 | brain | 20.18 | No changes | |
| R1 high dose | #93 | brain | 21.20 | PB2: M/V 202M PB1: E/G 178G NS1: D/G 209G |
| Chicken Group (Transmission Stage) and Individual Identifier | Clinical Specimen | Ct Value | Genetic Polymorphisms | |
|---|---|---|---|---|
| D0 high dose | #28 | heart | 17.14 | No changes |
| R1 high dose | #45 | heart | 18.52 | No changes |
| brain | 18.01 | |||
| R2 high dose | #53 | pancreas | 23.76 | PB1: E75G NP: A234V |
| brain | 23.62 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seekings, A.H.; Warren, C.J.; Thomas, S.S.; Lean, F.Z.X.; Selden, D.; Mollett, B.C.; van Diemen, P.M.; Banyard, A.C.; Slomka, M.J. Different Outcomes of Chicken Infection with UK-Origin H5N1-2020 and H5N8-2020 High-Pathogenicity Avian Influenza Viruses (Clade 2.3.4.4b). Viruses 2023, 15, 1909. https://doi.org/10.3390/v15091909
Seekings AH, Warren CJ, Thomas SS, Lean FZX, Selden D, Mollett BC, van Diemen PM, Banyard AC, Slomka MJ. Different Outcomes of Chicken Infection with UK-Origin H5N1-2020 and H5N8-2020 High-Pathogenicity Avian Influenza Viruses (Clade 2.3.4.4b). Viruses. 2023; 15(9):1909. https://doi.org/10.3390/v15091909
Chicago/Turabian StyleSeekings, Amanda H., Caroline J. Warren, Saumya S. Thomas, Fabian Z. X. Lean, David Selden, Benjamin C. Mollett, Pauline M. van Diemen, Ashley C. Banyard, and Marek J. Slomka. 2023. "Different Outcomes of Chicken Infection with UK-Origin H5N1-2020 and H5N8-2020 High-Pathogenicity Avian Influenza Viruses (Clade 2.3.4.4b)" Viruses 15, no. 9: 1909. https://doi.org/10.3390/v15091909
APA StyleSeekings, A. H., Warren, C. J., Thomas, S. S., Lean, F. Z. X., Selden, D., Mollett, B. C., van Diemen, P. M., Banyard, A. C., & Slomka, M. J. (2023). Different Outcomes of Chicken Infection with UK-Origin H5N1-2020 and H5N8-2020 High-Pathogenicity Avian Influenza Viruses (Clade 2.3.4.4b). Viruses, 15(9), 1909. https://doi.org/10.3390/v15091909







