The DACH1 Gene Transcriptional Activation and Protein Degradation Mediated by Transactivator Tas of Prototype Foamy Virus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture, Plasmids, and Transfection
2.2. PFV Collection, Quantification, and Infection
2.3. Quantitative PCR
2.4. Dual-Luciferase Reporter Assay
2.5. Electrophoretic Mobility Gel Shift Assay (EMSA)
2.6. Chromatin Immunoprecipitation Assay
2.7. RNA Interference
2.8. SUMOylation Site-Directed Mutation Assays
2.9. Co-Immunoprecipitation
2.10. Western Blot Analysis
2.11. Statistical Analysis
3. Results
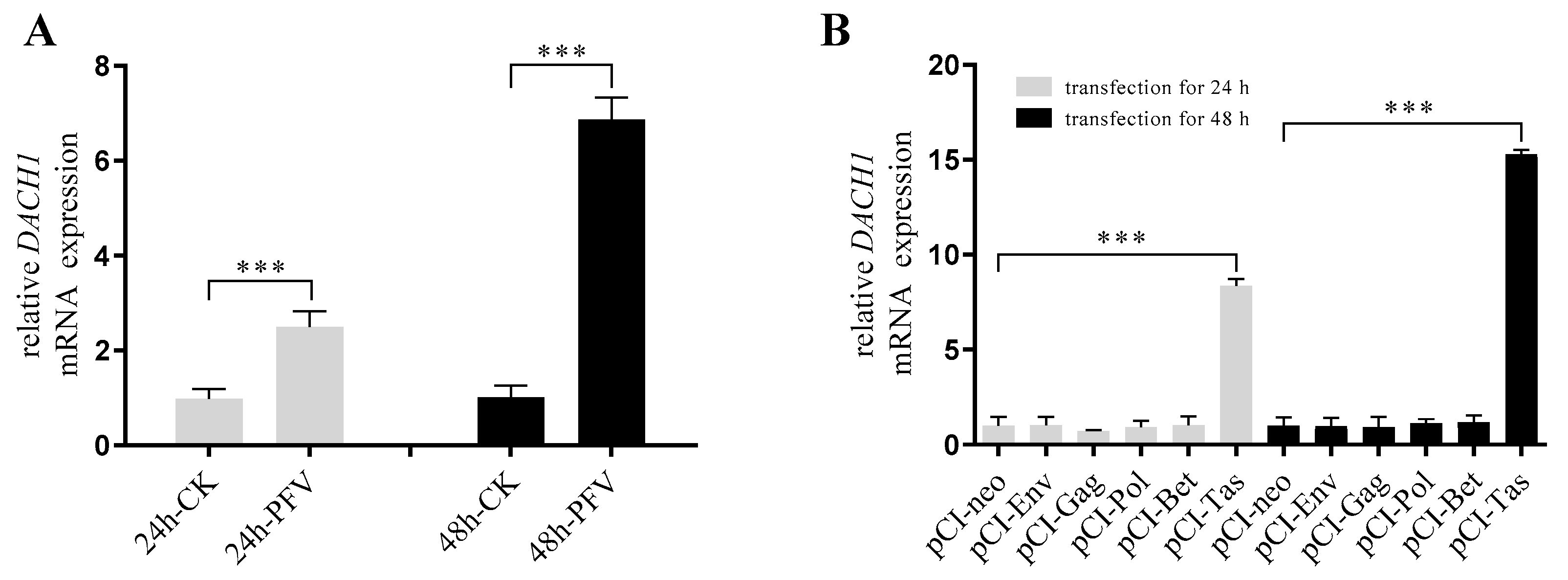
3.1. PFV and Tas Can Activate the Expression of DACH1 Transcription Level
3.2. Tas Directly Regulates DACH1 Gene Transcription through a Binding Site in the DACH1 Promoter Region
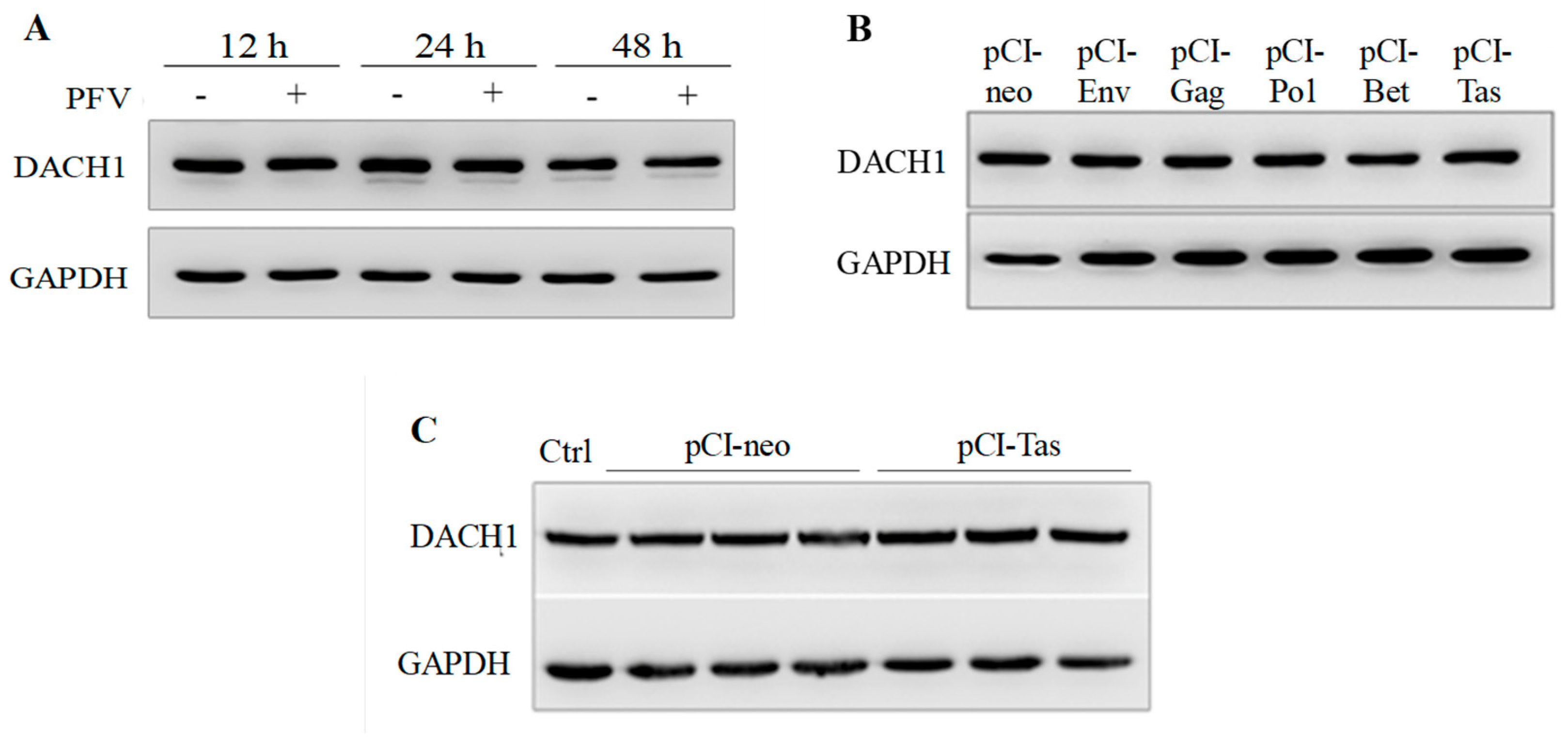
3.3. The Protein Expression of DACH1 Was Not Influenced by PFV or Tas
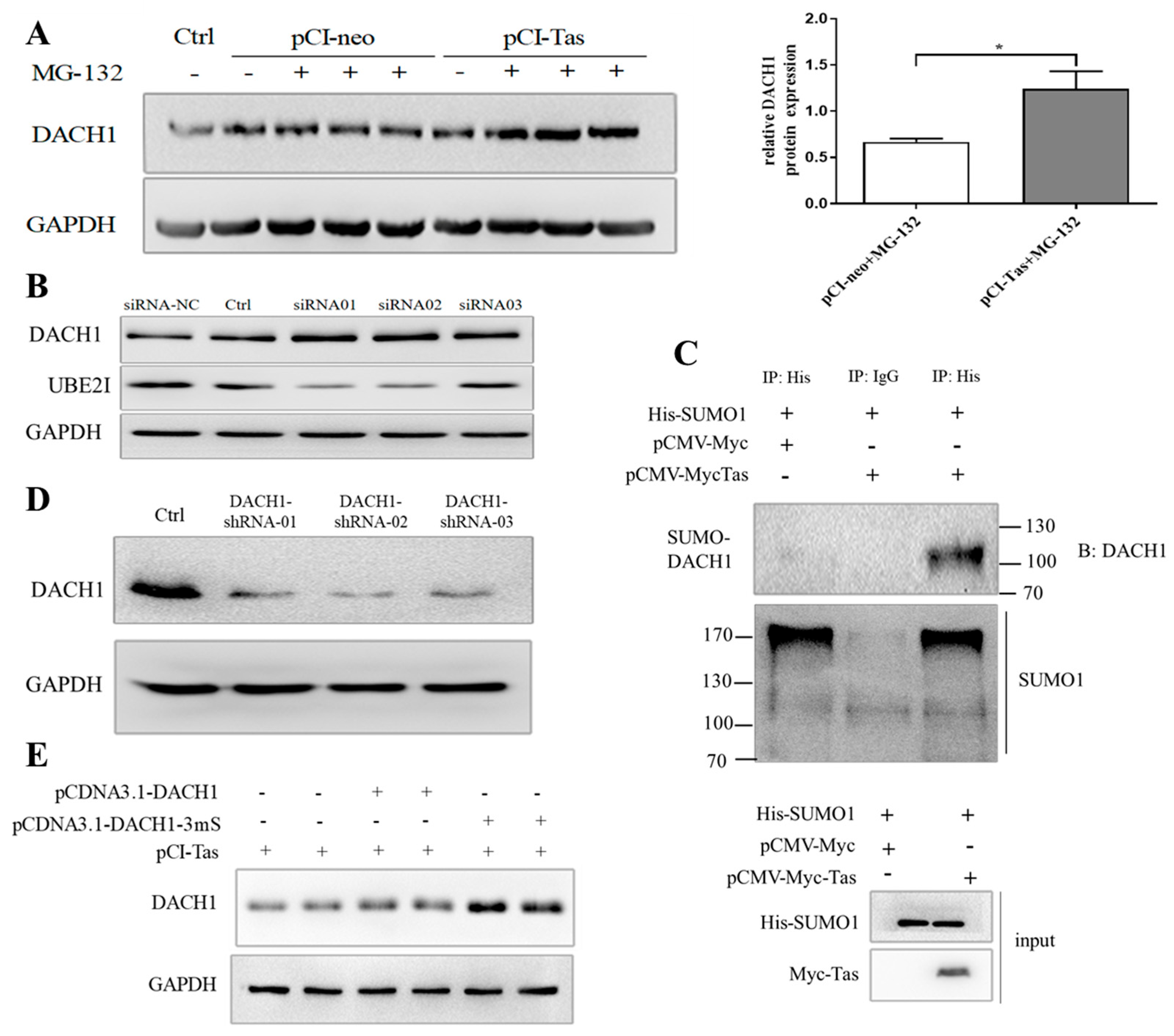
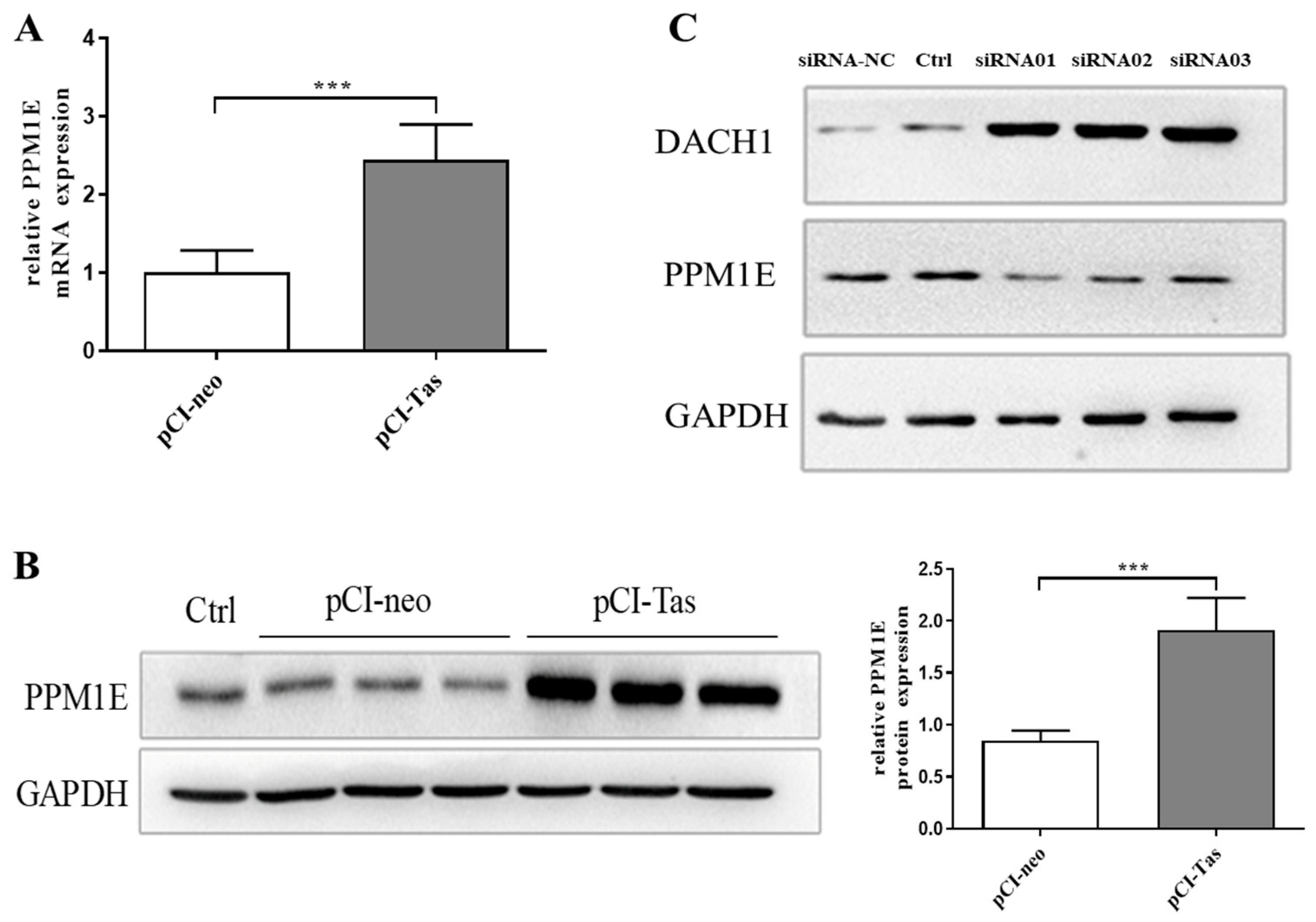
3.4. Tas Promoted DACH1 in SUMO-Mediated Protease Degradation Depending on PPM1E
3.5. DACH1 Overexpression Inhibits PFV Replication
4. Discussion
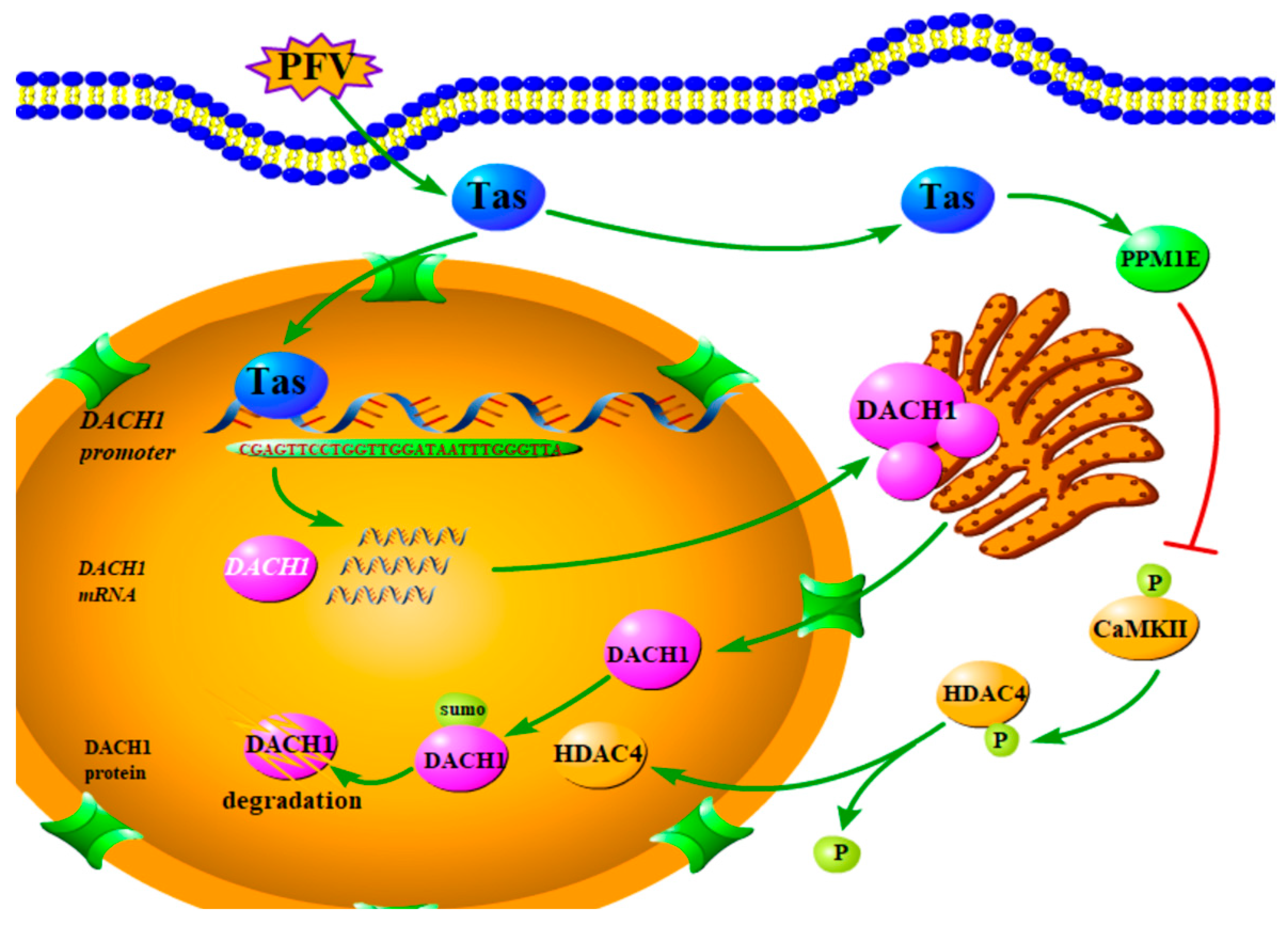
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rethwilm, A.; Baunach, G.; Netzer, K.O.; Maurer, B.; Borisch, B.; ter Meulen, V. Infectious DNA of the human spumaretrovirus. Nucleic Acids Res. 1990, 18, 733–738. [Google Scholar] [CrossRef] [PubMed]
- Kane, M.; Mele, V.; Liberatore, R.A.; Bieniasz, P.D. Inhibition of spumavirus gene expression by PHF11. PLoS Pathog. 2020, 16, e1008644. [Google Scholar] [CrossRef] [PubMed]
- Saïb, A. Non-primate foamy viruses. Curr. Top. Microbiol. Immunol. 2003, 277, 197–211. [Google Scholar] [PubMed]
- Xu, S.; Yang, W.; Yuan, P.; Yan, J.; Tang, Y.; Zheng, Y.; Li, Z.; Sun, Y.; Han, S.; Yin, J.; et al. The Long-Noncoding RNA lnc-NONH Enhances the Early Transcription of Prototype Foamy Virus Via Upregulating Expression of miR-34c-5p and Tas Protein. Intervirology 2019, 62, 156–163. [Google Scholar] [CrossRef]
- Kang, S.Y.; Ahn, D.G.; Lee, C.; Lee, Y.S.; Shin, C.G. Functional nucleotides of U5 LTR determining substrate specificity of prototype foamy virus integrase. J. Microbiol. Biotechnol. 2008, 18, 1044–1049. [Google Scholar]
- Wei, J.; Sun, Y.; Wang, T.T.; Zhu, G.; Liu, W.H.; He, X.H.; Li, Z. The Regulation of Prototype Foamy Virus 5’Long Terminal Repeats and Internal Promoter by Endogenous Transcription Factors. Intervirology 2022, 65, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Wang, S.; Du, T.; Liu, L.; Chen, X.; Yan, J.; Han, S.; Peng, B.; He, X.; Liu, W. ZNF219, a novel transcriptional repressor, inhibits transcription of the prototype foamy virus by interacting with the viral LTR promoter. Virus Res. 2023, 334, 199161. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Chen, L.; Tang, Y.; Yuan, P.; Yan, J.; Zheng, Y.; Huang, L.; Li, Z.; Sun, Y.; Han, S.; et al. Lnc-RP5 Regulates the miR-129-5p/Notch1/PFV Internal Promoter Axis to Promote the Expression of the Prototype Foamy Virus Transactivator Tas. Virol. Sin. 2020, 35, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, C.; Tuo, X.; Chai, K.; Xu, Y.; Qiao, W.; Tan, J. Prototype foamy virus downregulates RelB expression to facilitate viral replication. FEBS Open Bio 2020, 10, 2137–2148. [Google Scholar] [CrossRef]
- Pinto-Santini, D.M.; Stenbak, C.R.; Linial, M.L. Foamy virus zoonotic infections. Retrovirology 2017, 14, 55. [Google Scholar] [CrossRef]
- Bodem, J. Regulation of foamy viral transcription and RNA export. Adv. Virus Res. 2011, 81, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Bodem, J.; Krausslich, H.G.; Rethwilm, A. Acetylation of the foamy virus transactivator Tas by PCAF augments promoter-binding affinity and virus transcription. J. Gen. Virol. 2007, 88, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Regad, T.; Saib, A.; Lallemand-Breitenbach, V.; Pandolfi, P.P.; de Thé, H.; Chelbi-Alix, M.K. PML mediates the interferon-induced antiviral state against a complex retrovirus via its association with the viral transactivator. EMBO J. 2001, 20, 3495–3505. [Google Scholar] [CrossRef]
- Hu, X.; Yang, W.; Liu, R.; Geng, Y.; Qiao, W.; Tan, J. N-Myc interactor inhibits prototype foamy virus by sequestering viral Tas protein in the cytoplasm. J. Virol. 2014, 88, 7036–7044. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Zhang, R.G.; Braunstein, S.E.; Joachimiak, A.; Cvekl, A.; Hegde, R.S. Structure of the retinal determination protein Dachshund reveals a DNA binding motif. Structure 2002, 10, 787–795. [Google Scholar] [CrossRef]
- Popov, V.M.; Wu, K.; Zhou, J.; Powell, M.J.; Mardon, G.; Wang, C.; Pestell, R.G. The Dachshund gene in development and hormone-responsive tumorigenesis. Trends Endocrinol. Metab. 2010, 21, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Han, N.; Zhou, S.; Zhou, R.; Yuan, X.; Xu, H.; Zhang, C.; Yin, T.; Wu, K. The DACH/EYA/SIX gene network and its role in tumor initiation and progression. Int. J. Cancer 2016, 138, 1067–1075. [Google Scholar] [CrossRef]
- Cheng, Q.; Ning, D.; Chen, J.; Li, X.; Chen, X.P.; Jiang, L. SIX1 and DACH1 influence the proliferation and apoptosis of hepatocellular carcinoma through regulating p53. Cancer Biol. Ther. 2018, 19, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Aman, S.; Li, Y.; Cheng, Y.; Yang, Y.; Lv, L.; Li, B.; Xia, K.; Li, S.; Wu, H. DACH1 inhibits breast cancer cell invasion and metastasis by down-regulating the transcription of matrix metalloproteinase 9. Cell Death Discov. 2021, 7, 351. [Google Scholar] [CrossRef]
- Han, N.; Yuan, X.; Wu, H.; Xu, H.; Chu, Q.; Guo, M.; Yu, S.; Chen, Y.; Wu, K. DACH1 inhibits lung adenocarcinoma invasion and tumor growth by repressing CXCL5 signaling. Oncotarget 2015, 6, 5877–5888. [Google Scholar] [CrossRef] [PubMed]
- Chu, Q.; Han, N.; Yuan, X.; Nie, X.; Wu, H.; Chen, Y.; Guo, M.; Yu, S.; Wu, K. DACH1 inhibits cyclin D1 expression, cellular proliferation and tumor growth of renal cancer cells. J. Hematol. Oncol. 2014, 7, 73. [Google Scholar] [CrossRef]
- Nan, F.; Lu, Q.; Zhou, J.; Cheng, L.; Popov, V.M.; Wei, S.; Kong, B.; Pestell, R.G.; Lisanti, M.P.; Jiang, J.; et al. Altered expression of DACH1 and cyclin D1 in endometrial cancer. Cancer Biol. Ther. 2009, 8, 1534–1539. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Wu, K.; Herman, J.G.; Brock, M.V.; Fuks, F.; Yang, L.; Zhu, H.; Li, Y.; Yang, Y.; Guo, M. Epigenetic regulation of DACH1, a novel Wnt signaling component in colorectal cancer. Epigenetics 2013, 8, 1373–1383. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wu, K.; Yan, W.; Hu, L.; Yuan, J.; Dong, Y.; Li, Y.; Jing, K.; Yang, Y.; Guo, M. Epigenetic silencing of DACH1 induces loss of transforming growth factor-beta1 antiproliferative response in human hepatocellular carcinoma. Hepatology 2013, 58, 2012–2022. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Wu, K.; Cai, S.; Zhang, W.; Zhou, J.; Wang, J.; Ertel, A.; Li, Z.; Rui, H.; Quong, A.; et al. Dachshund binds p53 to block the growth of lung adenocarcinoma cells. Cancer Res. 2013, 73, 3262–3274. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Yang, Y.; Wang, C.; Davoli, M.A.; D’Amico, M.; Li, A.; Cveklova, K.; Kozmik, Z.; Lisanti, M.P.; Russell, R.G.; et al. DACH1 inhibits transforming growth factor-beta signaling through binding Smad4. J. Biol. Chem. 2003, 278, 51673–51684. [Google Scholar] [CrossRef]
- Wu, L.; Herman, J.G.; Brock, M.V.; Wu, K.; Mao, G.; Yan, W.; Nie, Y.; Liang, H.; Zhan, Q.; Li, W.; et al. Silencing DACH1 promotes esophageal cancer growth by inhibiting TGF-beta signaling. PLoS ONE 2014, 9, e95509. [Google Scholar] [CrossRef]
- Lu, Y.; Tang, K.; Wang, S.; Tian, Z.; Fan, Y.; Li, B.; Wang, M.; Zhao, J.; Xie, J. Dach1 deficiency drives alveolar epithelium apoptosis in pulmonary fibrosis via modulating C-Jun/Bim activity. Transl. Res. 2023, 257, 54–65. [Google Scholar] [CrossRef]
- Li, Z.; Yang, P.; Liu, H.; Li, W.X. A new indicator cell line to monitor human foamy virus infection and stability in vitro. Intervirology 2002, 45, 79–84. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Lee, J.W.; Kim, H.S.; Kim, S.; Hwang, J.; Kim, Y.H.; Lim, G.Y.; Sohn, W.J.; Yoon, S.R.; Kim, J.Y.; Park, T.S.; et al. DACH1 regulates cell cycle progression of myeloid cells through the control of cyclin D, Cdk 4/6 and p21Cip1. Biochem. Biophys. Res. Commun. 2012, 420, 91–95. [Google Scholar] [CrossRef]
- Campbell, M.; Renshaw-Gegg, L.; Renne, R.; Luciw, P.A. Characterization of the internal promoter of simian foamy viruses. J. Virol. 1994, 68, 4811–4820. [Google Scholar] [CrossRef] [PubMed]
- Mergia, A.; Pratt-Lowe, E.; Shaw, K.E.; Renshaw-Gegg, L.W.; Luciw, P.A. cis-acting regulatory regions in the long terminal repeat of simian foamy virus type 1. J. Virol. 1992, 66, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Kido, K.; Doerks, A.; Lochelt, M.; Flugel, R.M. Identification and functional characterization of an intragenic DNA binding site for the spumaretroviral trans-activator in the human p57Kip2 gene. J. Biol. Chem. 2002, 277, 12032–12039. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Blair, W.S.; Cullen, B.R. Identification and functional characterization of a high-affinity Bel-1 DNA binding site located in the human foamy virus internal promoter. J. Virol. 1998, 72, 504–511. [Google Scholar] [CrossRef]
- Liu, Q.Q.; Zhou, Y.Q.; Liu, H.Q.; Qiu, W.H.; Liu, H.; Hu, T.Y.; Xu, Q.; Lv, Y.M.; Wu, K.M. Decreased DACH1 expression in glomerulopathy is associated with disease progression and severity. Oncotarget 2016, 7, 86547–86560. [Google Scholar] [CrossRef]
- Ozcan, L.; Ghorpade, D.S.; Zheng, Z.; de Souza, J.C.; Chen, K.; Bessler, M.; Bagloo, M.; Schrope, B.; Pestell, R.; Tabas, I. Hepatocyte DACH1 Is Increased in Obesity via Nuclear Exclusion of HDAC4 and Promotes Hepatic Insulin Resistance. Cell Rep. 2016, 15, 2214–2225. [Google Scholar] [CrossRef]
- Ishida, A.; Sueyoshi, N.; Kameshita, I. Functions and dysfunctions of Ca(2+)/calmodulin-dependent protein kinase phosphatase (CaMKP/PPM1F) and CaMKP-N/PPM1E. Arch. Biochem. Biophys. 2018, 640, 83–92. [Google Scholar] [CrossRef]
- Kitani, T.; Okuno, S.; Nakamura, Y.; Tokuno, H.; Takeuchi, M.; Fujisawa, H. Post-translational excision of the carboxyl-terminal segment of CaM kinase phosphatase N and its cytosolic occurrence in the brain. J. Neurochem. 2006, 96, 374–384. [Google Scholar] [CrossRef]
- Onouchi, T.; Sueyoshi, N.; Ishida, A.; Kameshita, I. Phosphorylation and activation of nuclear Ca2+/calmodulin-dependent protein kinase phosphatase (CaMKP-N/PPM1E) by Ca2+/calmodulin-dependent protein kinase I (CaMKI). Biochem. Biophys. Res. Commun. 2012, 422, 703–709. [Google Scholar] [CrossRef]
- Zhang, J.; Ren, X.; Wang, B.; Cao, J.; Tian, L.; Liu, M. Effect of DACH1 on proliferation and invasion of laryngeal squamous cell carcinoma. Head Face Med. 2018, 14, 20. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Cheng, Q.; Wang, Z.; Yuan, P.; Li, Z.; Sun, Y.; Han, S.; Yin, J.; Peng, B.; He, X.; et al. Human Pirh2 is a novel inhibitor of prototype foamy virus replication. Viruses 2015, 7, 1668–1684. [Google Scholar] [CrossRef] [PubMed]
- Meiering, C.D.; Rubio, C.; May, C.; Linial, M.L. Cell-type-specific regulation of the two foamy virus promoters. J. Virol. 2001, 75, 6547–6557. [Google Scholar] [CrossRef] [PubMed]
- Reiss, Y.; Gur, E.; Ravid, T. Releasing the Lockdown: An Emerging Role for the Ubiquitin-Proteasome System in the Breakdown of Transient Protein Inclusions. Biomolecules 2020, 10, 1168. [Google Scholar] [CrossRef]
- Ding, W.X.; Yin, X.M. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy 2008, 4, 141–150. [Google Scholar] [CrossRef]
- Cheng, X.; Xiong, R.; Li, Y.; Li, F.; Zhou, X.; Wang, A. Sumoylation of Turnip mosaic virus RNA Polymerase Promotes Viral Infection by Counteracting the Host NPR1-Mediated Immune Response. Plant Cell 2017, 29, 508–525. [Google Scholar] [CrossRef]
- Kubota, T.; Matsuoka, M.; Chang, T.H.; Tailor, P.; Sasaki, T.; Tashiro, M.; Kato, A.; Ozato, K. Virus infection triggers SUMOylation of IRF3 and IRF7, leading to the negative regulation of type I interferon gene expression. J. Biol. Chem. 2008, 283, 25660–25670. [Google Scholar] [CrossRef]
- Sandler, N.G.; Bosinger, S.E.; Estes, J.D.; Zhu, R.T.; Tharp, G.K.; Boritz, E.; Levin, D.; Wijeyesinghe, S.; Makamdop, K.N.; del Prete, G.Q.; et al. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature 2014, 511, 601–605. [Google Scholar] [CrossRef]
- Tripathi, V.; Chatterjee, K.S.; Das, R. Non-covalent Interaction With SUMO Enhances the Activity of Human Cytomegalovirus Protein IE1. Front. Cell Dev. Biol. 2021, 9, 662522. [Google Scholar] [CrossRef]
- Wilson, V.G. Viral Interplay with the Host Sumoylation System. Adv. Exp. Med. Biol. 2017, 963, 359–388. [Google Scholar] [CrossRef]
- Imbert, F.; Leavitt, G.; Langford, D. SUMOylation and Viral Infections of the Brain. Pathogens 2022, 11, 818. [Google Scholar] [CrossRef] [PubMed]
- Zamborlini, A.; Coiffic, A.; Beauclair, G.; Delelis, O.; Paris, J.; Koh, Y.; Magne, F.; Giron, M.L.; Tobaly-Tapiero, J.; Deprez, E.; et al. Impairment of human immunodeficiency virus type-1 integrase SUMOylation correlates with an early replication defect. J. Biol. Chem. 2011, 286, 21013–21022. [Google Scholar] [CrossRef]
- Gurer, C.; Berthoux, L.; Luban, J. Covalent modification of human immunodeficiency virus type 1 p6 by SUMO-1. J. Virol. 2005, 79, 910–917. [Google Scholar] [CrossRef]
- Jie, W.; Rui-Fen, Z.; Zhong-Xiang, H.; Yan, W.; Wei-Na, L.; Yong-Ping, M.; Jing, S.; Jing-Yi, C.; Wan-Hong, L.; Xiao-Hua, H.; et al. Inhibition of cell proliferation by Tas of foamy viruses through cell cycle arrest or apoptosis underlines the different mechanisms of virus-host interactions. Virulence 2022, 13, 342–354. [Google Scholar] [CrossRef] [PubMed]
- Ko, C.; Lee, S.; Windisch, M.P.; Ryu, W.S. DDX3 DEAD-box RNA helicase is a host factor that restricts hepatitis B virus replication at the transcriptional level. J. Virol. 2014, 88, 13689–13698. [Google Scholar] [CrossRef] [PubMed]
- Zhai, T.; Wu, C.; Wang, N.; Shi, B.; Li, J.; Chen, R.; Dong, J.; Zhang, Y.; Zhou, E.M.; Nan, Y. Development of a monoclonal antibody against swine leukocyte antigen (SLA)-DR alpha chain and evaluation of SLA-DR expression in bone marrow-derived dendritic cells after PRRSV infection. Vet. Immunol. Immunopathol. 2019, 211, 19–24. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, S.; Lu, J.; Yuan, D.; He, L.; Qin, P.; Tan, H.; Xu, L. MicroRNA-363-3p promote the development of acute myeloid leukemia with RUNX1 mutation by targeting SPRYD4 and FNDC3B. Medicine 2021, 100, e25807. [Google Scholar] [CrossRef]
- Lee, J.W.; Kim, H.S.; Hwang, J.; Kim, Y.H.; Lim, G.Y.; Sohn, W.J.; Yoon, S.R.; Kim, J.Y.; Park, T.S.; Oh, S.H.; et al. Regulation of HOXA9 activity by predominant expression of DACH1 against C/EBPalpha and GATA-1 in myeloid leukemia with MLL-AF9. Biochem. Biophys. Res. Commun. 2012, 426, 299–305. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Wei, J.; Song, J.; Hu, Z.; Zhang, R.; Li, Z.; Sun, Y. The DACH1 Gene Transcriptional Activation and Protein Degradation Mediated by Transactivator Tas of Prototype Foamy Virus. Viruses 2023, 15, 1899. https://doi.org/10.3390/v15091899
Ma Y, Wei J, Song J, Hu Z, Zhang R, Li Z, Sun Y. The DACH1 Gene Transcriptional Activation and Protein Degradation Mediated by Transactivator Tas of Prototype Foamy Virus. Viruses. 2023; 15(9):1899. https://doi.org/10.3390/v15091899
Chicago/Turabian StyleMa, Yongping, Jie Wei, Jing Song, Zhongxiang Hu, Ruifen Zhang, Zhi Li, and Yan Sun. 2023. "The DACH1 Gene Transcriptional Activation and Protein Degradation Mediated by Transactivator Tas of Prototype Foamy Virus" Viruses 15, no. 9: 1899. https://doi.org/10.3390/v15091899
APA StyleMa, Y., Wei, J., Song, J., Hu, Z., Zhang, R., Li, Z., & Sun, Y. (2023). The DACH1 Gene Transcriptional Activation and Protein Degradation Mediated by Transactivator Tas of Prototype Foamy Virus. Viruses, 15(9), 1899. https://doi.org/10.3390/v15091899



