Mannose-Binding Lectins as Potent Antivirals against SARS-CoV-2
Abstract
:1. Introduction
2. Materials and Methods
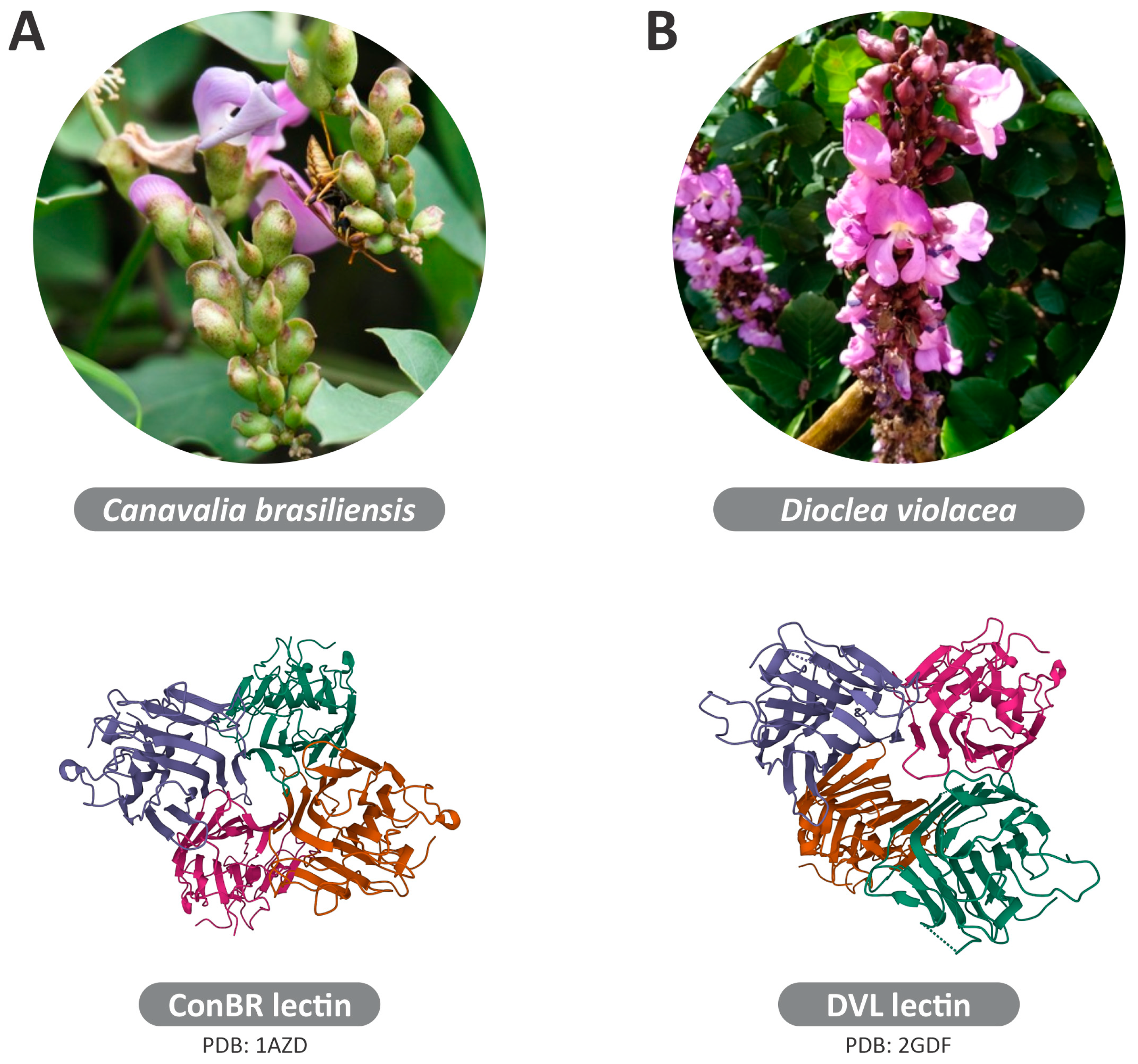
2.1. Extraction and Purification of ConBR and DVL
2.2. Cell Culture
2.3. Cell Viability
2.4. Virus Rescue and Titration
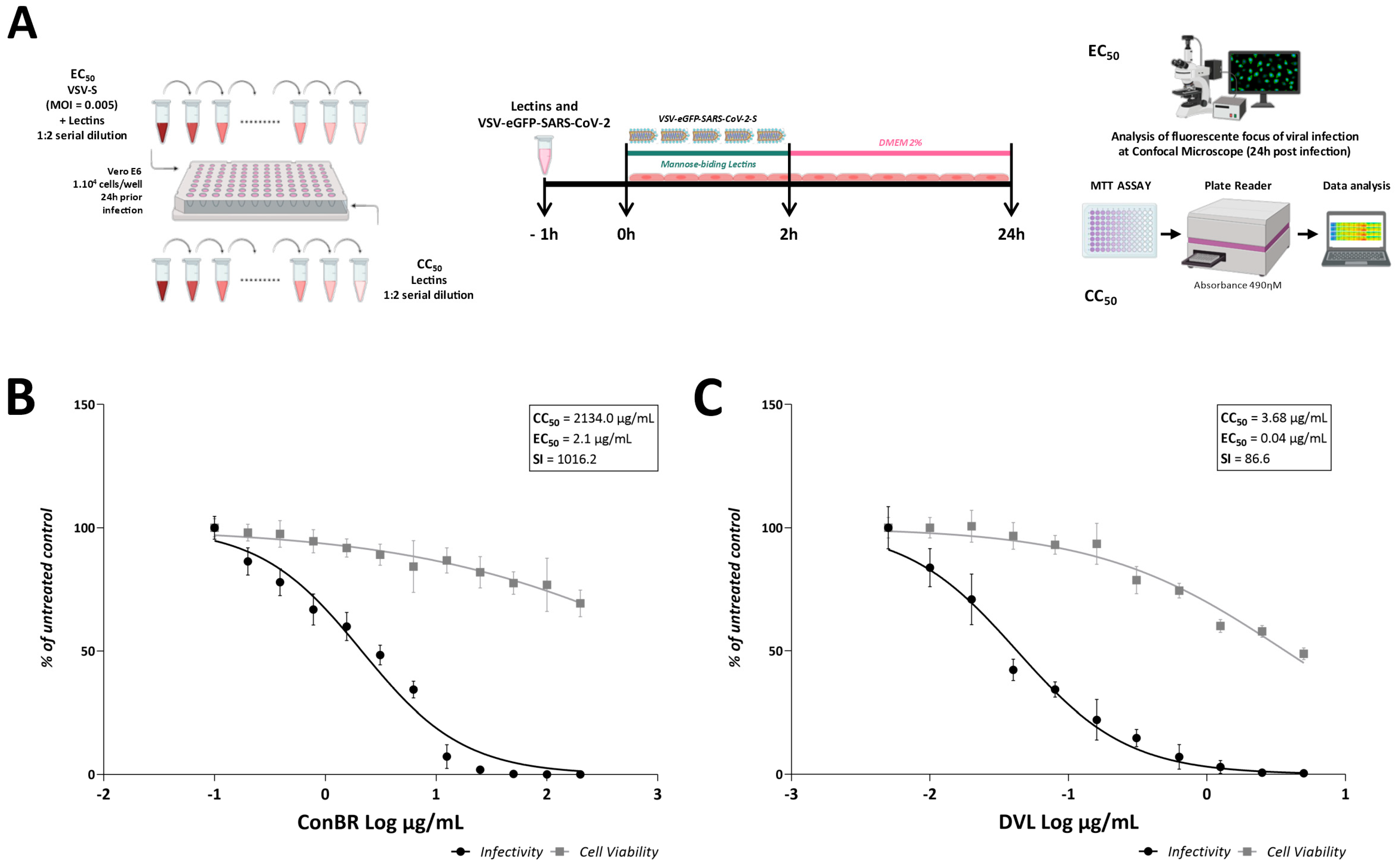
2.5. Antiviral Assays with VSV-SARS-CoV-2 and SARS-CoV-2WT
2.6. Antiviral Assays with SARS-CoV-2WT and Variants Gamma and Omicron Measured by Quantitative PCR
2.7. Mannose-Biding Lectins Blocking Assay
2.8. RNA Extraction and cDNA Synthesis
2.9. Determination of Viral Load by Real-Time PCR
2.10. Protein Structures
2.11. Protein–Protein Docking
2.12. Attenuated Total Reflection (ATR) Coupled to Fourier Transform Infrared (FTIR) Analysis
2.13. Statistical Analysis
3. Results
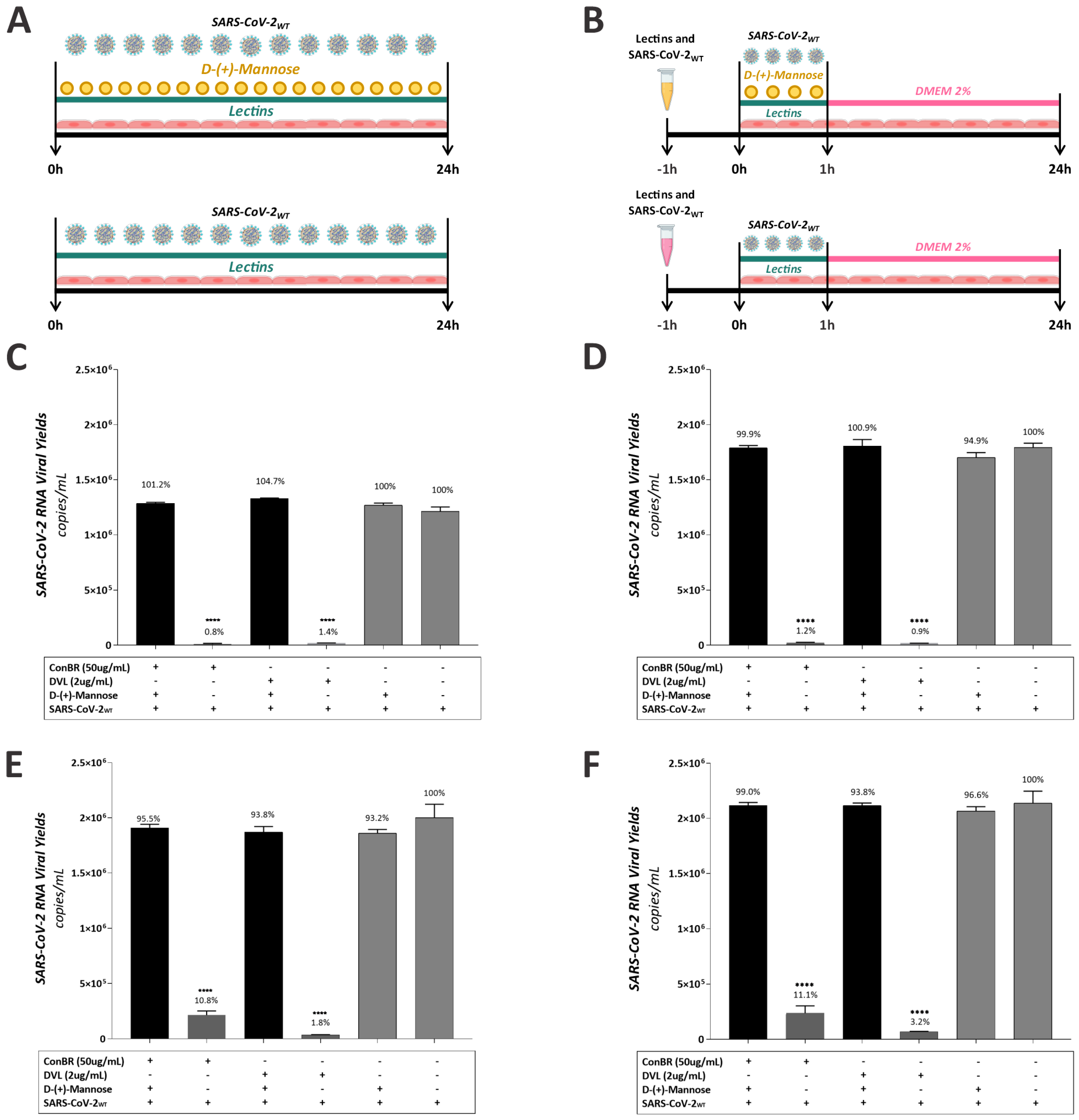
3.1. ConBR and DVL Block SARS-CoV-2 Entry to the Host Cells
3.2. ConBR and DVL Are Potent Inhibitors of SARS-CoV-2WT, but Also Variants Omicron and Gamma
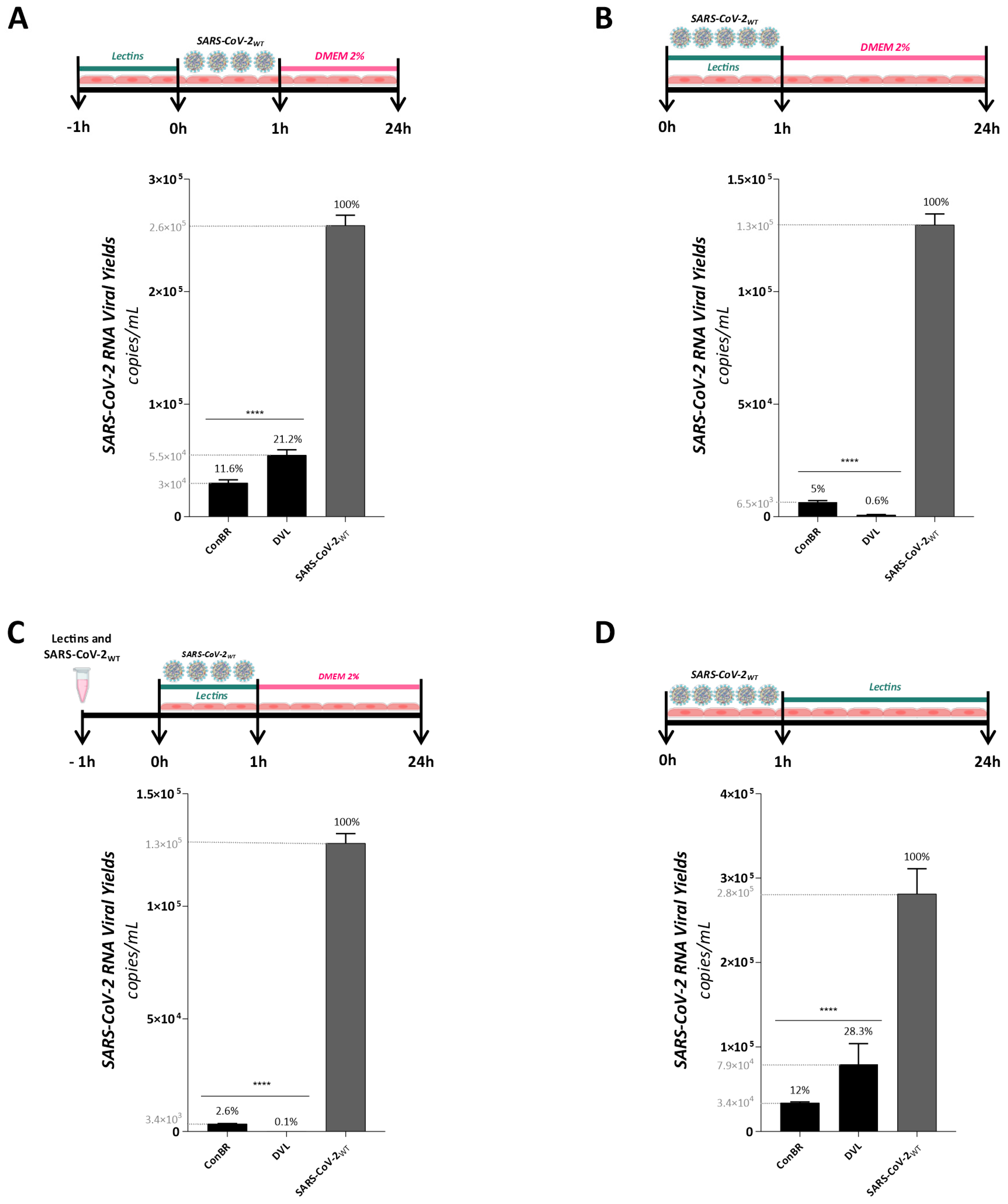
3.3. Multiple Effects of ConBR and DVL on the Replicative Cycle of SARS-CoV-2WT
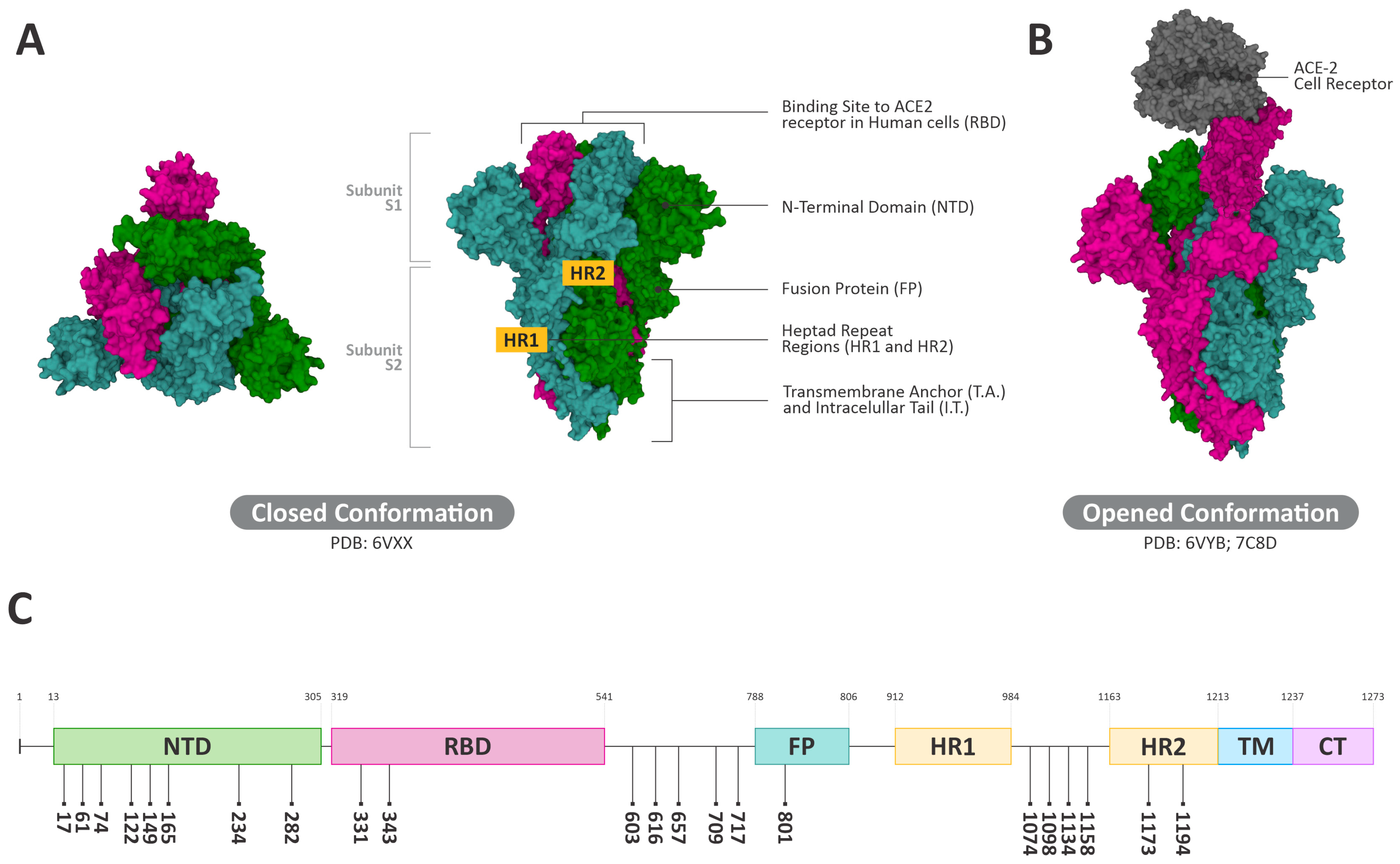
3.4. Interactions of ConBR and DVL with VSV-eGFP-SARS-CoV-2-S
3.5. Insights on the Role of Mannose-Biding Lectins against SARS-CoV-2 Infections
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Cucinotta, D.; Vanelli, M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020, 91, 157–160. [Google Scholar] [CrossRef]
- WHO. WHO Coronavirus Disease (COVID-19) Dashboard; WHO: Geneva, Switzerland, 2023. [Google Scholar]
- Fiolet, T.; Kherabi, Y.; MacDonald, C.-J.; Ghosn, J.; Peiffer-Smadja, N. Comparing COVID-19 Vaccines for Their Characteristics, Efficacy and Effectiveness against SARS-CoV-2 and Variants of Concern: A Narrative Review. Clin. Microbiol. Infect. 2022, 28, 202–221. [Google Scholar] [CrossRef]
- Kirtipal, N.; Bharadwaj, S.; Kang, S.G. From SARS to SARS-CoV-2, Insights on Structure, Pathogenicity and Immunity Aspects of Pandemic Human Coronaviruses. Infect. Genet. Evol. 2020, 85, 104502. [Google Scholar] [CrossRef]
- Aleem, A.; Kothadia, J.P. Remdesivir. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Tian, L.; Pang, Z.; Li, M.; Lou, F.; An, X.; Zhu, S.; Song, L.; Tong, Y.; Fan, H.; Fan, J. Molnupiravir and Its Antiviral Activity Against COVID-19. Front. Immunol. 2022, 13, 855496. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, N.C.; Kim, Y.; Liu, H.; Galasiti Kankanamalage, A.C.; Eckstrand, C.; Groutas, W.C.; Bannasch, M.; Meadows, J.M.; Chang, K.-O. Efficacy of a 3C-like Protease Inhibitor in Treating Various Forms of Acquired Feline Infectious Peritonitis. J. Feline Med. Surg. 2018, 20, 378–392. [Google Scholar] [CrossRef] [PubMed]
- Brayer, S.W.; Reddy, K.R. Ritonavir-Boosted Protease Inhibitor Based Therapy: A New Strategy in Chronic Hepatitis C Therapy. Expert Rev. Gastroenterol. Hepatol. 2015, 9, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Hamdoun, S.; Chen, R.; Yang, L.; Ip, C.K.; Qu, Y.; Li, R.; Jiang, H.; Yang, Z.; Chung, S.K.; et al. Identification of Natural Compounds as SARS-CoV-2 Entry Inhibitors by Molecular Docking-Based Virtual Screening with Bio-Layer Interferometry. Pharmacol. Res. 2021, 172, 105820. [Google Scholar] [CrossRef]
- de Andrade Santos, I.; Grosche, V.R.; Bergamini, F.R.G.; Sabino-Silva, R.; Jardim, A.C.G. Antivirals Against Coronaviruses: Candidate Drugs for SARS-CoV-2 Treatment? Front. Microbiol. 2020, 11, 1818. [Google Scholar] [CrossRef]
- Wang, M.-Y.; Zhao, R.; Gao, L.-J.; Gao, X.-F.; Wang, D.-P.; Cao, J.-M. SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Front. Cell. Infect. Microbiol. 2020, 10, 587269. [Google Scholar] [CrossRef]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell Entry Mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.-Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.-X.; Gong, L.; et al. CD147-Spike Protein Is a Novel Route for SARS-CoV-2 Infection to Host Cells. Signal Transduct. Target. Ther. 2020, 5, 283. [Google Scholar] [CrossRef] [PubMed]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 Facilitates SARS-CoV-2 Cell Entry and Infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, C.; Xu, X.; Xu, W.; Liu, S. Structural and Functional Properties of SARS-CoV-2 Spike Protein: Potential Antivirus Drug Development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef] [PubMed]
- Fraser, B.J.; Beldar, S.; Seitova, A.; Hutchinson, A.; Mannar, D.; Li, Y.; Kwon, D.; Tan, R.; Wilson, R.P.; Leopold, K.; et al. Structure and Activity of Human TMPRSS2 Protease Implicated in SARS-CoV-2 Activation. Nat. Chem. Biol. 2022, 18, 963–971. [Google Scholar] [CrossRef] [PubMed]
- Condor Capcha, J.M.; Lambert, G.; Dykxhoorn, D.M.; Salerno, A.G.; Hare, J.M.; Whitt, M.A.; Pahwa, S.; Jayaweera, D.T.; Shehadeh, L.A. Generation of SARS-CoV-2 Spike Pseudotyped Virus for Viral Entry and Neutralization Assays: A 1-Week Protocol. Front. Cardiovasc. Med. 2021, 7, 618651. [Google Scholar] [CrossRef]
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef]
- Watanabe, Y.; Allen, J.D.; Wrapp, D.; McLellan, J.S.; Crispin, M. Site-Specific Glycan Analysis of the SARS-CoV-2 Spike. Science 2020, 369, 330–333. [Google Scholar] [CrossRef]
- Walls, A.C.; Xiong, X.; Park, Y.-J.; Tortorici, M.A.; Snijder, J.; Quispe, J.; Cameroni, E.; Gopal, R.; Dai, M.; Lanzavecchia, A.; et al. Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion. Cell 2019, 176, 1026–1039.e15. [Google Scholar] [CrossRef]
- Zhao, X.; Chen, H.; Wang, H. Glycans of SARS-CoV-2 Spike Protein in Virus Infection and Antibody Production. Front. Mol. Biosci. 2021, 8, 629873. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Qin, S.; Dai, L.; Tian, Z. The Glycosylation in SARS-CoV-2 and Its Receptor ACE2. Signal Transduct. Target. Ther. 2021, 6, 396. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Behura, A.; Mawatwal, S.; Kumar, A.; Naik, L.; Mohanty, S.S.; Manna, D.; Dokania, P.; Mishra, A.; Patra, S.K.; et al. Structure-Function and Application of Plant Lectins in Disease Biology and Immunity. Food Chem. Toxicol. 2019, 134, 110827. [Google Scholar] [CrossRef] [PubMed]
- Coelho, L.C.B.B.; dos Santos Silva, P.M.; de Oliveira, W.F.; de Moura, M.C.; Pontual, E.V.; Gomes, F.S.; Paiva, P.M.G.; Napoleão, T.H.; dos Santos Correia, M.T. Lectins as Antimicrobial Agents. J. Appl. Microbiol. 2018, 125, 1238–1252. [Google Scholar] [CrossRef] [PubMed]
- Moreira, R.A.; Cavada, B.S. Lectin from Canavalia Brasiliensis Characterization and Behavior during Germination. Biol. Plant. 1984, 26, 113–120. [Google Scholar] [CrossRef]
- Moreira, R.D.A.; Cordeiro, E.D.F.; Ramos, V.; Grangeiro, T.B.; Martins, J.L.; De Oliveira, T.A.; Cavada, B.S.; De Ciências, C.; Federal, U. Isolation and partial characterization of a lectin from seeds of Dioclea violacea. Rev. Bras. Fisiol. Veg. 1996, 8, 23–29. [Google Scholar]
- Lam, S.K.; Ng, T.B. Lectins: Production and Practical Applications. Springer. Appl. Microbiol. Biotechnol. 2011, 89, 45. [Google Scholar] [CrossRef]
- Silva, N.R.G.; de Araújo, F.N. Antibacterial Activity of Plant Lectins: A Review. Brazilian Arch. Biol. Technol. 2021, 64, 2021. [Google Scholar] [CrossRef]
- Jandú, J.J.B.; Moraes Neto, R.N.; Zagmignan, A.; de Sousa, E.M.; Brelaz-de-Castro, M.C.A.; dos Santos Correia, M.T.; da Silva, L.C.N. Targeting the Immune System with Plant Lectins to Combat Microbial Infections. Front. Pharmacol. 2017, 8, 671. [Google Scholar] [CrossRef]
- Ahmed, M.N.; Jahan, R.; Nissapatorn, V.; Wilairatana, P.; Rahmatullah, M. Plant Lectins as Prospective Antiviral Biomolecules in the Search for COVID-19 Eradication Strategies. Biomed. Pharmacother. 2022, 146, 112507. [Google Scholar] [CrossRef]
- Konozy, E.; Osman, M.; Dirar, A. Plant Lectins as Potent Anti-Coronaviruses, Anti-Inflammatory, Antinociceptive and Antiulcer Agents. Saudi J. Biol. Sci. 2022, 29, 103301. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, D.P.; Cavalcanti, E.S.B.; de Morais, S.M.; Pinto, C.C.C.; Montes, R.A.; dos Santos, L.M.B.; Furtado, M.L.; da Silva, E.S. Perfil Fitoquímico E Potencial Antioxidante De Extratos Etanólicos Da Espécie Bauhinia Monandra Kurz (Fabaceae)/Phytochemical Profile and Antioxidant Potential of Ethanol Extracts of the Species Bauhinia Monandra Kurz (Fabaceae). Braz. J. Dev. 2020, 6, 86551–86564. [Google Scholar] [CrossRef]
- Shaheen, M.; Borsanyiova, M.; Mostafa, S.; Bopegamage, S.; El Esnawy, N. In Vitro and In Vivo Evaluation of Bauhinia Variegata Extracts to Prevent Coxsackievirus B3 Infection. J. Proteom. Bioinform. 2017, 10, 73–78. [Google Scholar] [CrossRef]
- Huang, L.; Adachi, T.; Shimizu, Y.; Goto, Y.; Toyama, J.; Tanaka, H.; Akashi, R.; Sawaguchi, A.; Iwata, H.; Haga, T. Characterization of Lectin Isolated from Momordica Charantia Seed as a B Cell Activator. Immunol. Lett. 2008, 121, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Lee-Huang, S.; Huang, P.L.; Chen, H.C.; Huang, P.L.; Bourinbaiar, A.; Huang, H.I.; Kung, H. fu Anti-HIV and Anti-Tumor Activities of Recombinant MAP30 from Bitter Melon. Gene 1995, 161, 151–156. [Google Scholar] [CrossRef]
- Praseno, P. Antiviral Activity of Momordica Charantia: A Preliminary Study on in Vitro Anti Herpes Simplex Virus. J. Med. Sci. 1997, 29, 3. [Google Scholar]
- Pongthanapisith, V.; Ikuta, K.; Puthavathana, P.; Leelamanit, W. Antiviral Protein of Momordica Charantia L. Inhibits Different Subtypes of Influenza A. Evid.-Based Complement. Altern. Med. 2013, 2013, 729081. [Google Scholar] [CrossRef]
- Taghizadeh, S.F.; Azizi, M.; Asili, J.; Madarshahi, F.S.; Rakhshandeh, H.; Fujii, Y. Therapeutic Peptides of Mucuna Pruriens L.: Anti-Genotoxic Molecules against Human Hepatocellular Carcinoma and Hepatitis C Virus. Food Sci. Nutr. 2021, 9, 2908–2914. [Google Scholar] [CrossRef]
- Pawar, H.A.; Lalitha, K.G. Isolation, Purification and Characterization of Galactomannans as an Excipient from Senna Tora Seeds. Int. J. Biol. Macromol. 2014, 65, 167–175. [Google Scholar] [CrossRef]
- Wen, C.C.; Shyur, L.F.; Jan, J.T.; Liang, P.H.; Kuo, C.J.; Arulselvan, P.; Wu, J.B.; Kuo, S.C.; Yang, N.S. Traditional Chinese Medicine Herbal Extracts of Cibotium Barometz, Gentiana Scabra, Dioscorea Batatas, Cassia Tora, and Taxillus Chinensis Inhibit SARS-CoV Replication. J. Tradit. Complement. Med. 2011, 1, 41–50. [Google Scholar] [CrossRef]
- Batista, K.L.R.; Silva, C.R.; Santos, V.F.; Silva, R.C.; Roma, R.R.; Santos, A.L.E.; Pereira, R.O.; Delatorre, P.; Rocha, B.A.M.; Soares, A.M.S.; et al. Structural Analysis and Anthelmintic Activity of Canavalia Brasiliensis Lectin Reveal Molecular Correlation between the Carbohydrate Recognition Domain and Glycans of Haemonchus Contortus. Mol. Biochem. Parasitol. 2018, 225, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Santos, V.F.; Araújo, A.C.J.; Silva, A.L.F.; Almeida, D.V.; Freitas, P.R.; Santos, A.L.E.; Rocha, B.A.M.; Garcia, W.; Leme, A.M.; Bondan, E.; et al. Dioclea Violacea Lectin Modulates the Gentamicin Activity against Multi-Resistant Strains and Induces Nefroprotection during Antibiotic Exposure. Int. J. Biol. Macromol. 2020, 146, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Ammerman, N.C.; Beier-Sexton, M.; Azad, A.F. Growth and Maintenance of Vero Cell Lines. Curr. Protoc. Microbiol. 2008, 11, A.4E.1–A.4E.7. [Google Scholar] [CrossRef] [PubMed]
- Kamiloglu, S.; Sari, G.; Ozdal, T.; Capanoglu, E. Guidelines for Cell Viability Assays. Food Front. 2020, 1, 332–349. [Google Scholar] [CrossRef]
- Prism, G. Compute EC Anything from EC 50. Available online: https://www.graphpad.com/quickcalcs/ECanything2/ (accessed on 30 July 2023).
- Case, J.B.; Rothlauf, P.W.; Chen, R.E.; Liu, Z.; Zhao, H.; Kim, A.S.; Bloyet, L.M.; Zeng, Q.; Tahan, S.; Droit, L.; et al. Neutralizing Antibody and Soluble ACE2 Inhibition of a Replication-Competent VSV-SARS-CoV-2 and a Clinical Isolate of SARS-CoV-2. Cell Host Microbe 2020, 28, 475–485.e5. [Google Scholar] [CrossRef] [PubMed]
- Case, J.B.; Bailey, A.L.; Diamond, M.S.; Kim, A.S.; Chen, R.E. Growth, Detection, Quantification, and Inactivation of SARS-CoV-2. Virol. J. 2020, 548, 39–48. [Google Scholar] [CrossRef]
- Chiu, W.; Verschueren, L.; Van den Eynde, C.; Buyck, C.; De Meyer, S.; Jochmans, D.; Bojkova, D.; Ciesek, S.; Cinatl, J.; De Jonghe, S.; et al. Development and Optimization of a High-Throughput Screening Assay for in Vitro Anti-SARS-CoV-2 Activity: Evaluation of 5676 Phase 1 Passed Structures. J. Med. Virol. 2022, 94, 3101–3111. [Google Scholar] [CrossRef]
- Song, J.; Yeo, S.-G.; Hong, E.-H.; Lee, B.-R.; Kim, J.-W.; Kim, J.; Jeong, H.; Kwon, Y.; Kim, H.; Lee, S.; et al. Antiviral Activity of Hederasaponin B from Hedera Helix against Enterovirus 71 Subgenotypes C3 and C4a. Biomol. Ther. 2014, 22, 41–46. [Google Scholar] [CrossRef]
- Uppal, T.; Tuffo, K.; Khaiboullina, S.; Reganti, S.; Pandori, M.; Verma, S.C. Screening of SARS-CoV-2 Antivirals through a Cell-Based RNA-Dependent RNA Polymerase (RdRp) Reporter Assay. Cell Insight 2022, 1, 100046. [Google Scholar] [CrossRef]
- Toni, L.S.; Garcia, A.M.; Jeffrey, D.A.; Jiang, X.; Stauffer, B.L.; Miyamoto, S.D.; Sucharov, C.C. Optimization of Phenol-Chloroform RNA Extraction. MethodsX 2018, 5, 599–608. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). CDC’s Influenza SARS-CoV-2 Multiplex Assay. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/virus-requests.html (accessed on 22 August 2022).
- Bezerra, E.H.S.; Rocha, B.A.M.; Nagano, C.S.; de Arruda Bezerra, G.; de Moura, T.R.; Bezerra, M.J.B.; Benevides, R.G.; Sampaio, A.H.; Assreuy, A.M.S.; Delatorre, P.; et al. Structural Analysis of ConBr Reveals Molecular Correlation between the Carbohydrate Recognition Domain and Endothelial NO Synthase Activation. Biochem. Biophys. Res. Commun. 2011, 408, 566–570. [Google Scholar] [CrossRef] [PubMed]
- Bezerra, M.J.B.; Rodrigues, N.V.F.C.; Pires, A.D.F.; Bezerra, G.A.; Nobre, C.B.; Alencar, K.L.D.L.; Soares, P.M.G.; Do Nascimento, K.S.; Nagano, C.S.; Martins, J.L.; et al. Crystal Structure of Dioclea Violacea Lectin and a Comparative Study of Vasorelaxant Properties with Dioclea Rostrata Lectin. Int. J. Biochem. Cell Biol. 2013, 45, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, H.; Jégouzo, S.A.F.; Rex, M.J.; Drickamer, K.; Weis, W.I.; Taylor, M.E. Mechanism of Pathogen Recognition by Human Dectin-2. J. Biol. Chem. 2017, 292, 13402–13414. [Google Scholar] [CrossRef]
- Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.8; Schrodinger LLC: New York, NY, USA, 2015. [Google Scholar]
- Dominguez, C.; Boelens, R.; Bonvin, A.M.J.J. HADDOCK: A Protein-Protein Docking Approach Based on Biochemical or Biophysical Information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, U.E.A.; Santos, I.A.; Grosche, V.R.; Fernandes, R.S.; de Godoy, A.S.; Torres, J.D.A.; Freire, M.C.L.C.; de Moraes Roso Mesquita, N.C.; Guevara-Vega, M.; Nicolau-Junior, N.; et al. Imidazonaphthyridine Effects on Chikungunya Virus Replication: Antiviral Activity by Dependent and Independent of Interferon Type 1 Pathways. Virus Res. 2023, 324, 199029. [Google Scholar] [CrossRef]
- de Oliveira, D.M.; de Andrade Santos, I.; Martins, D.O.S.; Gonçalves, Y.G.; Cardoso-Sousa, L.; Sabino-Silva, R.; Von Poelhsitz, G.; de Faria Franca, E.; Nicolau-Junior, N.; Pacca, C.C.; et al. Organometallic Complex Strongly Impairs Chikungunya Virus Entry to the Host Cells. Front. Microbiol. 2020, 11, 608924. [Google Scholar] [CrossRef]
- Santos, I.A.; Shimizu, J.F.; de Oliveira, D.M.; Martins, D.O.S.; Cardoso-Sousa, L.; Cintra, A.C.O.; Aquino, V.H.; Sampaio, S.V.; Nicolau-Junior, N.; Sabino-Silva, R.; et al. Chikungunya Virus Entry Is Strongly Inhibited by Phospholipase A2 Isolated from the Venom of Crotalus Durissus Terrificus. Sci. Rep. 2021, 11, 8717. [Google Scholar] [CrossRef]
- Santos, I.A.; dos Santos Pereira, A.K.; Guevara-Vega, M.; de Paiva, R.E.F.; Sabino-Silva, R.; Bergamini, F.R.G.; Corbi, P.P.; Jardim, A.C.G. Repurposing Potential of Rimantadine Hydrochloride and Development of a Promising Platinum(II)-Rimantadine Metallodrug for the Treatment of Chikungunya Virus Infection. Acta Trop. 2022, 227, 106300. [Google Scholar] [CrossRef]
- Harcourt, J.; Tamin, A.; Lu, X.; Kamili, S.; Sakthivel, S.K.; Murray, J.; Queen, K.; Tao, Y.; Paden, C.R.; Zhang, J.; et al. Isolation and Characterization of SARS-CoV-2 from the First US COVID-19 Patient. BioRxiv 2020. [Google Scholar] [CrossRef]
- Lei, Y.; Hannoufa, A.; Christensen, D.; Shi, H.; Prates, L.; Yu, P. Molecular Structural Changes in Alfalfa Detected by ATR-FTIR Spectroscopy in Response to Silencing of TT8 and HB12 Genes. Int. J. Mol. Sci. 2018, 19, 1046. [Google Scholar] [CrossRef]
- Yoshida, S.; Miyazaki, M.; Sakai, K.; Takeshita, M.; Yuasa, S.; Sato, A.; Kobayashi, T.; Watanabe, S.; Okuyama, H. Fourier Transform Infrared Spectroscopic Analysis of Rat Brain Microsomal Membranes Modified by Dietary Fatty Acids: Possible Correlation with Altered Learning Behavior. Biospectroscopy 1997, 3, 281–290. [Google Scholar] [CrossRef]
- Shetty, G.; Kendall, C.; Shepherd, N.; Stone, N.; Barr, H. Raman Spectroscopy: Elucidation of Biochemical Changes in Carcinogenesis of Oesophagus. Br. J. Cancer 2006, 94, 1460–1464. [Google Scholar] [CrossRef] [PubMed]
- Wood, B.R.; Quinn, M.A.; Tait, B.; Ashdown, M.; Hislop, T.; Romeo, M.; McNaughton, D. FTIR Microspectroscopic Study of Cell Types and Potential Confounding Variables in Screening for Cervical Malignancies. Biospectroscopy 1998, 4, 75–91. [Google Scholar] [CrossRef]
- Dovbeshko, G. FTIR Spectroscopy Studies of Nucleic Acid Damage. Talanta 2000, 53, 233–246. [Google Scholar] [CrossRef]
- Yang, Y.; Sulé-Suso, J.; Sockalingum, G.D.; Kegelaer, G.; Manfait, M.; El Haj, A.J. Study of Tumor Cell Invasion by Fourier Transform Infrared Microspectroscopy. Biopolymers 2005, 78, 311–317. [Google Scholar] [CrossRef]
- Paluszkiewicz, C.; Kwiatek, W.M. Analysis of Human Cancer Prostate Tissues Using FTIR Microspectroscopy and SRIXE Techniques. J. Mol. Struct. 2001, 565–566, 329–334. [Google Scholar] [CrossRef]
- Dovbeshko, G.I.; Chegel, V.I.; Gridina, N.Y.; Repnytska, O.P.; Shirshov, Y.M.; Tryndiak, V.P.; Todor, I.M.; Solyanik, G.I. Surface Enhanced IR Absorption of Nucleic Acids from Tumor Cells: FTIR Reflectance Study. Biopolymers 2002, 67, 470–486. [Google Scholar] [CrossRef]
- Eckel, R.; Huo, H.; Guan, H.-W.; Hu, X.; Che, X.; Huang, W.-D. Characteristic Infrared Spectroscopic Patterns in the Protein Bands of Human Breast Cancer Tissue. Vib. Spectrosc. 2001, 27, 165–173. [Google Scholar] [CrossRef]
- Agarwal, R.; Tandon, P.; Gupta, V.D. Phonon Dispersion in Poly(Dimethylsilane). J. Organomet. Chem. 2006, 691, 2902–2908. [Google Scholar] [CrossRef]
- Barnas, E.; Skret-Magierlo, J.; Skret, A.; Kaznowska, E.; Depciuch, J.; Szmuc, K.; Łach, K.; Krawczyk-Marć, I.; Cebulski, J. Simultaneous FTIR and Raman Spectroscopy in Endometrial Atypical Hyperplasia and Cancer. Int. J. Mol. Sci. 2020, 21, 4828. [Google Scholar] [CrossRef]
- Kar, S.; Katti, D.R.; Katti, K.S. Fourier Transform Infrared Spectroscopy Based Spectral Biomarkers of Metastasized Breast Cancer Progression. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 208, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.P.; Aguiar, E.M.; Cardoso-Sousa, L.; Caixeta, D.C.; Guedes, C.C.; Siqueira, W.L.; Maia, Y.C.P.; Cardoso, S.V.; Sabino-Silva, R. Differential Molecular Signature of Human Saliva Using ATR-FTIR Spectroscopy for Chronic Kidney Disease Diagnosis. Braz. Dent. J. 2019, 30, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Cavada, B.S.; Osterne, V.J.S.; Lossio, C.F.; Pinto-Junior, V.R.; Oliveira, M.V.; Silva, M.T.L.; Leal, R.B.; Nascimento, K.S. One Century of ConA and 40 years of ConBr Research: A Structural Review. Int. J. Biol. Macromol. 2019, 134, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Ramos, E.L.P.; Santana, S.S.; Silva, M.V.; Santiago, F.M.; Mineo, T.W.P.; Mineo, J.R. Lectins from Synadenium Carinatum (ScLL) and Artocarpus Heterophyllus (ArtinM) Are Able to Induce Beneficial Immunomodulatory Effects in a Murine Model for Treatment of Toxoplasma Gondii Infection. Front. Cell. Infect. Microbiol. 2016, 6, 164. [Google Scholar] [CrossRef]
- Holanda, M.L.; Melo, V.M.M.; Silva, L.M.C.M.; Amorim, R.C.N.; Pereira, M.G.; Benevides, N.M.B. Differential Activity of a Lectin from Solieria Filiformis against Human Pathogenic Bacteria. Braz. J. Med. Biol. Res. 2005, 38, 1769–1773. [Google Scholar] [CrossRef]
- Auth, J.; Fröba, M.; Große, M.; Rauch, P.; Ruetalo, N.; Schindler, M.; Morokutti-Kurz, M.; Graf, P.; Dolischka, A.; Prieschl-Grassauer, E.; et al. Lectin from Triticum Vulgaris (WGA) Inhibits Infection with SARS-CoV-2 and Its Variants of Concern Alpha and Beta. Int. J. Mol. Sci. 2021, 22, 10205. [Google Scholar] [CrossRef]
- Liu, Y.-M.; Shahed-Al-Mahmud, M.; Chen, X.; Chen, T.-H.; Liao, K.-S.; Lo, J.M.; Wu, Y.-M.; Ho, M.-C.; Wu, C.-Y.; Wong, C.-H.; et al. A Carbohydrate-Binding Protein from the Edible Lablab Beans Effectively Blocks the Infections of Influenza Viruses and SARS-CoV-2. Cell Rep. 2020, 32, 108016. [Google Scholar] [CrossRef]
- Sheehan, S.A.; Hamilton, K.L.; Retzbach, E.P.; Balachandran, P.; Krishnan, H.; Leone, P.; Lopez-Gonzalez, M.; Suryavanshi, S.; Kumar, P.; Russo, R.; et al. Evidence That Maackia Amurensis Seed Lectin (MASL) Exerts Pleiotropic Actions on Oral Squamous Cells with Potential to Inhibit SARS-CoV-2 Infection and COVID-19 Disease Progression. Exp. Cell Res. 2021, 403, 112594. [Google Scholar] [CrossRef]
- Wang, W.; Li, Q.; Wu, J.; Hu, Y.; Wu, G.; Yu, C.; Xu, K.; Liu, X.; Wang, Q.; Huang, W.; et al. Lentil Lectin Derived from Lens Culinaris Exhibit Broad Antiviral Activities against SARS-CoV-2 Variants. Emerg. Microbes Infect. 2021, 10, 1519–1529. [Google Scholar] [CrossRef]
- Spira, B. The Impact of the Highly Virulent SARS-CoV-2 Gamma Variant on Young Adults in the State of São Paulo: Was It Inevitable? Cureus 2022, 14, 26486. [Google Scholar] [CrossRef]
- McMahan, K.; Giffin, V.; Tostanoski, L.H.; Chung, B.; Siamatu, M.; Suthar, M.S.; Halfmann, P.; Kawaoka, Y.; Piedra-Mora, C.; Jain, N.; et al. Reduced Pathogenicity of the SARS-CoV-2 Omicron Variant in Hamsters. Med 2022, 3, 262–268.e4. [Google Scholar] [CrossRef]
- Keyaerts, E.; Vijgen, L.; Pannecouque, C.; Van Damme, E. Plant lectins are potent inhibitors of coronaviruses by interfering with two targets in the viral replication cycle. Antivir. Res. 2007, 75, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.W.; Shin, J.S.; Park, S.-J.; Jung, E.; Park, Y.-G.; Lee, J.; Kim, S.J.; Park, H.-J.; Lee, J.-H.; Park, S.-M.; et al. Antiviral Activity and Safety of Remdesivir against SARS-CoV-2 Infection in Human Pluripotent Stem Cell-Derived Cardiomyocytes. Antivir. Res. 2020, 184, 104955. [Google Scholar] [CrossRef] [PubMed]
- Cox, R.M.; Wolf, J.D.; Lieber, C.M.; Sourimant, J.; Lin, M.J.; Babusis, D.; DuPont, V.; Chan, J.; Barrett, K.T.; Lye, D.; et al. Oral Prodrug of Remdesivir Parent GS-441524 Is Efficacious against SARS-CoV-2 in Ferrets. Nat. Commun. 2021, 12, 6415. [Google Scholar] [CrossRef]
- Franco, E.J.; Drusano, G.L.; Hanrahan, K.C.; Warfield, K.L.; Brown, A.N. Combination Therapy with UV-4B and Molnupiravir Enhances SARS-CoV-2 Suppression. Viruses 2023, 15, 1175. [Google Scholar] [CrossRef]
- Lokhande, K.B.; Apte, G.R.; Shrivastava, A.; Singh, A.; Pal, J.K.; Swamy, K.V.; Gupta, R.K. Sensing the Interactions between Carbohydrate-Binding Agents and N-Linked Glycans of SARS-CoV-2 Spike Glycoprotein Using Molecular Docking and Simulation Studies. J. Biomol. Struct. Dyn. 2022, 40, 3880–3898. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, W.; Mao, Y.; Chen, Y.; Wang, S.; Zhong, Y.; Su, T.; Gong, M.; Du, D.; Lu, X.; et al. Site-Specific N-Glycosylation Characterization of Recombinant SARS-CoV-2 Spike Proteins. Mol. Cell. Proteom. 2021, 20, 100058. [Google Scholar] [CrossRef]
- Chikhale, R.V.; Gurav, S.S.; Patil, R.B.; Sinha, S.K.; Prasad, S.K.; Shakya, A.; Shrivastava, S.K.; Gurav, N.S.; Prasad, R.S. SARS-CoV-2 Host Entry and Replication Inhibitors from Indian Ginseng: An in-Silico Approach. J. Biomol. Struct. Dyn. 2021, 39, 4510–4521. [Google Scholar] [CrossRef] [PubMed]
- Barton, C.; Kouokam, J.C.; Lasnik, A.B.; Foreman, O.; Cambon, A.; Brock, G.; Montefiori, D.C.; Vojdani, F.; McCormick, A.A.; O’Keefe, B.R.; et al. Activity of and Effect of Subcutaneous Treatment with the Broad-Spectrum Antiviral Lectin Griffithsin in Two Laboratory Rodent Models. Antimicrob. Agents Chemother. 2014, 58, 120–127. [Google Scholar] [CrossRef]
- Meuleman, P.; Albecka, A.; Belouzard, S.; Vercauteren, K.; Verhoye, L.; Wychowski, C.; Leroux-Roels, G.; Palmer, K.E.; Dubuisson, J. Griffithsin Has Antiviral Activity against Hepatitis C Virus. Antimicrob. Agents Chemother. 2011, 55, 5159–5167. [Google Scholar] [CrossRef]
- Takebe, Y.; Saucedo, C.J.; Lund, G.; Uenishi, R.; Hase, S.; Tsuchiura, T.; Kneteman, N.; Ramessar, K.; Tyrrell, D.L.J.; Shirakura, M.; et al. Antiviral Lectins from Red and Blue-Green Algae Show Potent In Vitro and In Vivo Activity against Hepatitis C Virus. PLoS ONE 2013, 8, e64449. [Google Scholar] [CrossRef] [PubMed]
- Ishag, H.Z.A.; Li, C.; Huang, L.; Sun, M.; Wang, F.; Ni, B.; Malik, T.; Chen, P.; Mao, X. Griffithsin Inhibits Japanese Encephalitis Virus Infection In Vitro and In Vivo. Arch. Virol. 2013, 158, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Harb, A.; Fakhreddine, M.; Zaraket, H.; Saleh, F.A. Three-Dimensional Cell Culture Models to Study Respiratory Virus Infections Including COVID-19. Biomimetics 2021, 7, 3. [Google Scholar] [CrossRef]
- Do, T.N.D.; Donckers, K.; Vangeel, L.; Chatterjee, A.K.; Gallay, P.A.; Bobardt, M.D.; Bilello, J.P.; Cihlar, T.; De Jonghe, S.; Neyts, J.; et al. A Robust SARS-CoV-2 Replication Model in Primary Human Epithelial Cells at the Air Liquid Interface to Assess Antiviral Agents. Antivir. Res. 2021, 192, 105122. [Google Scholar] [CrossRef]
- Mitchell, C.A.; Ramessar, K.; O’Keefe, B.R. Antiviral Lectins: Selective Inhibitors of Viral Entry. Antivir. Res. 2017, 142, 37–54. [Google Scholar] [CrossRef]
- Dimitrijevic, R.; Stojanovic, M.; Micic, M.; Dimitrijevic, L.; Gavrovic-Jankulovic, M. Recombinant Banana Lectin as Mucosal Immunostimulator. J. Funct. Foods 2012, 4, 636–641. [Google Scholar] [CrossRef]
- Nixon, B.; Stefanidou, M.; Mesquita, P.M.M.; Fakioglu, E.; Segarra, T.; Rohan, L.; Halford, W.; Palmer, K.E.; Herold, B.C. Griffithsin Protects Mice from Genital Herpes by Preventing Cell-to-Cell Spread. J. Virol. 2013, 87, 6257–6269. [Google Scholar] [CrossRef] [PubMed]
- Ziółkowska, N.E.; O’Keefe, B.R.; Mori, T.; Zhu, C.; Giomarelli, B.; Vojdani, F.; Palmer, K.E.; McMahon, J.B.; Wlodawer, A. Domain-Swapped Structure of the Potent Antiviral Protein Griffithsin and Its Mode of Carbohydrate Binding. Structure 2006, 14, 1127–1135. [Google Scholar] [CrossRef]
- O’Keefe, B.R.; Giomarelli, B.; Barnard, D.L.; Shenoy, S.R.; Chan, P.K.S.; McMahon, J.B.; Palmer, K.E.; Barnett, B.W.; Meyerholz, D.K.; Wohlford-Lenane, C.L.; et al. Broad-Spectrum In Vitro Activity and In Vivo Efficacy of the Antiviral Protein Griffithsin against Emerging Viruses of the Family Coronaviridae. J. Virol. 2010, 84, 2511–2521. [Google Scholar] [CrossRef]
- Girard, L.; Birse, K.; Holm, J.B.; Gajer, P.; Humphrys, M.S.; Garber, D.; Guenthner, P.; Noël-Romas, L.; Abou, M.; McCorrister, S.; et al. Impact of the Griffithsin Anti-HIV Microbicide and Placebo Gels on the Rectal Mucosal Proteome and Microbiome in Non-Human Primates. Sci. Rep. 2018, 8, 8059. [Google Scholar] [CrossRef]
- Franzén Boger, M.; Benhach, N.; Hasselrot, T.; Brand, R.M.; Rohan, L.C.; Wang, L.; McGowan, I.; Edick, S.; Ho, K.; Meyn, L.; et al. A Topical Rectal Douche Product Containing Q-Griffithsin Does Not Disrupt the Epithelial Border or Alter CD4+ Cell Distribution in the Human Rectal Mucosa. Sci. Rep. 2023, 13, 7547. [Google Scholar] [CrossRef] [PubMed]



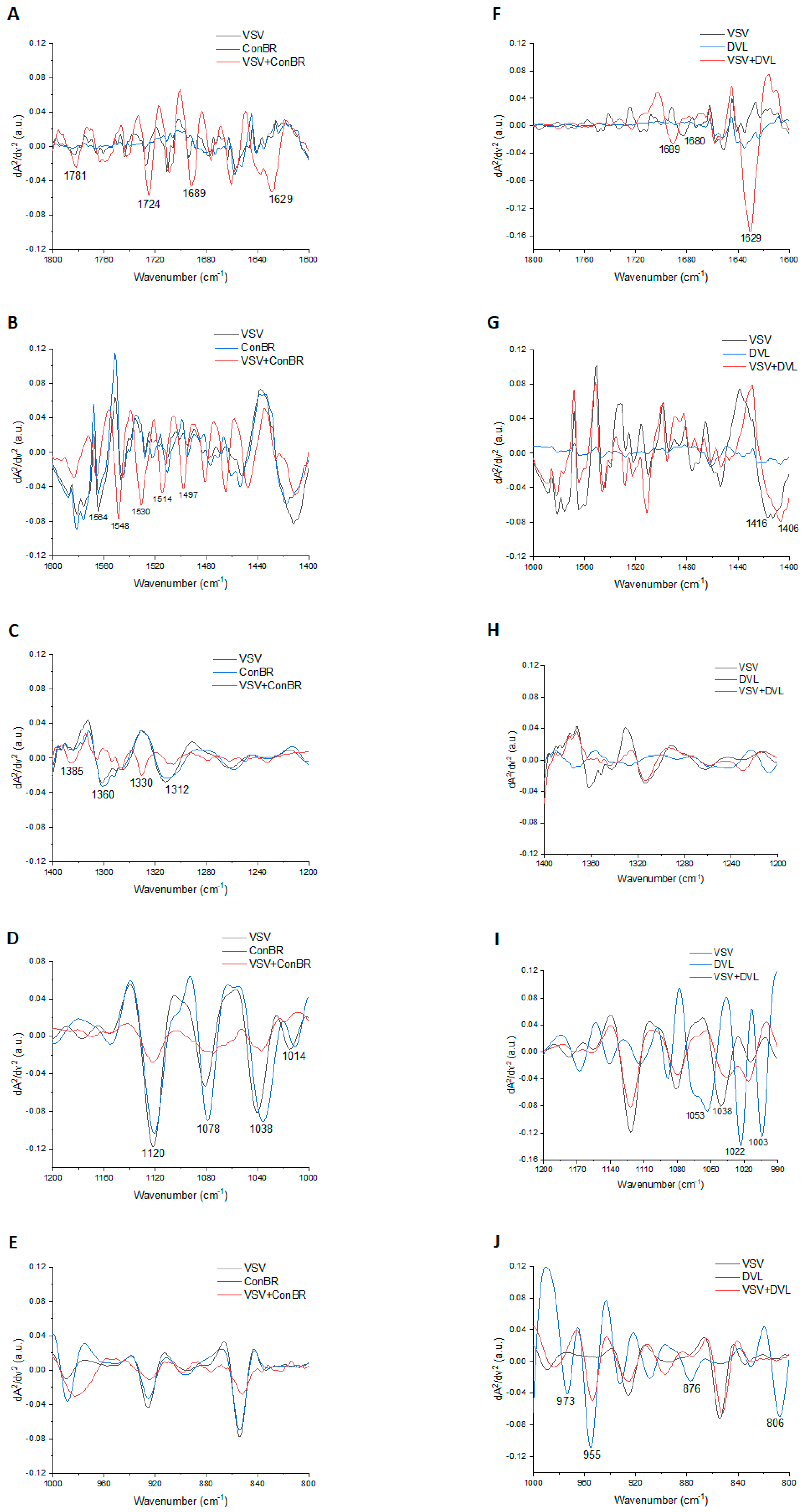

| Lectins | Peak | Tentative Assignment | Reference |
|---|---|---|---|
| ConBR | 1781 | Carbonyl C=O ester stretching region | [65] |
| 1724 | C=O stretching band mode of the fatty acid ester | [66] | |
| 1689 | Amide I (disordered structure-non-hydrogen bonded) | [67] | |
| 1629 | Amide I region | [68] | |
| 1564 | Ring base | [69] | |
| 1548 | Amide II | [70] | |
| 1530 | Stretching C=N, C=C | [69] | |
| 1514 | Amide II | [71] | |
| 1497 | C=C, deformation C-H | [69] | |
| 1385 | Deformation C-H | [69] | |
| 1360 | Deformation C-H | [69] | |
| 1330 | CH2 wagging | [70] | |
| 1312 | Amide III band components of proteins | [70] | |
| 1120 | Mannose-6-phosphate | [66] | |
| 1078 | Phosphate I in RNA | [72] | |
| 1038 | Stretching C-O ribose | [69] | |
| 1012 | Stretching C-O deoxyribose | [69] | |
| DVL | 1689 | Amide I (disordered structure-non-hydrogen bonded) | [67] |
| 1680 | Unordered random coils and turns of amide I | [73] | |
| 1629 | Amide I region | [68] | |
| 1416 | Deformation C-H, N-H, stretching C-N | [69] | |
| 1406 | CH3 asymmetric deformation | [74] | |
| 1053 | νC-O and δC-O of carbohydrates | [75] | |
| 1038 | Stretching C-O ribose | [69] | |
| 1022 | Glycogen | [68] | |
| 1003 | Carbohydrate residues attached to collagen and amide III vibration | [76] | |
| 973 | OCH3 (polysaccharides, pectin) | [67] | |
| 955 | Phospholipids/carbohydrates | [77] | |
| 876 | (A-form helix) conformation | [72] | |
| 806 | (A-form helix) conformation | [72] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grosche, V.R.; Souza, L.P.F.; Ferreira, G.M.; Guevara-Vega, M.; Carvalho, T.; Silva, R.R.d.S.; Batista, K.L.R.; Abuna, R.P.F.; Silva, J.S.; Calmon, M.d.F.; et al. Mannose-Binding Lectins as Potent Antivirals against SARS-CoV-2. Viruses 2023, 15, 1886. https://doi.org/10.3390/v15091886
Grosche VR, Souza LPF, Ferreira GM, Guevara-Vega M, Carvalho T, Silva RRdS, Batista KLR, Abuna RPF, Silva JS, Calmon MdF, et al. Mannose-Binding Lectins as Potent Antivirals against SARS-CoV-2. Viruses. 2023; 15(9):1886. https://doi.org/10.3390/v15091886
Chicago/Turabian StyleGrosche, Victória Riquena, Leandro Peixoto Ferreira Souza, Giulia Magalhães Ferreira, Marco Guevara-Vega, Tamara Carvalho, Romério Rodrigues dos Santos Silva, Karla Lilian Rodrigues Batista, Rodrigo Paolo Flores Abuna, João Santana Silva, Marília de Freitas Calmon, and et al. 2023. "Mannose-Binding Lectins as Potent Antivirals against SARS-CoV-2" Viruses 15, no. 9: 1886. https://doi.org/10.3390/v15091886
APA StyleGrosche, V. R., Souza, L. P. F., Ferreira, G. M., Guevara-Vega, M., Carvalho, T., Silva, R. R. d. S., Batista, K. L. R., Abuna, R. P. F., Silva, J. S., Calmon, M. d. F., Rahal, P., da Silva, L. C. N., Andrade, B. S., Teixeira, C. S., Sabino-Silva, R., & Jardim, A. C. G. (2023). Mannose-Binding Lectins as Potent Antivirals against SARS-CoV-2. Viruses, 15(9), 1886. https://doi.org/10.3390/v15091886









