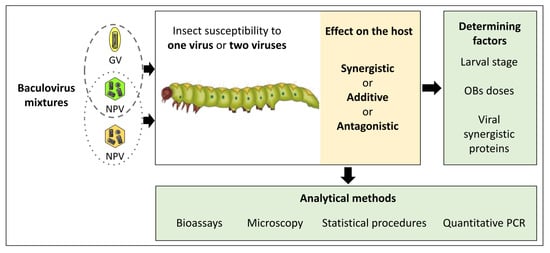
Effects of Mixed Baculovirus Infections in Biological Control: A Comprehensive Historical and Technical Analysis
Abstract
1. Introduction
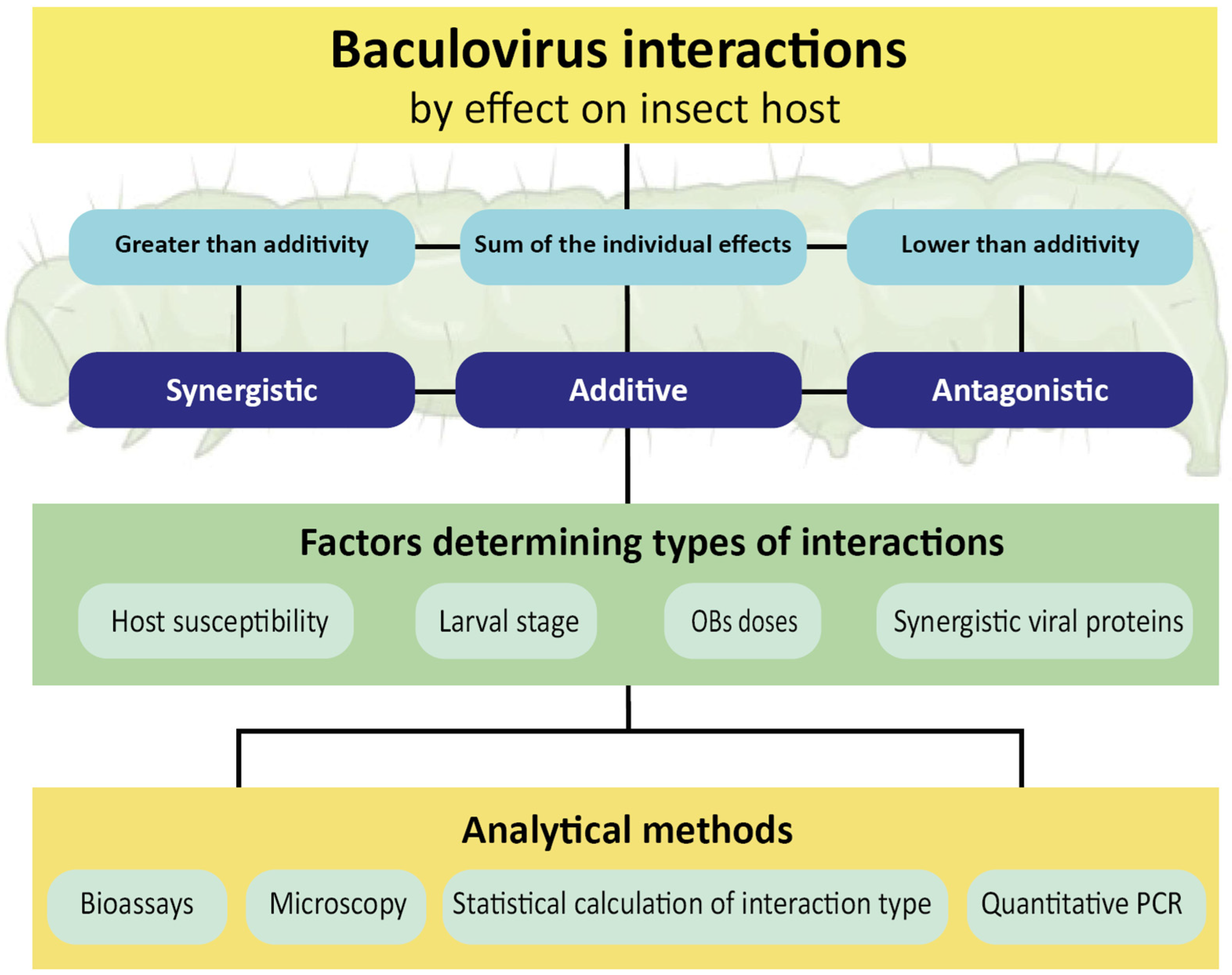
2. Baculovirus Interactions In Vivo
2.1. NPV and GV Mixtures
2.2. Synergistic Factors
2.3. NPV Mixtures
2.4. Unknown Synergistic Factor
3. Strategies to Evaluate Combinations of NPV and GVs: Technical Aspects
3.1. Quantification of Viral Inocula
| Host | Instar | NPV | NPV Group | GV | GV Clade | GV Infects the Host | Enhancin Genes (GV) | Effect on Time to Kill | Effect on Virulence | Relative Potency | Overall Effect (Reported) | Calculated Effect (This Work) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. unipuncta | third, 4th, fifth, sixth | PsunNPV | ND | PsunGV | a | yes | 3 | ND | increased mortality | ND (1) | synergistic | synergistic/additive (5) | [22] |
| C. fumiferana | third, fourth | CfMNPV (6) | I | CfGV (6) | b | yes | 0 | not affected | increased mortality | - | interference/not synergistic | antagonistic | [26] |
| T. ni | fourth | TnNPV | II | TnGV | a | yes | 3 | delayed | reduced mortality | - | additive | [27] | |
| P. unipuncta | fifth | PsunNPV | ND | PsunGV | a | yes | 3 | ND | reduced ID50 | 56.3 (2) | synergistic | [35] | |
| H. armigera | second/third | HearNPV | II | HearGV (6) | a | yes | 4 | delayed | reduced mortality | - | interference | antagonistic | [28] |
| P. separata | fifth | PsunNPV | PsunGV | a | yes | 3 | ND | reduced LD50 | 15,523.3 (2, 3) | enhancement | [36] | ||
| S. litura | fifth | SpliNPV | II | PsunGV | a | no | 3 | ND | reduced LD50 | 11.5 (2, 3) | enhancement | [36] | |
| B. mori | third | BmNPV | I | PsunGV | a | no | 3 | ND | no effect | - | no effect | [36] | |
| X. c-nigrum | fourth, fifth | XcenNPV | ND | XcenGV | a | yes | 4 | ND | increased ID50/reduced ID50 | 0.9/240.2 (2, 3) | not enhanced/enhanced | [32] | |
| H. zea | first, second | HzSNPV | II | HearGV (6) | a | yes | 4 | delayed | ND | - | interference | [18] | |
| L. dispar | second | LdMNPV | II | HearGV (6) | a | no | 4 | reduced LT50 | reduced LD50 | 286.4 | enhancement | [37] | |
| L. dispar | second | LdMNPV | II | SpfrGV (6) | a | no | 2 | not affected | reduced LD50 | 13.1 | enhancement | [37] | |
| L. dispar | second, third, fourth | LdMNPV | II | HearGV (6) | a | no | 4 | Reduced LT50 | increased mortality | ND (1) | enhancement | synergistic/additive (5) | [80] |
| T. ni | first | AcMNPV | I | TnGV (6) | a | yes | 3 | ND | reduced LC50 | 10.7 (2) | synergistic | [39] | |
| S. litura | fifth | SpltNPV | II | XcenGV | a | no | 4 | not affected | reduced LC50 | 6.48 | synergistic | [40] | |
| H. armigera | second, third, fourth, fifth | HearNPV | II | HearGV (6) | a | yes | 4 | delayed | increased/reduced | - | no enhancement | antagonistic | [31] |
| S. littoralis | third | SpliNPV | II | SpliGV | a | yes | ND | delayed | increased LD50 | 0.2 | antagonistic | [33] | |
| H. armigera | second, third, fourth, fifth | HearNPV | II | SpltGV (6) | a | no | ND | reduced | reduced LC50 | 13.32 (4) | synergistic | [42] | |
| H. armigera | second, third, fourth, fifth | HearNPV | II | AgseGV | a | no | 1 | not affected | ND | - | neutral | [42] | |
| H. armigera | second, third, fourth, fifth | HearNPV | II | PlxyGV (6) | a | no | 0 | not affected | ND | - | neutral | [42] | |
| H. armigera | second, third, fourth, fifth | HearNPV | II | AjGV | ND | no | ND | not affected | ND | - | neutral | [42] | |
| H. armigera | second, third, fourth, fifth | HearNPV | II | CiGV | ND | no | ND | not affected | ND | - | neutral | [42] | |
| A. gemmatalis | third | AgMNPV | I | EpapGV | b | no | 0 | reduced ST | increased mortality | ND (1) | enhancement | synergistic | [41] |
| A. segetum | neonates | AgseNPV-B | II | AgseGV | a | yes | 1 | ND | not affected mortality | - | additive | additive | [34] |
| S. frugiperda | second | SfMNPV | II | SpfGrV (6) | a | yes | 2 | reduced MTD | reduced LC50 | 11.4 | enhancement | [24] | |
| S. ornithgalli | neonates | SporNPV | II | SporGV | a | yes | 0 | ND | increased mortality | 3.06 | synergistic | synergistic | [25] |
| Host | NPV | GV Proteins | Effect on Time to Kill | Overall Effect (Reported) | Reference |
|---|---|---|---|---|---|
| P. unipuncta | PsunNPV | PsunGV | ND | enhancement | [35] |
| T. ni | AcMNPV | TnGV | ND | enhancement | [38] |
| M. Brassicae | MbMNPV | XcenGV | reduced | enhancement | [43,46] |
| H. armigera | MbMNPV | XcenGV | reduced | enhancement | [44,45] |
| A. nigrisigna | MbMNPV | XcenGV | reduced | enhancement | [47] |
| S. frugiperda | SfMNPV | SpfrGV | ND | enhancement | [24] |
| Host | Instar | NPV | NPV Group | GV | GV Clade | GV Infects the Host | Enhancin Genes (GV) | Effect on Time to Kill | Effect on Virulence | Relative Potency | Overall Effect (Reported) | Calculated Effect (This Work) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. segetum | neonates | AgseNPV-B | II | AgseGV | a | yes | 1 | ND | not affected mortality | ND (1) | additive | additive | [34] |
| H. armigera | second, third, fourth, fifth | HearNPV | II | AgseGV | a | no | 1 | not affected | ND | - | neutral | [42] | |
| H. armigera | second, third | HearNPV | II | HearGV (5) | a | yes | 4 | delayed | reduced mortality | - | interference | antagonistic | [28] |
| H. zea | first, second | HzSNPV | II | HearGV (5) | a | yes | 4 | delayed | n/d | - | interference | [18] | |
| H. armigera | second, third, fourth, fifth | HearNPV | II | HearGV (5) | a | yes | 4 | delayed | increased/reduced | - | no enhancement | antagonistic | [31] |
| L. dispar | second | LdMNPV | II | HearGV (5) | a | no | 4 | reduced LT50 | reduced LD50 | 286.4 | enhancement | [37] | |
| L. dispar | second, third, fourth | LdMNPV | II | HearGV (5) | a | no | 4 | Reduced LT50 | Reduced LC50 | ND (1) | increased mortality | synergistic/additive (4) | [80] |
| P. unipuncta | third, fourth, fifth, sixth | PsunNPV | ND | PsunGV | a | yes | 3 | ND | increased mortality | ND (1) | synergistic | synergistic/additive (4) | [22] |
| P. unipuncta | fifth | PsunNPV | ND | PsunGV | a | yes | 3 | ND | reduced ID50 | 56.3 (2) | synergistic | [35] | |
| P. separata | fifth | PsunNPV | PsunGV | a | yes | 3 | ND | reduced LD50 | 15,523.3 (2, 3) | enhancement | [36] | ||
| S. litura | fifth | SpltNPV (5) | II | PsunGV | a | no | 3 | ND | reduced LD50 | 11.5 (2, 3) | enhancement | [36] | |
| B. mori | third | BmNPV | I | PsunGV | a | no | 3 | ND | no effect | - | no effect | [36] | |
| L. dispar | second | LdMNPV | II | SpfrGV (5) | a | no | 2 | not affected | reduced LD50 | 13.1 | enhancement | [37] | |
| S. frugiperda | second | SfMNPV | II | SpfrGV (5) | a | yes | 2 | reduced MTD | reduced LC50 | 11.4 | enhancement | [24] | |
| T. ni | fourth | TnNPV | II | TnGV | a | yes | 3 | delayed | reduced mortality | - | additive | [27] | |
| T. ni | first | AcMNPV | I | TnGV | a | yes | 3 | ND | reduced LC50 | 10.7 (2) | synergistic | [39] | |
| X. c-nigrum | fourth, fifth | XcenNPV | ND | XcenGV | a | yes | 4 | ND | increased ID50/reduced ID50 | 0.9/240.2 (2, 3) | not enhanced/enhanced | [32] | |
| S. litura | fifth | SpltNPV (5) | II | XcenGV | a | no | 4 | not affected | reduced LC50 | 6.48 | synergistic | [40] |
| Host | Instar | NPV 1 | NPV 1 Group | NPV 2 | NPV 2 Group | NPV 2 Infects the Host | Effect on Time to Kill | Effect on Virulence | Overall Effect (Reported) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Pseudaletia unipuncta | fifth | TNPV (Typical typical NPV) | ND | HNPV (Hypertrophy hypertrophy NPV) | ND | yes | ND | decreased | interference | [60] |
| Trichoplusia nu | first | AcMNPV | I | TnSNPV | II | yes | ND | decreased LD50 | synergistic | [39] |
| Pseudoplusia includens | third | ThorMNPV | I | ThorSNPV | II | yes | ND | no effect | neutralistic | [63] |
| Choristoneura fumiferana | ND | CfMNPV | I | CfDEFMNPV | II | yes | ND | increased * | synergistic | [65] |
| Helicoverpa zea | second | HzSNPV | II | LdMNPV | II | no | ND | decreased LC50 | synergistic | [66] |
| Helicoverpa zea | second | AcMNPV | I | LdMNPV | II | no | ND | decreased LC50 | synergistic | [66] |
| Helicoverpa zea | second | AfMNPV | I | LdMNPV | II | no | ND | decreased LC50 | synergistic | [66] |
| Helicoverpa zea | second | GmMNPV | I | LdMNPV | II | No | ND | decreased LC50 | synergistic | [66] |
| Helicoverpa zea | second | HearMNPV | II | LdMNPV | II | no | ND | decreased LC50 | synergistic | [66] |
| Helicoverpa zea | second | PxMNPV | I | LdMNPV | II | no | ND | decreased LC50 | synergistic | [66] |
| Helicoverpa zea | second | RoMNPV | I | LdMNPV | II | no | ND | decreased LC50 | synergistic | [66] |
| Spodoptera exigua | second | SeMNPV | II | LdMNPV | II | no | ND | decreased LC50 | synergistic | [66] |
| Spodoptera exigua | second | AcMNPV | I | LdMNPV | II | no | ND | decreased LC50 | synergistic | [66] |
| Spodoptera exigua | second | AfMNPV | I | LdMNPV | II | no | ND | decreased LC50 | synergistic | [66] |
| Spodoptera exigua | second | GmMNPV | I | LdMNPV | II | no | ND | decreased LC50 | synergistic | [66] |
| Spodoptera exigua | second | HearMNPV | II | LdMNPV | II | no | ND | decreased LC50 | synergistic | [66] |
| Spodoptera exigua | second | PxMNPV | I | LdMNPV | II | no | ND | decreased LC50 | synergistic | [66] |
| Spodoptera exigua | second | RoMNPV | I | LdMNPV | II | No | ND | decreased LC50 | synergistic | [66] |
| Spodoptera frugiperda | second | SfMNPV | II | LdMNPV | II | no | ND | decreased LC50 | synergistic | [66] |
| Spodoptera frugiperda | second | AcMNPV | I | LdMNPV | II | no | ND | decreased LC50 | synergistic | [66] |
| Spodoptera frugiperda | second | AfMNPV | I | LdMNPV | II | no | ND | decreased LC50 | synergistic | [66] |
| Spodoptera frugiperda | second | GmMNPV | I | LdMNPV | II | no | ND | decreased LC50 | synergistic | [66] |
| Spodoptera frugiperda | second | HearMNPV | II | LdMNPV | II | no | ND | decreased LC50 | synergistic | [66] |
| Spodoptera frugiperda | second | PxMNPV | I | LdMNPV | II | no | ND | decreased LC50 | synergistic | [66] |
| Spodoptera frugiperda | second | RoMNPV | I | LdMNPV | II | no | ND | decreased LC50 | synergistic | [66] |
| Rachiplusia nu | fourth | AcMNPV | I | RanuNPV | I | yes | reduced | slightly increased mortality | ND ** | [69] |
3.2. Amount of Each Virus OB in the Viral Mix
3.3. Effects of Administering NPV and GV at Different Times
3.4. Selection of Larval Instar to Evaluate Synergism
3.5. Mathematical Determination of the Interaction Effect
3.6. Progeny Analysis and Diagnostics
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ODV | Occlusion-derived viruses |
| OB | Occlusion body |
| BV | Budded virus |
| NPV | Nucleopolyhedrovirus |
| GV | Granulovirus |
| MTD | Mean time to death |
| LD10 | Lethal dose 10 |
| LD25 | Lethal dose 25 |
| LD50 | Lethal dose 50 |
| LD95 | Lethal dose 95 |
| LC50 | Lethal concentration 50 |
| LT50 | Lethal time 50 |
| ID50 | Infectious dose 50 |
| dpi | Days post-infection |
| GVP | Granulovirus proteins |
| VEF | Viral enhancing factor |
| PM | Peritrophic membrane |
| CM | Calculated mortality |
| EM | Expected mortality |
| RP | Relative potency |
| PCR | Polymerase chain reaction |
| qPCR | Quantitative polymerase chain reaction |
| df | Degree of freedom |
| ANOVA | Analysis of variance |
| ND | Not determined |
References
- DaPalma, T.; Doonan, B.P.; Trager, N.M.; Kasman, L.M. A Systematic Approach to Virus–Virus Interactions. Virus Res. 2010, 149, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Muñoz, S.L. Viral Co-infection Is Shaped by Host Ecology and Virus–Virus Interactions across Diverse Microbial Taxa and Environments. Virus Evol. 2017, 3, vex011. [Google Scholar] [CrossRef] [PubMed]
- Cobián Güemes, A.G.; Youle, M.; Cantú, V.A.; Felts, B.; Nulton, J.; Rohwer, F. Viruses as Winners in the Game of Life. Annu. Rev. Virol. 2016, 3, 197–214. [Google Scholar] [CrossRef] [PubMed]
- Theze, J.; Takatsuka, J.; Nakai, M.; Arif, B.; Herniou, E.A. Gene Acquisition Convergence between Entomopoxviruses and Baculoviruses. Viruses 2015, 7, 1960–1974. [Google Scholar] [CrossRef] [PubMed]
- Göertz, G.P.; Vogels, C.B.F.; Geertsema, C.; Koenraadt, C.J.M.; Pijlman, G.P. Mosquito Co-Infection with Zika and Chikungunya Virus Allows Simultaneous Transmission without Affecting Vector Competence of Aedes Aegypti. PLoS Negl. Trop. Dis. 2017, 11, e0005654. [Google Scholar] [CrossRef] [PubMed]
- Hess, R.T.; Summers, M.D.; Falcon, L.A. A Mixed Virus Infection in Midgut Cells of Autographa californica and Trichoplusia Ni Larvae. J. Ultrastruct. Res. 1978, 65, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.W.; Lynn, D.E. Baculovirus Interactions In Vitro and In Vivo. Adv. Appl. Microbiol. 2009, 68, 217–239. [Google Scholar] [CrossRef]
- Otálora, P.C.; Rivero, L.V. Interacciones de los Virus Entomopatógenos y su Efecto sobre la Actividad Biológica. Rev. Fac. Cienc. Básicas 2011, 7, 220–239. [Google Scholar] [CrossRef]
- Williams, T.; Virto, C.; Murillo, R.; Caballero, P. Covert Infection of Insects by Baculoviruses. Front. Microbiol. 2017, 8, 1337. [Google Scholar] [CrossRef]
- Szewczyk, B.; Moscardi, F.; de Castro, M.E.B.; de Souza, M.L.; Moscardi, M.L. Baculovirus Biopesticides; INTECH Open Access Publisher: London, UK, 2011; ISBN 978-953-307-532-7. [Google Scholar]
- Moscardi, F. Assessment of the Application of Baculoviruses for Control of Lepidoptera. Annu. Rev. Entomol. 1999, 44, 257–289. [Google Scholar] [CrossRef]
- Blissard, G.W.; Theilmann, D.A. Baculovirus Entry and Egress from Insect Cells. Annu. Rev. Virol. 2018, 5, 113–139. [Google Scholar] [CrossRef] [PubMed]
- Rohrmann, G.F. Baculovirus Molecular Biology, 4th ed.; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2019.
- Harrison, R.L.; Herniou, E.A.; Jehle, J.A.; Theilmann, D.A.; Burand, J.P.; Becnel, J.J.; Krell, P.J.; van Oers, M.M.; Mowery, J.D.; Bauchan, G.R.; et al. ICTV Virus Taxonomy Profile: Baculoviridae. J. Gen. Virol. 2018, 99, 1185–1186. [Google Scholar] [CrossRef] [PubMed]
- Miele, S.A.B.; Garavaglia, M.J.; Belaich, M.N.; Ghiringhelli, P.D. Baculovirus: Molecular Insights on Their Diversity and Conservation. Int. J. Evol. Biol. 2011, 2011, 15. [Google Scholar] [CrossRef] [PubMed]
- Koppenhöfer, A.M.; Kaya, H.K. Additive and Synergistic Interaction between Entomopathogenic Nematodes AndBacillus Thuringiensisfor Scarab Grub Control. Biol. Control 1997, 8, 131–137. [Google Scholar] [CrossRef]
- Levine, S.L.; Borgert, C.J. Review and Recommendations on Criteria to Evaluate the Relevance of Pesticide Interaction Data for Ecological Risk Assessments. Chemosphere 2018, 209, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Hackett, K.J.; Boore, A.; Deming, C.; Buckley, E.; Camp, M.; Shapiro, M. Helicoverpa Armigera Granulovirus Interference with Progression of H. Zea Nucleopolyhedrovirus Disease in H. Zea Larvae. J. Invertebr. Pathol. 2000, 75, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Paillot, A. Nouveau Type de Maladies a Polyedres Ou Polyedries Observe Chez Les Chenilles d’Euxoa(Agrotis) Segetum Schiff. Compt. Rend. Acad. Sci. 1936, 202, 254–256. [Google Scholar]
- Steinhaus, E.A. New Records of Insect-Virus Diseases. Hilgardia 1957, 26, 417–430. [Google Scholar] [CrossRef][Green Version]
- Tanada, Y. Description and Characteristics of a Granulosis Virus of the Imported Cabbageworm. Proc. Hawaii. Entomol. Soc. 1953, XV, 235–260. [Google Scholar]
- Tanada, Y. Some Factors Affecting the Susceptibility of the Armyworm to Virus Infections. J. Econ. Entomol. 1956, 49, 52–57. [Google Scholar] [CrossRef]
- Tanada, Y. Synergism between Two Viruses of the Armyworm, Pseudaletia Unipuncta (Haworth) (Lepidoptera: Noctuidae). J. Insect. Pathol. 1959, 1, 215–231. [Google Scholar]
- Cuartas-Otálora, P.E.; Gómez-Valderrama, J.A.; Ramos, A.E.; Barrera-Cubillos, G.P.; Villamizar-Rivero, L.F. Bio-Insecticidal Potential of Nucleopolyhedrovirus and Granulovirus Mixtures to Control the Fall Armyworm Spodoptera Frugiperda (J.E. Smith, 1797) (Lepidoptera: Noctuidae). Viruses 2019, 11, 684. [Google Scholar] [CrossRef] [PubMed]
- Barrera, G.P.; Villamizar, L.F.; Araque, G.A.; Gómez, J.A.; Guevara, E.J.; Cerrudo, C.S.; Belaich, M.N. Natural Co-infection between Novel Species of Baculoviruses in Spodoptera Ornithogalli Larvae. Viruses 2021, 13, 2520. [Google Scholar] [CrossRef] [PubMed]
- Bird, F.T. Polyhedrosis and Granulosis Viruses Causing Single and Double Infections in the Spruce Budworm, Choristoneura Fumiferana Clemens. J. Insect Pathol. 1959, 1, 406–430. [Google Scholar]
- Lowe, R.E.; Paschke, J.D. Simultaneous Infection with the Nucleopolyhedrosis and Granulosis Viruses of Trichoplusia Ni. J. Invertebr. Pathol. 1968, 12, 86–92. [Google Scholar] [CrossRef]
- Whitlock, V.H. Simultaneous Treatments of Heliothis Armigera with a Nuclear Polyhedrosis and a Granulosis Virus. J. Invertebr. Pathol. 1977, 29, 297–303. [Google Scholar] [CrossRef]
- Lowe, R.E.; Paschke, J.D. Pathology of a Double Viral Infection of Trichoplusia Ni. J. Invertebr. Pathol. 1968, 12, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Paschke, J.D.; Lowe, R.E.; Giese, R.L. Bioassay of the Nucleopolyhedrosis and Granulosis Viruses of Trichoplusia Ni. J. Invertebr. Pathol. 1968, 10, 327–334. [Google Scholar] [CrossRef]
- Jeyarani, S.; Karuppuchamy, P. Investigations on the Enhancing Efficacy of Granulovirus on Nucleopolyhedrovirus of Helicoverpa Armigera (Hubner). J. Biopestic. 2010, 3, 172–176. [Google Scholar]
- Goto, C. Enhancement of a Nuclear Polyhedrosis Virus (NPV) Infection by a Granulosis Virus (GV) Isolated from the Spotted Cutworm, Xestia c-Nigrum L.: Lepidoptera: Noctuidae. Appl. Entomol. Zool. 1990, 25, 135–137. [Google Scholar] [CrossRef]
- Hatem, A.E.; Shalaby, H.; Fargalla, F.; Vargas-Osuna, E. Combination Effects of Spodoptera Littoralis Nuclear Polyhedrosis and Granulous Virus against Larvae of the Cotton Leafworm. World Rural Obs. 2012, 4, 10–16. [Google Scholar]
- Wennmann, J.T.; Köhler, T.; Gueli Alletti, G.; Jehle, J.A. Mortality of Cutworm Larvae Is Not Enhanced by Agrotis segetum Granulovirus and Agrotis segetum Nucleopolyhedrovirus B Coinfection Relative to Single Infection by Either Virus. Appl. Environ. Microbiol. 2015, 81, 2893–2899. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tanada, Y.; Hukuhara, T. Enhanced Infection of a Nuclear-Polyhedrosis Virus in Larvae of the Armyworm, Pseudaletia Unipuncta, by a Factor in the Capsule of a Granulosis Virus. J. Invertebr. Pathol. 1971, 17, 116–126. [Google Scholar] [CrossRef]
- Hukuhara, T.; Tamura, K.; Zhu, Y.; Abe, H.; Tanada, Y. Synergistic Factor Shows Specificity in Enhancing Nuclear Polyhedrosis Virus Infections. Appl. Entomol. Zool. 1987, 22, 235–236. [Google Scholar] [CrossRef][Green Version]
- Shapiro, M. Effect of Two Granulosis Viruses on the Activity of the Gypsy Moth (Lepidoptera: Lymantriidae) Nuclear Polyhedrosis Virus. J. Econ. Entomol. 2000, 93, 1633–1637. [Google Scholar] [CrossRef] [PubMed]
- Derksen, A.C.; Granados, R.R. Alteration of a Lepidopteran Peritrophic Membrane by Baculoviruses and Enhancement of Viral Infectivity. Virology 1988, 167, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Lara-Reyna, J.; Del Rincon-Castro, M.C.; Ibarra, J.E. Synergism between the Nucleopolyhedroviruses of Autographa californica and Trichoplusia Ni. Acta Virol. 2003, 47, 189–194. [Google Scholar] [PubMed]
- Guo, H.F.; Fang, J.C.; Wang, J.P.; Zhong, W.F.; Liu, B.S. Interaction of Xestia C-Nigrum Granulovirus with Peritrophic Matrix and Spodoptera Litura Nucleopolyhedrovirus in Spodoptera Litura. J. Econ. Entomol. 2007, 100, 20–25. [Google Scholar] [CrossRef]
- Biedma, M.E.; Salvador, R.; Ferrelli, M.L.; Sciocco-Cap, A.; Romanowski, V. Effect of the Interaction between Anticarsia Gemmatalis Multiple Nucleopolyhedrovirus and Epinotia Aporema Granulovirus, on A. Gemmatalis (Lepidoptera: Noctuidae) Larvae. Biol. Control 2015, 91, 17–21. [Google Scholar] [CrossRef]
- Jeyarani, S.; Sathiah, N.; Karuppuchamy, P. In Vivo Enhancement of Nucleopolyhedrovirus Infection in Helicoverpa Armigera (Hübner) by the Granulovirus of Spodoptera Litura Fabricius. J. Biol. Control 2012, 26, 234–239. [Google Scholar]
- Mukawa, S.; Goto, C. Enhancement of Nucleopolyhedrovirus Infectivity against Mamestra Brassicae (Lepidoptera: Noctuidae) by Proteins Derived from Granulovirus and a Fluorescent Brightener. J. Econ. Entomol. 2007, 100, 1075–1083. [Google Scholar] [CrossRef] [PubMed]
- Mukawa, S.; Goto, C. In Vivo Characterization of Two Granuloviruses in Larvae of Mythimna separata (Lepidoptera: Noctuidae). J. Gen. Virol. 2008, 89, 915–921. [Google Scholar] [CrossRef] [PubMed]
- Mukawa, S.; Goto, C. Mamestra Brassicae Nucleopolyhedrovirus Infection and Enhancing Effect of Proteins Derived from Xestia c-Nigrum Granulovirus in Larvae of Mamestra Brassicae and Helicoverpa Armigera (Lepidoptera: Noctuidae) on Cabbage. J. Econ. Entomol. 2010, 103, 257–264. [Google Scholar] [CrossRef]
- Goto, C.; Mukawa, S.; Mitsunaga, T. Two Year Field Study to Evaluate the Efficacy of Mamestra Brassicae Nucleopolyhedrovirus Combined with Proteins Derived from Xestia C-Nigrum Granulovirus. Viruses 2015, 7, 1062–1078. [Google Scholar] [CrossRef] [PubMed]
- Mukawa, S.; Goto, C. Enhancing Effect of Proteins Derived from Xestia C-Nigrum Granulovirus on Mamestra Brassicae Nucleopolyhedrovirus Infection in Larvae of Autographa Nigrisigna (Lepidoptera: Noctuidae) on Cabbage. Appl. Entomol. Zool. 2011, 46, 55–63. [Google Scholar] [CrossRef]
- Tanada, Y.; Himeno, M.; Omi, E.M. Isolation of a Factor, from the Capsule of a Granulosis Virus, Synergistic for a Nuclear-Polyhedrosis Virus of the Armyworm. J. Invertebr. Pathol. 1973, 21, 31–40. [Google Scholar] [CrossRef]
- Hara, S.; Tanada, Y.; Omi, E.M. Isolation and Characterization of a Synergistic Enzyme from the Capsule of a Granulosis Virus of the Armyworm, Pseudaletia Unipuncta. J. Invertebr. Pathol. 1976, 27, 115–124. [Google Scholar] [CrossRef]
- Gallo, L.G.; Corsaro, B.G.; Hughes, P.R.; Granados, R.R. In Vivo Enhancement of Baculovirus Infection by the Viral Enhancing Factor of a Granulosis Virus of the Cabbage Looper, Trichoplusia Ni (Lepidoptera: Noctuidae). J. Invertebr. Pathol. 1991, 58, 203–210. [Google Scholar] [CrossRef]
- Lepore, L.S.; Roelvink, P.R.; Granados, R.R. Enhancin, the Granulosis Virus Protein That Facilitates Nucleopolyhedrovirus (NPV) Infections, Is a Metalloprotease. J. Invertebr. Pathol. 1996, 68, 131–140. [Google Scholar] [CrossRef]
- Wang, P.; Granados, R.R. An Intestinal Mucin Is the Target Substrate for a Baculovirus Enhancin. Proc. Natl. Acad. Sci. USA 1997, 94, 6977–6982. [Google Scholar] [CrossRef]
- Tanada, Y. A Synopsis of Studies on the Synergistic Property of an Insect Baculovirus: A Tribute to Edward A. Steinhaus. J. Invertebr. Pathol. 1985, 45, 125–138. [Google Scholar] [CrossRef]
- Hukuhara, T.; Zhu, Y. Enhancement of the in Vitro Infectivity of a Nuclear Polyhedrosis Virus by a Factor in the Capsule of a Granulosis Virus. J. Invertebr. Pathol. 1989, 54, 71–78. [Google Scholar] [CrossRef]
- Kozuma, K.; Hukuhara, T. Fusion Characteristics of a Nuclear Polyhedrosis Virus in Cultured Cells: Time Course and Effect of a Synergistic Factor and PH. J. Invertebr. Pathol. 1994, 63, 63–67. [Google Scholar] [CrossRef]
- Slavicek, J.M.; Popham, H.J. The Lymantria Dispar Nucleopolyhedrovirus Enhancins Are Components of Occlusion-Derived Virus. J. Virol. 2005, 79, 10578–10588. [Google Scholar] [CrossRef]
- Slavicek, J.M. Baculovirus Enhancins and Their Role in Viral Pathogenicity. In Molecular Virology; INTECH Open Access Publisher: London, UK, 2012. [Google Scholar] [CrossRef]
- Cuartas, P.; Barrera, G.; Belaich, M.; Barreto, E.; Ghiringhelli, P.; Villamizar, L. The Complete Sequence of the First Spodoptera Frugiperda Betabaculovirus Genome: A Natural Multiple Recombinant Virus. Viruses 2015, 7, 394–421. [Google Scholar] [PubMed]
- Ferrelli, M.L.; Pidre, M.L.; Ghiringhelli, P.D.; Torres, S.; Fabre, M.L.; Masson, T.; Cédola, M.T.; Sciocco-Cap, A.; Romanowski, V. Genomic Analysis of an Argentinean Isolate of Spodoptera Frugiperda Granulovirus Reveals That Various Baculoviruses Code for Lef-7 Proteins with Three F-Box Domains. PLoS ONE 2018, 13, e0202598. [Google Scholar] [CrossRef] [PubMed]
- Ritter, K.S.; Tanada, Y. Interference between Two Nuclear Polyhedrosis Viruses of the Armyworm, Pseudaletia Unipuncta [Lep.: Noctuidae]. Entomophaga 1978, 23, 349–359. [Google Scholar] [CrossRef]
- Del Rincon-Castro, M.C.; Ibarra, J.E. Caracterización de Cepas Silvestres de Virus de Poliedrosis Nuclear Aisladas de TRichoplusia Ni (Lepidoptera: Noctuidae) En El Centro de Mexico. Vedalia 1995, 2, 7–15. [Google Scholar]
- Willis, L.G.; Seipp, R.; Stewart, T.M.; Erlandson, M.A.; Theilmann, D.A. Sequence Analysis of the Complete Genome of Trichoplusia Ni Single Nucleopolyhedrovirus and the Identification of a Baculoviral Photolyase Gene. Virology 2005, 338, 209–226. [Google Scholar] [CrossRef]
- Cheng, X.W.; Carner, G.R.; Lange, M.; Jehle, J.A.; Arif, B.M. Biological and Molecular Characterization of a Multicapsid Nucleopolyhedrovirus from Thysanoplusia Orichalcea (L.) (Lepidoptera: Noctuidae). J. Invertebr. Pathol. 2005, 88, 126–135. [Google Scholar] [CrossRef]
- Lauzon, H.A.; Jamieson, P.B.; Krell, P.J.; Arif, B.M. Gene Organization and Sequencing of the Choristoneura Fumiferana Defective Nucleopolyhedrovirus Genome. J. Gen. Virol. 2005, 86, 945–961. [Google Scholar] [CrossRef] [PubMed]
- de Jong, J.G.; Lauzon, H.A.; Dominy, C.; Poloumienko, A.; Carstens, E.B.; Arif, B.M.; Krell, P.J. Analysis of the Choristoneura Fumiferana Nucleopolyhedrovirus Genome. J. Gen. Virol. 2005, 86, 929–943. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, M.S. The Gypsy Moth (Lepidoptera: Lymantriidae) Nucleopolyhedrovirus as a Synergist for Baculoviruses against Beet Armyworm, Fall Armyworm and Corn Earworm (Lepidoptera: Noctuidae). J. Agric. Urban Entomol. 2006, 23, 243–251. [Google Scholar]
- Rodríguez, V.A.; Belaich, M.N.; Quintana, G.; Cap, A.S.; Ghiringhelli, P.D. Isolation and Characterization of a Nucleopolyhedrovirus from Rachiplusia Nu (Guenée) (Lepidoptera: Noctuidae). Int. J. Virol. Mol. Biol. 2012, 1, 28–34. [Google Scholar]
- Jakubowicz, V.; Taibo, C.B.; Sciocco-Cap, A.; Arneodo, J.D. Biological and Molecular Characterization of Rachiplusia Nu Single Nucleopolyhedrovirus, a Promising Biocontrol Agent against the South American Soybean Pest Rachiplusia Nu. J. Invertebr. Pathol. 2019, 166, 107211. [Google Scholar] [CrossRef] [PubMed]
- Decker-Franco, C.; Taibo, C.B.; Di Rienzo, J.A.; Alfonso, V.; Arneodo, J.D. Comparative Pathogenesis of Generalist AcMNPV and Specific RanuNPV in Larvae of Rachiplusia Nu (Lepidoptera: Noctuidae) Following Single and Mixed Inoculations. J. Econ. Entomol. 2021, 114, 1009–1014. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Fang, W.; Fan, R.; Zhang, L.; Lei, C.; Zhang, J.; Nian, W.; Dou, T.; An, S.; Zhou, L.; et al. Granulovirus GP37 Facilitated ODVs Cross Insect Peritrophic Membranes and Fuse with Epithelia. Toxins 2019, 11, 145. [Google Scholar] [CrossRef]
- Gross, C.H.; Wolgamot, G.M.; Russell, R.L.; Pearson, M.N.; Rohrmann, G.F. A 37-Kilodalton Glycoprotein from a Baculovirus of Orgyia Pseudotsugata Is Localized to Cytoplasmic Inclusion Bodies. J. Virol. 1993, 67, 469–475. [Google Scholar] [CrossRef]
- Liu, X.; Yang, G.; Qiu, B.; Tian, P. Molecular cloning of enhancin gene from Helicoverpa armigera granulosis virus and its expression in E. coli. Wei Sheng Wu Xue Bao 2000, 40, 379–383. [Google Scholar]
- Masson, T.; Fabre, M.L.; Ferrelli, M.L.; Pidre, M.L.; Romanowski, V. Protein Composition of the Occlusion Bodies of Epinotia Aporema Granulovirus. PLoS ONE 2019, 14, e0207735. [Google Scholar] [CrossRef]
- Wang, M.; Hu, Z. Cross-Talking between Baculoviruses and Host Insects towards a Successful Infection. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180324. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, N.; Setoyama, Y.; Chiba, M.; Kimata, K.; Watanabe, H. Baculovirus Envelope Protein ODV-E66 Is a Novel Chondroitinase with Distinct Substrate Specificity. J. Biol. Chem. 2011, 286, 29026–29034. [Google Scholar] [CrossRef] [PubMed]
- Hou, D.; Kuang, W.; Luo, S.; Zhang, F.; Zhou, F.; Chen, T.; Zhang, Y.; Wang, H.; Hu, Z.; Deng, F.; et al. Baculovirus ODV-E66 Degrades Larval Peritrophic Membrane to Facilitate Baculovirus Oral Infection. Virology 2019, 537, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Hawtin, R.E.; Arnold, K.; Ayres, M.D.; Zanotto, P.M.; Howard, S.C.; Gooday, G.W.; Chappell, L.H.; Kitts, P.A.; King, L.A.; Possee, R.D. Identification and Preliminary Characterization of a Chitinase Gene in the Autographa californica Nuclear Polyhedrosis Virus Genome. Virology 1995, 212, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Ishimwe, E.; Hodgson, J.J.; Clem, R.J.; Passarelli, A.L. Reaching the Melting Point: Degradative Enzymes and Protease Inhibitors Involved in Baculovirus Infection and Dissemination. Virology 2015, 479–480, 637–649. [Google Scholar] [CrossRef]
- Eberle, K.E.; Wennmann, J.T.; Kleespies, R.G.; Jehle, A. Basic Techniques in Insect Virology. In Manual of Techniques in Invertebrate Pathology, 2nd ed.; Lacey, L.A., Ed.; Academic Press: London, UK, 2012; pp. 15–74. [Google Scholar]
- Webb, R.E.; Shapiro, M.; Thorpe, K.W.; Peiffer, R.A.; Fuester, R.W.; Valenti, M.A.; White, G.B.; Podgwaite, J.D. Potentiation by a Granulosis Virus of Gypchek, the Gypsy Moth (Lepidoptera: Lymantriidae) Nuclear Polyhedrosis Virus Product. J. Entomol. Sci. 2001, 36, 169–176. [Google Scholar] [CrossRef]
- Tanada, Y.; Hukuhara, T. A Nonsynergistic Strain of a Granulosis Virus of the Armyworm, Pseudaletia Unipuncta. J. Invertebr. Pathol. 1968, 12, 263–268. [Google Scholar] [CrossRef]
- Busvine, J.R. A Critical Review of the Techniques for Testing Insecticides. In A Critical Review of the Techniques for Testing Insecticides, 2nd ed.; Commonwealth Institute of Entomology: London, UK, 1971. [Google Scholar]
- Wennmann, J.T.; Jehle, J.A. Detection and Quantitation of Agrotis Baculoviruses in Mixed Infections. J. Virol. Methods 2014, 197, 39–46. [Google Scholar] [CrossRef]
- Espinel-Correal, C.; Lopez-Ferber, M.; Zeddam, J.L.; Villamizar, L.; Gomez, J.; Cotes, A.M.; Lery, X. Experimental Mixtures of Phthorimaea Operculella Granulovirus Isolates Provide High Biological Efficacy on Both Phthorimaea Operculella and Tecia Solanivora (Lepidoptera: Gelechiidae). J. Invertebr. Pathol. 2012, 110, 375–381. [Google Scholar] [CrossRef]
- Fan, J.; Wennmann, J.T.; Wang, D.; Jehle, J.A. Single Nucleotide Polymorphism (SNP) Frequencies and Distribution Reveal Complex Genetic Composition of Seven Novel Natural Isolates of Cydia Pomonella Granulovirus. Virology 2020, 541, 32–40. [Google Scholar] [CrossRef]
- Hinsberger, A.; Blachère-Lopez, C.; Knox, C.; Moore, S.; Marsberg, T.; Lopez-Ferber, M. CpGV-M Replication in Type I Resistant Insects: Helper Virus and Order of Ingestion Are Important. Viruses 2021, 13, 1695. [Google Scholar] [CrossRef] [PubMed]
- Jehle, J.A.; Fritsch, E.; Huber, J.; Backhaus, H. Intra-Specific and Inter-Specific Recombination of Tortricid-Specific Granuloviruses during Co-Infection in Insect Larvae. Arch. Virol. 2003, 148, 1317–1333. [Google Scholar] [CrossRef] [PubMed]
- Sciocco-Cap, A.; Parola, A.D.; Goldberg, A.V.; Ghiringhelli, P.D.; Romanowski, V. Characterization of a Granulovirus Isolated from Epinotia Aporema Wals. (Lepidoptera: Tortricidae) Larvae. Appl. Env. Microbiol. 2001, 67, 3702–3706. [Google Scholar] [CrossRef] [PubMed]
- Ferrelli, M.L.; Salvador, R.; Biedma, M.E.; Berretta, M.F.; Haase, S.; Sciocco-Cap, A.; Ghiringhelli, P.D.; Romanowski, V. Genome of Epinotia Aporema Granulovirus (EpapGV), a Polyorganotropic Fast Killing Betabaculovirus with a Novel Thymidylate Kinase Gene. BMC Genom. 2012, 13, 548. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.; Cory, J.S.; Theilmann, D.A.; Myers, J.H. Nucleopolyhedroviruses of Forest and Western Tent Caterpillars: Cross-Infectivity and Evidence for Activation of Latent Virus in High-Density Field Populations. Ecol. Entomol. 2003, 28, 41–50. [Google Scholar] [CrossRef]
- Kouassi, L.N.; Tsuda, K.; Goto, C.; Mukawa, S.; Sakamaki, Y.; Kusigemati, K.; Nakamura, M. Prevalence of Latent Virus in Spodoptera Litura (Fabricius) (Lepidoptera: Noctuidae) and Its Activation by a Heterologous Virus. Appl. Entomol. Zool. 2009, 44, 95–102. [Google Scholar] [CrossRef]
- Lacey, L.A.; Grzywacz, D.; Shapiro-Ilan, D.I.; Frutos, R.; Brownbridge, M.; Goettel, M.S. Insect Pathogens as Biological Control Agents: Back to the Future. J. Invertebr. Pathol. 2015, 132, 1–41. [Google Scholar] [CrossRef]
- Haase, S.; Sciocco-Cap, A.; Romanowski, V. Baculovirus Insecticides in Latin America: Historical Overview, Current Status and Future Perspectives. Viruses 2015, 7, 2230–2267. [Google Scholar] [CrossRef]
- Williams, T.; López-Ferber, M.; Caballero, P. Nucleopolyhedrovirus Coocclusion Technology: A New Concept in the Development of Biological Insecticides. Front. Microbiol. 2022, 12, 810026. [Google Scholar] [CrossRef]
- Arrizubieta, M.; Simón, O.; Torres-Vila, L.M.; Figueiredo, E.; Mendiola, J.; Mexia, A.; Caballero, P.; Williams, T. Insecticidal Efficacy and Persistence of a Co-Occluded Binary Mixture of Helicoverpa Armigera Nucleopolyhedrovirus (HearNPV) Variants in Protected and Field-Grown Tomato Crops on the Iberian Peninsula. Pest Manag. Sci. 2016, 72, 660–670. [Google Scholar] [CrossRef]
- Arrizubieta, M.; Simón, O.; Williams, T.; Caballero, P. A Novel Binary Mixture of Helicoverpa Armigera Single Nucleopolyhedrovirus Genotypic Variants Has Improved Insecticidal Characteristics for Control of Cotton Bollworms. Appl. Environ. Microbiol. 2015, 81, 3984–3993. [Google Scholar] [CrossRef]
- Beperet, I.; Simón, O.; López-Ferber, M.; van Lent, J.; Williams, T.; Caballero, P. Mixtures of Insect-Pathogenic Viruses in a Single Virion: Towards the Development of Custom-Designed Insecticides. Appl. Environ. Microbiol. 2021, 87, e02180-20. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrelli, M.L.; Salvador, R. Effects of Mixed Baculovirus Infections in Biological Control: A Comprehensive Historical and Technical Analysis. Viruses 2023, 15, 1838. https://doi.org/10.3390/v15091838
Ferrelli ML, Salvador R. Effects of Mixed Baculovirus Infections in Biological Control: A Comprehensive Historical and Technical Analysis. Viruses. 2023; 15(9):1838. https://doi.org/10.3390/v15091838
Chicago/Turabian StyleFerrelli, María Leticia, and Ricardo Salvador. 2023. "Effects of Mixed Baculovirus Infections in Biological Control: A Comprehensive Historical and Technical Analysis" Viruses 15, no. 9: 1838. https://doi.org/10.3390/v15091838
APA StyleFerrelli, M. L., & Salvador, R. (2023). Effects of Mixed Baculovirus Infections in Biological Control: A Comprehensive Historical and Technical Analysis. Viruses, 15(9), 1838. https://doi.org/10.3390/v15091838