Clinical Profile of SARS-CoV-2 Infection: Mechanisms of the Cellular Immune Response and Immunogenetic Markers in Patients from Brazil
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Participants
2.2. Data Collection of COVID-19 Patients
2.3. Inclusion and Exclusion Criteria
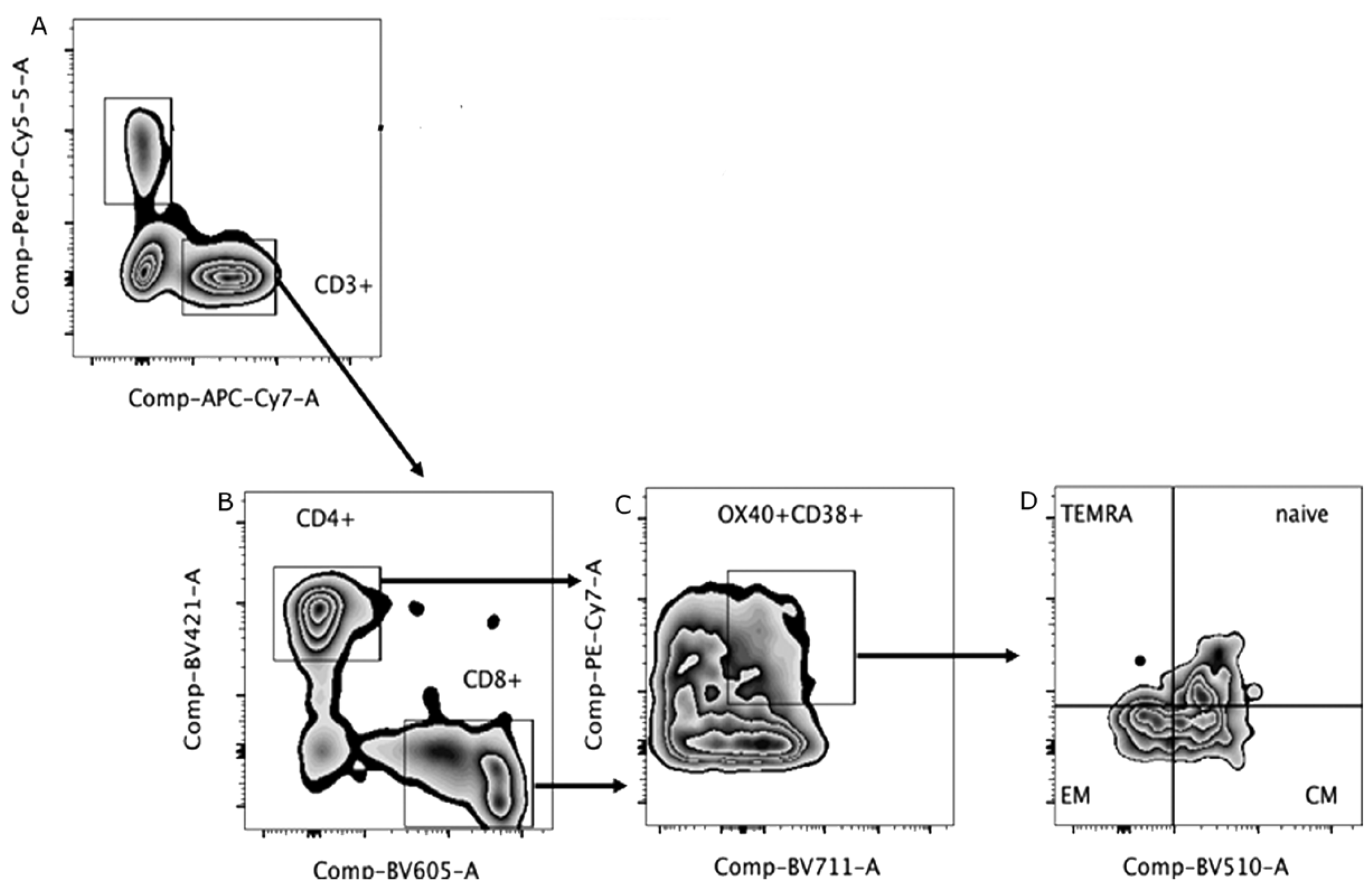
2.4. Immunophenotyping
2.5. Cytokine Detection
2.5.1. Immunospot Assay
2.5.2. Multiplex Micro Array
2.6. Statistical Analysis
3. Results
3.1. Investigation
| Medical Conditions | Severe | Non-Severe | Total | ||||
|---|---|---|---|---|---|---|---|
| (N = 31) | (N = 42) | (N = 73) | |||||
| n | % | N | % | n | % | p-Value * | |
| Diabetes Mellitus | 0.016 | ||||||
| Yes | 11 | 35.5 | 4 | 9.5 | 15 | 20.5 | |
| No | 20 | 64.5 | 38 | 90.5 | 58 | 79.5 | |
| Hipertension | 0.015 | ||||||
| Yes | 16 | 51.6 | 9 | 21.4 | 25 | 34.2 | |
| No | 15 | 48.4 | 33 | 78.6 | 48 | 65.8 | |
| Obesity | 0.8524 | ||||||
| Yes | 5 | 16.1 | 5 | 11.9 | 10 | 13.7 | |
| No | 26 | 83.9 | 37 | 88.1 | 63 | 86.3 | |
| Smoking (currently) | - | ||||||
| Yes | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | |
| No | 30 | 96.8 | 42 | 100.0 | 72 | 98.6 | |
| Unknown | 1 | 3.2 | 0 | 0.0 | 1 | 1.4 | |
| Ex-smoking | 0.3358 | ||||||
| Yes | 7 | 22.6 | 5 | 11.9 | 12 | 16.4 | |
| No | 23 | 74.2 | 37 | 88.1 | 60 | 82.2 | |
| Unknown | 1 | 3.2 | 0 | 0.0 | 1 | 1.4 | |
| Substance abuse ou misuse | - | ||||||
| Yes | 1 | 3.2 | 0 | 0.0 | 1 | 1.4 | |
| No | 30 | 96.8 | 42 | 100.0 | 72 | 98.6 | |
| Special Needs/Deficiency | - | ||||||
| Yes | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | |
| No | 31 | 100.0 | 42 | 100.0 | 73 | 100.0 | |
| Cardiovascular Disease | 0.7726 | ||||||
| Yes | 2 | 6.5 | 1 | 2.4 | 3 | 4.1 | |
| No | 29 | 93.5 | 41 | 97.6 | 70 | 95.9 | |
| Chronic Kidney Disease | - | ||||||
| Yes | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | |
| No | 31 | 100.0 | 42 | 100.0 | 73 | 100.0 | |
| Chronic Liver Disease | - | ||||||
| Yes | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | |
| No | 31 | 100.0 | 42 | 100.0 | 73 | 100.0 | |
| Chronic Lung Disease | 1000 | ||||||
| Yes | 1 | 3.2 | 1 | 2.4 | 2 | 2.7 | |
| No | 30 | 96.8 | 41 | 97.6 | 71 | 97.3 | |
| Pulmonary tuberculosis being treated | - | ||||||
| Yes | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | |
| No | 31 | 100.0 | 42 | 100.0 | 73 | 100.0 | |
| Psicologic condiction | 0.7726 | ||||||
| Yes | 2 | 6.5 | 1 | 2.4 | 3 | 4.1 | |
| No | 29 | 93.5 | 41 | 97.6 | 70 | 95.9 | |
| Other chronic disease | 0.7183 | ||||||
| Yes | 2 | 6.5 | 5 | 11.9 | 7 | 9.6 | |
| No | 29 | 93.5 | 37 | 88.1 | 66 | 90.4 | |
| Other condiction | - | ||||||
| Yes | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | |
| No | 31 | 100.0 | 42 | 100.0 | 73 | 100.0 | |
3.2. Laboratory Assays
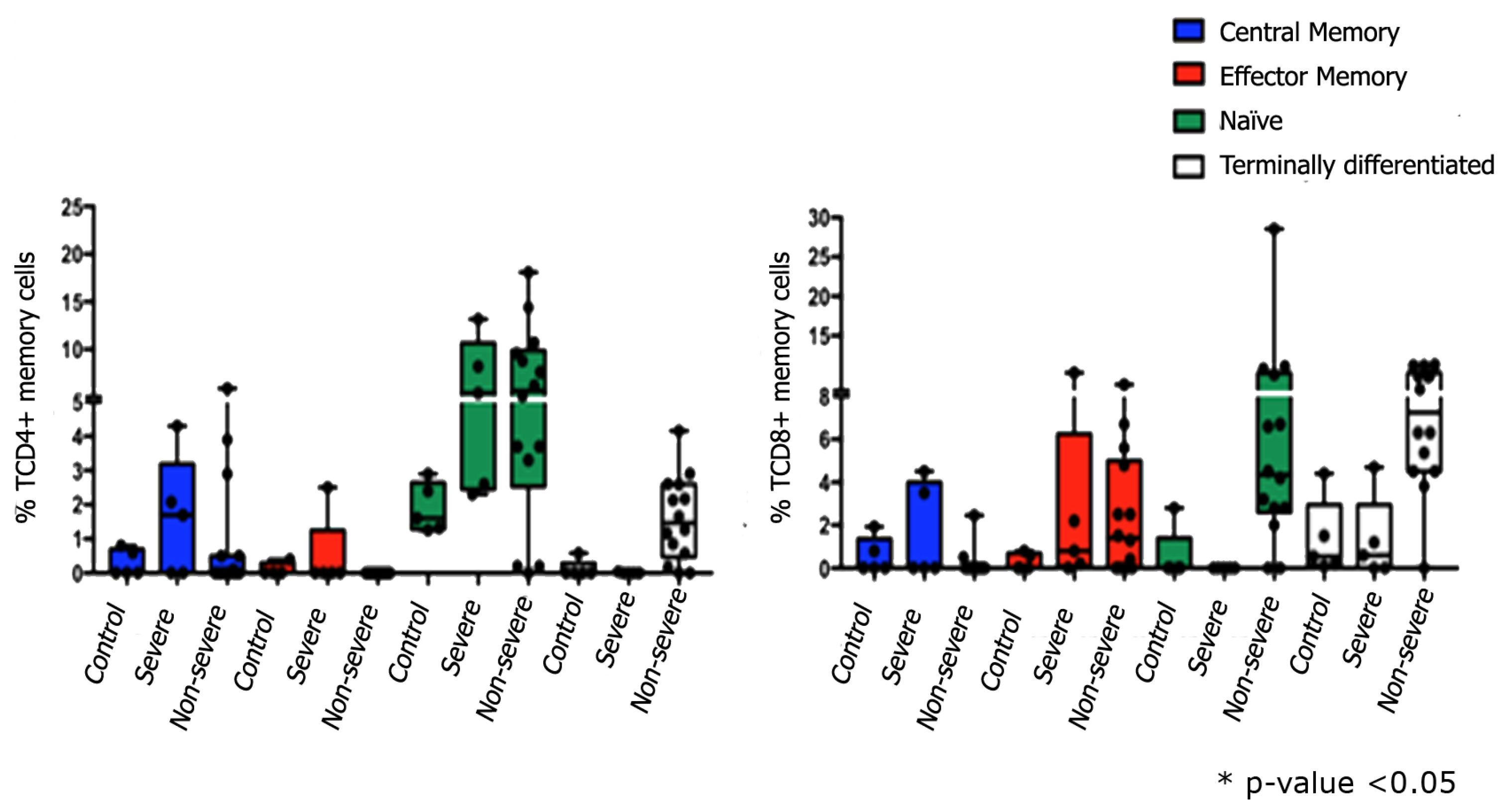
3.3. Immunophenotyping
3.4. Cytokine Detection
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sanders, J.M.; Monogue, M.L.; Jodlowski, T.Z.; Cutrell, J.B. Pharmacologic Treatments for Coronavirus Disease 2019 (COVID-19): A Review. JAMA 2020, 323, 1824–1836. [Google Scholar] [CrossRef]
- Jamilloux, Y.; Henry, T.; Belot, A.; Viel, S.; Fauter, M.; El Jammal, T.; Walzer, T.; François, B.; Sève, P. Should we stimulate or suppress immune responses in COVID-19? Cytokine and anti-cytokine interventions. Autoimmun. Rev. 2020, 19, 102567. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2019, 395, 497–506. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Asselta, R.; Paraboschi, E.M.; Mantovani, A.; Duga, S. ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID-19 severity in Italy. Aging 2020, 12, 10087–10098. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Announces COVID-19 Outbreak a Pandemic. Available online: https://www.euro.who.int/en/health-topics/health-emergencies/coronavirus-covid-19/news/news/2020/3/who-announces-covid-19-outbreak-a-pandemic (accessed on 18 June 2020).
- Straburzyński, M.; Nowaczewska, M.; Budrewicz, S.; Waliszewska-Prosół, M. COVID-19-related headache and sinonasal inflammation: A longitudinal study analysing the role of acute rhinosinusitis and ICHD-3 classification difficulties in SARS-CoV-2 infection. Cephalalgia 2022, 42, 218–228. [Google Scholar] [CrossRef]
- Fernandes, Q.; Inchakalodya, V.P.; Merhia, M.; Mestiria, S.; Taiba, N.; Moustafa Abo El-Ella, D.; Bedhiafi, T.; Raza, A.; Al-Zaidan, L.; Mohsen, M.O.; et al. Emerging COVID-19 variants and their impact on SARS-CoV-2 diagnosis, therapeutics and vacines. Ann. Med. 2022, 54, 524–540. [Google Scholar] [CrossRef]
- Ackermann, M.; Verleden, S.E.; Kuehnel, M.; Haverich, A.; Welte, T.; Laenger, F.; Vanstapel, A.; Werlein, C.; Stark, H.; Tzankov, A.; et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19. N. Engl. J. Med. 2020, 383, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Vardhana, S.A.; Wolchok, J.D. The many faces of the anti-COVID immune response. J. Exp. Med. 2020, 217, e20200678. [Google Scholar] [CrossRef]
- Saghazadeh, A.; Rezaei, N. Immune-epidemiological parameters of the novel coronavirus—A perspective. Exp. Rev. Clin. Immunol. 2020, 16, 465–470. [Google Scholar] [CrossRef]
- Mahmudpour, M.; Roozbeh, J.; Keshavarz, M.; Farrokhi, S.; Nabipour, I. COVID-19 cytokine storm: The anger of inflammation. Cytokine 2020, 133, 155151. [Google Scholar] [CrossRef] [PubMed]
- Melgaço, J.G.; Brito, E.; Cunha, D.; Azamor, T.; da Silva, A.M.V.; Tubarão, L.N.; Gonçalves, R.B.; Monteiro, R.Q.; Missailidis, S.; da Costa Neves, P.C.; et al. Cellular and Molecular Immunology Approaches for the Development of Immunotherapies against the New Coronavirus (SARS-cov-2): Challenges to Near-Future Breakthroughs. J. Immunol. Res. 2020, 2020, 8827670. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Alejo, E.; Tarancon-Diez, L.; de la Sierra Espinar-Buitrago, M.; Genebat, M.; Calderón, A.; Pérez-Cabeza, G.; Magro-Lopez, E.; Leal, M.; Muñoz-Fernández, M.Á. Persistent Exhausted T-Cell Immunity after Severe COVID-19: 6-Month Evaluation in a Prospective Observational Study. J. Clin. Med 2023, 12, 3539. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Qi, T.; Liu, L.; Ling, Y.; Qian, Z.; Li, T.; Li, F.; Xu, Q.; Zhang, Y.; Xu, S.; et al. Clinical progression of patients with COVID-19 in Shanghai, China. J. Infect. 2020, 80, e1–e6. [Google Scholar] [CrossRef]
- Vabret, N.; Britton, G.J.; Gruber, C.; Hegde, S.; Kim, J.; Kuksin, M.; Levantovsky, R.; Malle, L.; Moreira, A.; Park, M.D.; et al. Immunology of COVID-19: Current State of the Science. Immunity 2020, 52, 910–941. [Google Scholar] [CrossRef]
- Echaide, M.; Labiano, I.; Delgado, M.; Fernández de Lascoiti, A.; Ochoa, P.; Garnica, M.; Ramos, P.; Chocarro, L.; Fernández, L.; Arasanz, H.; et al. Immune Profiling Uncovers Memory T-Cell Responses with a Th17 Signature in Cancer Patients with Previous SARS-CoV-2 Infection Followed by mRNA Vaccination. Cancers 2022, 14, 4464. [Google Scholar] [CrossRef]
- Melo, A.K.G.; Milby, K.M.; Caparroz, A.L.M.A.; Pinto, A.C.P.N.; Santos, R.R.P.; Rocha, A.P.; Ferreira, G.A.; Souza, V.A.; Valadares, L.D.A.; Vieira, R.M.R.A.; et al. Biomarkers of cytokine storm as red flags for severe and fatal COVID-19 cases: A living systematic review and meta-analysis. PLoS ONE 2021, 16, e0253894. [Google Scholar] [CrossRef]
- Zhang, L.; Yan, X.; Fan, Q.; Liu, H.; Liu, X.; Liu, Z.; Zhang, Z. D-dimer levels on admission to predict in-hospital mortality in patients with Covid-19. J. Thromb. Haemost. 2020, 18, 1324–1329. [Google Scholar] [CrossRef]
- Tang, N.; Li, D.; Wang, X.; Sun, Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 2020, 18, 844–847. [Google Scholar] [CrossRef]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061. [Google Scholar] [CrossRef]
- Vargas-Vargas, M.; Cortés-Rojo, C. Ferritin levels and COVID-19. Rev. Panam. De Salud Pública 2020, 44, e72. [Google Scholar] [CrossRef]
- Stringer, D.; Braude, P.; Myint, P.K.; Evans, L.; Collins, J.T.; Verduri, A.; Quinn, T.J.; Vilches-Moraga, A.; Stechman, M.J.; Pearce, L.; et al. The role of C-reactive protein as a prognostic marker in COVID-19. Int. J. Epidemiol. 2021, 50, 420–429. [Google Scholar] [CrossRef]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-cov-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501. [Google Scholar] [CrossRef]
- del Valle-Mendoza, J.; Tarazona-Castro, Y.; Merino-Luna, A.; Carrillo-Ng, H.; Kym, S.; Aguilar-Luis, M.A.; del Valle, L.J.; Aquino-Ortega, R.; Martins-Luna, J.; Peña-Tuesta, I.; et al. Comparison of cytokines levels among COVID-19 patients living at sea level and high altitude. BMC Infect. Dis. 2022, 22, 96. [Google Scholar] [CrossRef]
- Cao, W.; Li, T. COVID-19: Towards understanding of pathogenesis. Cell Res. 2020, 30, 367–369. [Google Scholar] [CrossRef] [PubMed]
- Sebbar, E.H.; Choukri, M. Interleukin 6: A biomarker for COVID-19 progression. Proceedings 2023, 72, 3351–3355. [Google Scholar] [CrossRef] [PubMed]
- Prompetchara, E.; Ketloy, C.; Palaga, T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020, 38, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Henry, B.M.; de Oliveira, M.H.S.; Benoit, S.; Plebani, M.; Lippi, G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): A meta-analysis. Clin. Chem. Lab. Med. 2020, 58, 1021–1028. [Google Scholar] [CrossRef] [PubMed]


| Laboratory Analysis | Visit 1 | Visit 2 | Visit 3 | Visit 4 | ||||
|---|---|---|---|---|---|---|---|---|
| Severe | Non Severe | Severe | Non Severe | Severe | Non Severe | Severe | Non Severe | |
| Hemoglobin (g/dL) | ||||||||
| Minimum | 8.4 | 11.3 | 7.7 | 12 | 7.9 | 11.3 | 7.4 | 11.1 |
| Maximum | 16.7 | 16.6 | 12.4 | 16.2 | 15.3 | 15.9 | 15.7 | 16.1 |
| median | 13.5 | 14 | 10.1 | 13.9 | 12.5 | 13.4 | 13.1 | 13.7 |
| Average | 13.2 | 14.3 | 10.1 | 14 | 12.5 | 13.5 | 12.8 | 13.6 |
| Standard deviation | 1.9 | 1.3 | 3.3 | 1.1 | 1.9 | 1.2 | 1.7 | 1.3 |
| Hematocrit (%) | ||||||||
| Minimum | 26.6 | 34.7 | 25.6 | 37 | 26.2 | 34.6 | 22.6 | 34.9 |
| Maximum | 49.7 | 49.9 | 37.4 | 48.5 | 45.8 | 46.8 | 45.8 | 47.2 |
| median | 40.2 | 42.6 | 31.5 | 41.7 | 37.3 | 40.1 | 39.4 | 41.1 |
| Average | 40.1 | 42.8 | 31.5 | 41.9 | 37.8 | 40.5 | 38.7 | 41.1 |
| Standard deviation | 5.3 | 3.7 | 8.3 | 3.1 | 5.2 | 3.3 | 5.1 | 3.2 |
| Global leukocytes (/μL) | ||||||||
| Minimum | 3920 | 2560 | 6730 | 3240 | 5040 | 3400 | 5100 | 3030 |
| Maximum | 18500 | 9920 | 11920 | 10740 | 19040 | 12910 | 16860 | 10720 |
| median | 9070 | 4270 | 9325 | 5240 | 10250 | 5440 | 7115 | 5695 |
| Average | 9596.1 | 4576.1 | 9325 | 5739.3 | 10326 | 5805.6 | 7567.7 | 5758.3 |
| Standard deviation | 3198.4 | 1468.1 | 3669.9 | 1915.6 | 3752.1 | 1868.1 | 2553.6 | 1539.2 |
| Lymphocytes (/μL) | ||||||||
| Minimum | 396 | 726 | 740.3 | 907.4 | 617 | 1145.5 | 1180.2 | 1080 |
| Maximum | 3045 | 2171.5 | 2264 | 3494.4 | 3590 | 3526.4 | 3787 | 3541 |
| median | 1093.3 | 1434.5 | 1502.2 | 1785 | 1663.5 | 1796 | 1983 | 1846.5 |
| Average | 1204 | 1486.2 | 1502.2 | 1788.3 | 1718.6 | 1864.3 | 2119 | 1996.3 |
| Standard deviation | 591.4 | 375.1 | 1077.4 | 536.1 | 665.9 | 493 | 659.5 | 548.2 |
| Platelets (thousand//μL) | ||||||||
| Minimum | 142 | 110 | 162 | 135 | 138 | 165 | 44 | 143 |
| Maximum | 603 | 379 | 464 | 494 | 640 | 480 | 413 | 353 |
| median | 270 | 220 | 313 | 254 | 335 | 268 | 268 | 250.5 |
| Average | 304.4 | 219.2 | 313 | 266.8 | 339.8 | 282.5 | 257.5 | 242.1 |
| Standard deviation | 105.2 | 65.2 | 213.5 | 76.7 | 122.1 | 66 | 91.4 | 51.9 |
| LDH (IU/L) | ||||||||
| Minimum | 374.9 | 138.2 | 571.8 | 135.6 | 243 | 157.2 | 238.9 | 226.6 |
| Maximum | 2460.4 | 632.7 | 830.5 | 825 | 775.9 | 557.9 | 684.9 | 449.2 |
| median | 673.2 | 344.8 | 701.2 | 348.3 | 445.3 | 321.2 | 365.7 | 310.7 |
| Average | 777.1 | 366.7 | 701.2 | 358.4 | 462 | 335.4 | 373.7 | 326.6 |
| Standard deviation | 424.7 | 95.8 | 182.9 | 118.5 | 139.7 | 71.9 | 102.3 | 57.1 |
| Alkaline Phosphatase (IU/L) | ||||||||
| Minimum | 113 | 51.7 | 134 | 91 | 113 | 88 | 127 | 86 |
| Maximum | 418 | 90 | 245 | 357 | 505 | 361 | 351 | 287 |
| median | 195 | 170.5 | 189.5 | 182 | 166 | 175 | 170 | 168 |
| Average | 206.3 | 179 | 189.5 | 183.6 | 195.4 | 189.2 | 195.6 | 175 |
| Standard deviation | 75.9 | 51.7 | 78.5 | 54.5 | 85.3 | 65 | 57.3 | 50.6 |
| TGO/AST (UI/L) | ||||||||
| Minimum | 17 | 12 | 52 | 11 | 10 | 11 | 9 | 11 |
| Maximum | 219 | 116 | 55 | 110 | 65 | 62 | 33 | 63 |
| median | 52 | 24.5 | 53.5 | 21 | 21 | 20 | 17.5 | 18 |
| Average | 63.1 | 28.9 | 53.5 | 24.8 | 27.7 | 21.8 | 18.5 | 20.5 |
| Standard deviation | 52.6 | 16.5 | 2.1 | 15.8 | 16.2 | 9.3 | 6.1 | 9.2 |
| TGP/ALT (UI/L) | ||||||||
| Minimum | 11 | 12 | 47 | 9 | 13 | 10 | 9 | 10 |
| Maximum | 691 | 271 | 113 | 355 | 254 | 272 | 97 | 60 |
| median | 72 | 34.5 | 80 | 31 | 53 | 25 | 19 | 20.5 |
| Average | 91.8 | 39.7 | 80 | 41.2 | 64.4 | 37.6 | 24.2 | 23.8 |
| Standard deviation | 121.9 | 40.2 | 46.7 | 52.7 | 56.2 | 44.6 | 17.9 | 13.1 |
| Ultrasensitive C-reactive protein (mg/L) | ||||||||
| Minimum | 2.7 | 0.4 | 17.5 | 0.4 | 1.5 | 0.1 | 0.6 | 0.1 |
| Maximum | 228.4 | 127.8 | 142.2 | 200.9 | 169.9 | 18.4 | 253.2 | 15.1 |
| median | 61 | 3.8 | 79.8 | 1.6 | 7.6 | 1.4 | 3.8 | 1.3 |
| Average | 77.2 | 12 | 79.8 | 14.3 | 20.8 | 3.1 | 16 | 2.5 |
| Standard deviation | 67.2 | 23.4 | 88.2 | 36.9 | 35.2 | 4 | 53.2 | 2.8 |
| D-dimer (ng/mL) | ||||||||
| Minimum | 30 | 1.7 | 1325 | 25 | 25 | 25 | 25 | 25 |
| Maximum | 12968 | 1244 | 6360 | 1981 | 10636 | 3671 | 3040 | 25000 |
| median | 465 | 31 | 3842.5 | 56 | 420 | 30 | 401 | 30 |
| Average | 1451.7 | 178.3 | 3842.5 | 205.2 | 1441.8 | 270.3 | 719.1 | 744.8 |
| Standard deviation | 2734 | 236.3 | 3560.3 | 328 | 2401 | 575 | 903.3 | 3936 |
| Ferritin (ng/mL) | ||||||||
| Minimum | 88.2 | 24.3 | 1062.1 | 4.2 | 80.2 | 16.7 | 25.6 | 12.3 |
| Maximum | 4225 | 1137 | 1788 | 1620.5 | 1290 | 910.3 | 2864.2 | 365.4 |
| median | 1030.5 | 169.1 | 1425.1 | 151.8 | 598.1 | 160.2 | 213 | 103.6 |
| Average | 1300.6 | 270 | 1425.1 | 292.1 | 593.1 | 262.9 | 326.2 | 130.2 |
| Standard deviation | 1115.7 | 272.2 | 513.3 | 336.8 | 346 | 247.6 | 579.2 | 102.1 |
| Creatinine (mg/dL) | ||||||||
| Minimum | 0.5 | 0.5 | 1.1 | 0.5 | 0.4 | 0.5 | 0.5 | 0.6 |
| Maximum | 2.6 | 1.6 | 1.2 | 1.6 | 6 | 1.6 | 2.2 | 1.7 |
| median | 1.1 | 0.8 | 1.2 | 0.8 | 0.9 | 0.9 | 0.8 | 0.8 |
| Average | 1.1 | 0.8 | 1.2 | 0.8 | 1.2 | 0.9 | 0.9 | 0.9 |
| Standard deviation | 0.4 | 0.2 | 0.1 | 0.2 | 1 | 0.2 | 0.3 | 0.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pacheco, V.; Cuber Guimarães, R.; Corrêa-Moreira, D.; Magalhães, C.E.; Figueiredo, D.; Guttmann, P.; Trindade, G.F.; da Silva, J.F.A.; Ano Bom, A.P.D.; de Lourdes Maia, M.; et al. Clinical Profile of SARS-CoV-2 Infection: Mechanisms of the Cellular Immune Response and Immunogenetic Markers in Patients from Brazil. Viruses 2023, 15, 1609. https://doi.org/10.3390/v15071609
Pacheco V, Cuber Guimarães R, Corrêa-Moreira D, Magalhães CE, Figueiredo D, Guttmann P, Trindade GF, da Silva JFA, Ano Bom APD, de Lourdes Maia M, et al. Clinical Profile of SARS-CoV-2 Infection: Mechanisms of the Cellular Immune Response and Immunogenetic Markers in Patients from Brazil. Viruses. 2023; 15(7):1609. https://doi.org/10.3390/v15071609
Chicago/Turabian StylePacheco, Vanessa, Rosane Cuber Guimarães, Danielly Corrêa-Moreira, Carlos Eduardo Magalhães, Douglas Figueiredo, Patricia Guttmann, Gisela Freitas Trindade, Juliana Fernandes Amorim da Silva, Ana Paula Dinis Ano Bom, Maria de Lourdes Maia, and et al. 2023. "Clinical Profile of SARS-CoV-2 Infection: Mechanisms of the Cellular Immune Response and Immunogenetic Markers in Patients from Brazil" Viruses 15, no. 7: 1609. https://doi.org/10.3390/v15071609
APA StylePacheco, V., Cuber Guimarães, R., Corrêa-Moreira, D., Magalhães, C. E., Figueiredo, D., Guttmann, P., Trindade, G. F., da Silva, J. F. A., Ano Bom, A. P. D., de Lourdes Maia, M., Melgaço, J. G., da Costa Barros, T. A., da Silva, A. M. V., Group, C., & Oliveira, M. M. E. (2023). Clinical Profile of SARS-CoV-2 Infection: Mechanisms of the Cellular Immune Response and Immunogenetic Markers in Patients from Brazil. Viruses, 15(7), 1609. https://doi.org/10.3390/v15071609








